Abstract
Clubroot disease in canola (Brassica napus) continues to spread across the Canadian prairies. Growing resistant cultivars is considered the most economical means of controlling the disease. However, sources of resistance to clubroot in B. napus are very limited. In this study, we conducted interspecific crosses using a B. rapa line (T19) carrying race-specific resistance genes and two B. oleracea lines, ECD11 and JL04, carrying race non-specific QTLs. Employing embryo rescue and conventional breeding methods, we successfully resynthesized a total of eight B. napus lines, with four derived from T19 × ECD11 and four from T19 × JL04. Additionally, four semi-resynthesized lines were developed through crosses with a canola line (DH16516). Testing for resistance to eight significant races of Plasmodiophora brassicae was conducted on seven resynthesized lines and four semi-resynthesized lines. All lines exhibited high resistance to the strains. Confirmation of the presence of clubroot resistance genes/QTLs was performed in the resynthesized lines using SNP markers linked to race-specific genes in T19 and race non-specific QTLs in ECD11. The developed B. napus germplasms containing clubroot resistance are highly valuable for the development of canola cultivars resistant to clubroot.
Similar content being viewed by others
Introduction
Canada is the world's leading canola (Brassica napus) producer, and canola remains the most profitable agricultural crop in Canada. However, this success has been threatened by the emerging clubroot disease caused by Plasmodiophora brassicae Woronin1,2. Addressing this disease in canola is of utmost priority to mitigate potential losses and achieve the industry's goal of increasing average yields to 52 bushels/acre. This goal is crucial to meet the global market demand of 26 million tons by 2025, as opposed to the current annual production of 20 million tons3.
Clubroot has continued to spread across the Canadian prairies over the last decade4, posing a serious long-term threat to canola production. The disease has been reported to cause about a 10–15% yield loss globally5. Based on the Williams differential system6, five pathotypes of P. brassicae (pathotypes 2, 3, 5, 6, and 8) were previously identified, with pathotype 3 being the most prevalent on canola in the prairie region7. The first clubroot-resistant canola cultivar in western Canada was released in 2009, followed by the release of the first generation of resistant cultivars from various breeding companies starting in 2010. These cultivars showed strong resistance to the old pathotypes of P. brassicae present in Canada. However, resistance in Canadian canola cultivars was soon overcome by new strains of P. brassicae identified in canola fields. Strains of P. brassicae collected in Canada have been classified into more than 30 pathotypes based on reactions on the Canadian Clubroot Differential (CCD)2,8. This rapid breakdown of resistance highlights the vulnerability of qualitative resistance controlled by major genes. To achieve more durable resistance, it is essential to develop new sources of resistance that combine both quantitative and qualitative resistance.
Genetic mapping of clubroot resistance (CR) genes has been extensively conducted in Brassica species to address resistance to strains of P. brassicae collected in canola fields in western Canada. Our group has identified major resistance genes (Rcr1 to Rcr10) against both old and major new pathotypes such as 3A and 3D in Brassica species. We have developed robust SNP markers tightly linked to each of these resistance genes9,10,11,12,13,14,15,16,17. Recently, two major (Rcr11 and Rcr13) and one minor QTL (Rcr_C03-4ECD10) were identified for resistance to 12 pathotypes (3A, 2B, 5C, 3D, 8E, 5G, 3H, 8J, 5L, 3O, 8P, and 5X) of Plasmodiophora brassicae in the B. napus cultivar ECD10 through genotyping-by-sequencing18. Identification of major genes conferring resistance to Canadian pathotypes in Brassica species has been also conducted at the University of Manitoba19,20 and the University of Alberta21,22,23,24,25,26,27. However, these major genes typically provide qualitative resistance, which is considered race-specific and can be rapidly overcome by the pathogen.
B. napus, originating from hybridization between B. rapa and B. oleracea, stands out as the most significant canola species globally, particularly in Canada. As mentioned earlier, sources of resistance in the A-genome derived from B. rapa have been extensively identified and are actively employed in the development of resistant canola cultivars in Western Canada. However, resistance sources in the C-genome from B. oleracea have not been widely utilized in canola breeding programs. This is primarily due to the challenge of transferring resistance directly from B. oleracea to B. napus through interspecific crosses, owing to reproductive barriers. Moreover, the availability of clubroot resistance sources in B. oleracea is limited. Genetic analysis of the CR genes in B. oleracea reveals that they are quantitative traits primarily controlled by quantitative trait loci (QTL)28,29,30, suggesting the potential for more durable, race non-specific clubroot resistance.
Two B. oleracea lines, ECD11 and JL04, demonstrated resistance to new strains of the clubroot pathogen during the screening of a large B. oleracea collection in our research group. The cabbage cultivar ECD11 (B. oleracea subsp. capitata, Badger Shipper) exhibited resistance to 15 out of 17 pathotypes in the CCD Set8 and other new pathotypes2, suggesting that ECD11 may possess genes with a broad spectrum of resistance. Recently, race non-specific QTLs were identified in ECD1129. Additionally, the kale breeding line JL04, originating from China, exhibited robust resistance to clubroot and likely carries race non-specific resistance genes/QTLs. In this study, we conducted the resynthesis of B. napus lines by crossing B. oleracea lines with a B. rapa line (T19) carrying three major resistance genes (Rcr4, Rcr8 and Rcr9)11. Consequently, the resynthesized B. napus germplasms from this study harbor both quantitative and qualitative resistance genes.
The objectives of the current study were: (1) to resynthesize B. napus germplasms with qualitative resistance from A-genome species B. rapa and quantitative resistance from C-genome species B. oleracea, (2) to characterize the resynthesized germplasms for resistance to important races of P. brassicae identified in Western Canada, and (3) to confirm the presence of the genes/QTLs in the resynthesized germplasms using molecular markers.
Results
Development of the resynthesized and semi-resynthesized B. napus germplasms
In the cross set of T19 (AA) × ECD11 (CC), 137 ovaries were obtained from 221 pollinated flowers, resulting in an ovary setting rate of 61.9%. Thirteen F1 hybrid plants germinated from the transplanted ovaries on MS medium, and 10 plants survived after transplantation into the soil. Four F1 amphidiploid (AACC) plants (Re-4, Re-10, Re-11A, Re-11B) produced seeds following colchicine treatment (Fig. 1). In the reciprocal cross set of ECD11 × T19, 26 ovaries were obtained from 57 pollinated flowers, yielding an ovary setting rate of 45.6%. Three F1 hybrid plants germinated from the transplanted ovaries on MS medium, but only one plant survived after transplantation into the soil and could not produce seeds after colchicine treatment. Notably, a higher success silique set rate was observed using B. rapa as the female parent (61.9%) compared to B. oleracea (45.6%). Therefore, crosses with B. rapa as the female were exclusively employed in the T19 and JL04 crosses, utilizing 10 plants of T19 and 6 plants of JL04.
Schematic diagram showing the development process of clubroot resistant (CR) resynthesized (RS) and semi-resynthesized (SRS) B. napus (AACC). Reciprocal crosses were initiated between B. rapa (AA) line T19 and B. oleracea (CC) lines ECD11, and non-reciprocal crosses were made between B. rapa (AA) line T19 and B. oleracea (CC) lines JL04. Semi-resynthesized (SRS) B. napus (AACC) lines were created by crossing resynthesized (RS) F2 plants with doubled haploid (DH) B. napus canola line DH16516 (DHT).
The cross set for T19 (AA) × JL04 (CC) resulted in 230 ovaries from 269 pollinated flowers, achieving an ovary setting rate of 85.5%. Thirty-five hybrid plants germinated from the transplanted ovaries on MS medium, 34 plants survived after transplantation into the soil, and 4 F1 amphidiploid (AACC) plants (TJ1-B, TJ1-E, TJ1-G, TJ4) produced seeds after colchicine treatment (Fig. 1).
In total, 51 F1 hybrid plants germinated, with 45 surviving in the soil, and ultimately, 8 hybrid plants successfully produced seeds from a total of 547 pollinations across the three cross combinations (Table 1). The cross ability percentages for these three combinations (T19 × ECD11, ECD11 × T19, T19 × JL04) were 1.8%, 0.0%, and 1.5%, respectively.
Seed fertility of the developed resynthesized and semi-resynthesized plants
Hybridity of the F1 plants was confirmed through careful assessment of morphological characteristics such as vigorous growth, plant height, leaf size and shapes, flower size, and pollen viability. Eight F1 hybrid plants successfully produced fertile flowers after chromosome doubling with colchicine treatment. Upon self-pollination, F2 seeds were successfully generated, designating these plants as amphidiploid (AACC) B. napus (Fig. 2). To ensure the stability of the newly synthesized lines, selection of self-compatible plants and selfing were continued up to the F6 generation. Approximately 10 plants per line were grown from the T19 × ECD11 cross. Re-10, Re-11A and Re-11B exhibited higher seed set in advanced generations, while the Re-4 line consistently showed poor seed set. Similarly, among the four lines from the T19 × JL04 cross combination, TJ1-B, TJ1-E and TJ1-G demonstrated higher seed set up to the F4 generations (Table 2), while TJ4 exhibited poor seed set. Although resynthesized plants showed poor seed fertility until advanced generations, all semi-resynthesized plants exhibited normal seed fertility in their early generation, crossing with DH16516 (Fig. 1, Table 2).
Resynthesized Brassica napus (AACC) generations derived from interspecific hybridization between Brassica rapa (AA) parental line T19 and Brassica oleracea (CC) parental lines ECD11 and JL04. To double the chromosome numbers and restore pollen fertility in the resynthesized F1 (AC), colchicine treatment was applied to the roots/leaf axils of hybrid plants. Amphidiploids (AACC) were identified by the presence of fertile pollen and the development of fully formed stamens.
Characterization of resynthesized and semi-resynthesized B. napus lines with P. brassicae identified in Western Canada
Seven resynthesized and four semi-resynthesized F1 lines, which produced a substantial amount of seeds (Table 2), parental lines, and susceptible control (DH16516 and NRC11-24) were selected for inoculation with selected races of P. brassicae, as shown in Table S1.
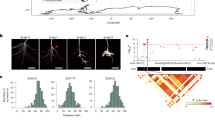
A total of 1,060 resynthesized and semi-resynthesized plants, along with parental lines and control plants, were tested against strains representing eight races of P. brassicae identified in Western Canada. All resynthesized lines exhibited high resistance against all eight strains/pathotypes (P.41-14/3H, 4-15/3A, CDCS/5G, LG02/5X, F.12-15/8J, Leduc C#37/8P, SK29, PSI11) (Figs. 3 and 4, Table S2): 3A (%DSI = 0.0–8.3), 5G (%DSI = 0.0), 3H (%DSI = 0.0–11.1), 8J (%DSI = 0.0–4.2), 8P (%DSI = 0.0), PSI11 (%DSI = 0.0–4.2), SK29 (%DSI = 0.0–4.8), and 5X (%DSI = 0.0). Similarly, semi-resynthesized lines also demonstrated resistance against all tested strains (Figs. 3 and 4, Table S2): 3A (%DSI = 0.0–27.8), 5G (%DSI = 4.2–28.2), 3H (%DSI = 0.0–14.6), 8J (%DSI = 0.0–25.0), 8P (%DSI = 0.0–13.3), PSI11 (%DSI = 0.0–8.3), SK29 (%DSI = 0.0–13.3), and 5X (%DSI = 0.0–12.5).
An evaluation of resynthesized (RS) Brassica napus (AACC) lines derived from two crosses, T19 × ECD11 and T19 × JL04, and semi-resynthesized (SRS) lines derived from DH16516 (DHT) × (T19 × ECD11) was conducted for clubroot resistance against 8 strains of Plasmodiophora brassicae. The root phenotypes of resynthesized lines, semi-resynthesized lines, parental lines (T19, ECD11, JL04) and susceptible (S) control lines DH16516 and NRC11-24 (N1) after six weeks of inoculation with strain SK29.
Evaluation of resynthesized Brassica napus (AACC) lines of different generations; Re-10, Re-11A and Re-11B lines were derived from the cross T19 × ECD11; lines TJ1-B1 and TJ1-G1 were derived from the cross T19 × JL04; and four semi-resynthesized lines derived from DH16516 (DHT) × (T19 × ECD11) for clubroot resistance. Distribution of disease severity indexes (DSIs) of resynthesized, semi-resynthesized, parental (T19, ECD11, JL04) and susceptible (S) control lines DH16516 and NRC11-24 (N1) against 8 strains: 3A, 5G, 3H, 8J, 8P, PSI11, SK29 and 5X, representing 8 races of Plasmodiophora brassicae.
The A-genome parent (T19) exhibited a high level of resistance against all eight strains (%DSI = 0.0), while resistance varied in C-genome parents ECD11 and JL04. ECD11 showed high resistance against 3A (%DSI = 2.8), 8J (%DSI = 4.8), 8P (%DSI = 6.7), SK29 (%DSI = 9.5), 5X (%DSI = 4.2), and moderate resistance against 5G (%DSI = 36.1), 3H (%DSI = 37.0), and susceptibility against PSI11 (%DSI = 61.9). JL04 showed high resistance against 3A (%DSI = 4.8), 5G, 3H, 8J, 5X (%DSI = 0.0), partial resistance against 8P (%DSI = 27.8), PSI11 (%DSI = 27.8), and SK29 (%DSI = 20.0).
DH16516 was highly susceptible against all eight strains (%DSI = 100), whereas NRC11-24 showed high resistance to 3H, 5G, 8J, PSI11 (%DSI = 0) and susceptibility against 3A (%DSI = 92.9), 5X (%DSI = 95.8), 8P (%DSI = 93.8), SK29 (%DSI = 100) (Fig. 4).
Confirmation of the presence of clubroot resistance genes/QTLs in the newly resynthesized B. napus lines
A total of 21 resynthesized plants (3 plants/line) from 7 resynthesized lines (Re-4, Re-10, Re-11A, Re-11B, TJ1-B, TJ1-E, TJ1-G) were selected to confirm the presence of identified clubroot resistance genes and QTLs: Rcr4, Rcr8, Rcr9 from A-genome and Rcr_C03-1, Rcr_C08-1 from C-genome (Table 3, Table S3). SNP markers associated with the resistance alleles were tested in the resynthesized lines from both crosses, T19 × ECD11 and T19 × JL04 (Table S3, Fig. S2). The results confirmed the presence of the resistance genes and QTLs in the resynthesized B. napus lines, also showing that JL04 may carry Rcr_C03-1 and Rcr_C08-1.
A-genome markers were designed based on the reference genome of B. rapa (sequence_v1.5.fa)31 downloaded from http://brassicadb.org/brad/downloadOverview.php, and the B. napus reference genome 'Darmor-bzh-v4.1’32 was obtained from https://www.genoscope.cns.fr/brassicanapus/data/. For C-genome markers, the reference genome of the DH line D134 (cabbage; B. oleracea var. capitata)33 was utilized and downloaded from https://db.cngb.org/search/?q=CNP0000469.
Discussion
Hybrid embryos encounter growth restrictions due to the absence of appropriate endosperm34,35,36,37. To overcome this limitation, techniques such as embryo, ovule, and ovary culture are commonly employed for embryo rescue in interspecific hybridization of Brassica species. Ovary culture proves effective when B. rapa is used as the female parent in crosses like B. rapa (♀) × B. oleracea (♂). However, it falls short in the reciprocal cross of B. oleracea (♀) × B. rapa (♂)37,38,39. In our study, we observed that B. rapa line T19 exhibited higher cross ability when used as a female parent, aligning with findings from previous research. Notably, there have been exceptions to this pattern, as reported in instances where resynthesized B. napus was successfully produced in both crossing directions using a B. oleracea line, CRGC3-1, as a parent40,41.
Resynthesized lines serve as valuable genetic resources in rapeseed breeding, contributing genetic diversity and crucial traits such as disease resistance (clubroot disease, sclerotinia stem rot, verticillium wilt, blackleg disease), yield, heterosis, insect resistance, pod shatter resistance, early flowering, and drought tolerance42. While some artificial rapeseeds were initially used solely as breeding materials due to low yields36,43,44, improved pollen and seed fertility have been achieved in advanced generations41,45,46. Consequently, selection lines from advanced generations with enhanced self-compatibility and seed fertility can be directly utilized for varietal development.
Resynthesized rapeseed lines exhibit self-incompatibility in the early generations, since they inherit functional S-alleles from both parent species, resulting in self-incompatibility in the offspring generations. The breakdown of self-incompatibility can occur due to not only loss-of-function mutations of alleles at the S-locus but also genetic interactions between specific S-alleles and unlinked modifiers47. In this study, although both parents of A and C genome used to produce the resynthesized lines were self-incompatible, self-compatible plants were selected during subsequent generations' selection processes and the self-compatibility trait improved in the advanced generations of the resynthesized lines. In semi-resynthesized F1 generation, it is expected that the S-alleles become heterozygous with non-functional and functional alleles provided by the respective parent, and there will be segregation of self-incompatible and self-compatible individuals in the F2 and subsequent generations of semi-resynthesized rapeseed lines. Unexpectedly, however, all our F1 semi-resynthesized rapeseed lines showed self-compatibility. Selection of the F2 and subsequent generations of semi-resynthesized rapeseed lines is underway; there is a need to select not only clubroot-resistant, but also self-compatible plants in the selection process as self-compatibility is an important breeding goal in rapeseed breeding.
In this study, earlier generations of resynthesized lines exhibited poor seed set, consistent with previous findings41,48,49. However, wide variation in seed set was observed in advanced generations, presenting opportunities for incorporation into canola breeding programs. Meiotic irregularities affecting genome stability contribute to low fertility in resynthesized B. napus50. These challenges can be addressed by selecting lines with improved seed fertility and by developing semi-resynthesized lines41. Consequently, four semi-resynthesized B. napus lines were developed by crossing resynthesized lines (Re-4, Re-10, Re-11A, and Re-11B) with a DH B. napus canola line, DH16516. While advanced generations of three resynthesized lines (Re-10, Re-11A, and Re-11B) showed good seed fertility, Re-4 did not. On the other hand, semi-resynthesized B. napus lines derived from all four resynthesized lines exhibited good seed fertility in their early generations. Thus, both selected resynthesized and all semi-resynthesized lines hold promise for integration into canola breeding programs.
The fertility of resynthesized lines is genotype-dependent, and due to frequent sterility in early generations, not all resynthesized lines may survive to advanced self-pollinated generations42. Notably, two resynthesized lines, Re-4 and TJ4, did not yield satisfactory self-pollinated seeds compared to the other six lines (Re-10, Re-11A, Re-11B, TJ1-B, TJ1-E, TJ1G). This variability in seed fertility not only exists among different lines but also among different plants of the same line, aligning with findings from previous studies41. Early generations of resynthesized B. napus often involve aneuploidy and gross chromosomal rearrangements. Notably, seed yield and pollen viability show an inverse correlation with increasing aneuploidy51.
In this study, our aim was to combine qualitative and quantitative resistance from B. rapa and B. oleracea into the B. napus resynthesized lines, resulting in lines that demonstrated a broad range of resistance to multiple races of P. brassicae. A previous study identified variation in quantitative traits, such as flowering time, attributed to structural rearrangements of chromosomes in resynthesized B. napus52. Interestingly, the stability of qualitative traits like self-incompatibility53 and clubroot resistance54 has often been observed in the self-pollinated progeny of resynthesized B. napus plants. While a loss of genomic regions carrying clubroot (CR) resistance (6–13%) has been reported during the development of self-pollination (S0 to S1 families) in resynthesized lines due to meiotic anomalies49, our current study did not observe any CR resistance loss in all homozygous resynthesized lines, as they maintained high resistance against all eight strains. Furthermore, four F1 semi-resynthesized lines were developed by crossing resynthesized lines with the clubroot-susceptible DH line DH16516. The resynthesized lines showed higher resistance (DSI = 0–11.1%) compared to the F1 semi-resynthesized B. napus lines (DSI = 0.0–28.2%). We observed some susceptible plants in the F1 semi-resynthesized B. napus lines; the reason for this is yet to be determined.
Meiotic anomalies and homoeologous pairing of chromosomes in the early generations of resynthesized Brassica allopolyploids can lead to structural rearrangements with the loss or gain of chromosomes50,51,55,56,57. An example is the European winter clubroot resistance (CR) canola cultivar ‘Mendel’, developed from a resynthesized B. napus line crossed from B. oleracea ECD15 × B. rapa ECD0454. ‘Mendel’ was initially expected to possess three dominant CR genes inherited from its diploid parent ECD0458,59,60. However, genetic mapping revealed that 'Mendel' had two dominant CR genes61, suggesting a potential loss of the other gene during the breeding process. Similar loss of CR genes has been reported in the breeding of a rutabaga line62. On the other hand, two B. rapa CR loci (Crr1 and Crr2) were efficiently transferred through the development of resynthesized B. napus lines and properly introduced into the recurrent parental lines using CR loci-linked markers63. There were 6, 4 and 2 genes encoding TIR-NBS-LRR class disease resistance proteins in the Rcr4, Rcr8, and Rcr9 flanking regions in T19, and developed SNP markers linked to these QTLs11. Recently, we first confirmed that clubroot resistance genes Rcr1, Rcr2, Rcr4, and CRa from B. rapa vegetables and the resistance gene from B. napus oilseed rape cv. “Mendel” on chromosome A03 were identical in their coding regions after cloning and functional verification of the clubroot resistance gene Rcr164. In our recent study, two race no-specific QTLs (Rcr_C03-1, Rcr_C08-1) were identified in ECD1129. There were 10 and 4 genes encoding TIR-NBS-LRR and CC-NBS-LRR class disease resistance proteins in the Rcr_C03-1 and Rcr_C08-1 flanking regions, and developed SNP markers linked to these QTLs. All targeted loci, including Rcr4, Rcr8, and Rcr9 in the resynthesized B. napus lines, were confirmed using SNP markers closely linked to the genes. C-genome CR QTLs (Rcr_C03-1, Rcr_C08-1) from ECD11 were also confirmed in the resynthesized lines using tightly linked SNP markers. However, no studies on genetic mapping for the identification of QTLs in JL04 have been carried out so far. Interestingly, some of the linked SNP markers for the QTLs in ECD11 were identified in the resynthesized lines from T19 × JL04. Further research on genetic mapping is needed to confirm these results and identify novel QTLs in JL04.
Conclusion
In this current study, we successfully developed eight resynthesized B. napus lines from T19 × ECD11 and JL04, achieving a broad spectrum of resistance from both qualitative and quantitative resistance backgrounds. All targeted loci were confirmed in the resynthesized lines using tightly linked markers. The germplasms and information obtained from this project are now available to canola breeders for rapid incorporation into their canola variety development programs (https://agriculture.canada.ca/en/science/technology-transfer-and-licensing-agriculture/evaluation-canola-breeding-lines-clubroot-resistance).
Materials and methods
Plant materials
A CR-resistant self-incompatible B. rapa breeding line, T19, derived from the German turnip cultivar ‘Pluto’, was developed at the Saskatoon Research and Development Centre, Agriculture and Agri-Food Canada (SRDC, AAFC), in Saskatoon, Saskatchewan, Canada. The seeds of the resistant B. rapa line (T19), crossed with two self-incompatible B. oleracea lines, ECD11 and JL04, were provided by Dr. G. R. Dixon (The University of Warwick, Wellesbourne, Warwick, UK), and Dr. Zhen Huang (Northwest A&F University, Yangling, Shaanxi, China), respectively.
Reciprocal crosses, embryo rescue and chromosome doubling
Seedlings of the parental B. rapa (T19) and B. oleracea (ECD11, JL04) were grown in a greenhouse at the SRDC, AAFC. Reciprocal crosses were initiated between B. rapa (AA) line T19 and B. oleracea (CC) lines ECD11 and JL04. The parental lines T19, ECD11, and JL04 were grown in the greenhouse for two weeks and then exposed to a 3-month vernalization period at a cold room (+ 4 °C) to synchronize flowering time. Reciprocal crosses were conducted using 6 plants of T19 × 3 plants of ECD11 and 10 plants of T19 × 6 plants of JL04. Emasculation was carried out on floral buds 1–2 days before anthesis. The emasculated buds were promptly dusted with fresh pollen grains collected from the male parents (B. oleracea ECD11 and JL04). Pollinated flowers were isolated in thin paper bags, and ovaries bearing ovules were collected at 16–20 days after pollination (DAP) for F1 embryo rescue.
The embryo rescue technique, adapted from the method described by Inomata38 with some modifications, involved the sterilization of ovary surfaces. Ovaries were sterilized with 70% ethanol for 3 min, followed by treatment with a 10% solution of sodium hypochlorite for 10 min and rinsed twice with sterile distilled water. The sterilized ovaries were placed on MS medium65, aseptically supplemented with 1% sucrose and 0.8% agar, adjusted to pH 5.8. Plastic petri dishes (9.0 cm × 1.5 cm) were used for the cultures, which were then placed in a growth chamber maintained at 22 °C with a 16-h photoperiod (7:00 am to 11:00 pm). After 20–30 days, regenerating embryos were transferred to basal MS agar medium in a plant tissue culture container (10.0 cm × 7.5 cm × 7.5 cm) for root and shoot development. Once the regenerated plantlets reached a height of 3–4 inches, they were removed from culture and transplanted into four-inch pots containing Osmocote potting mix (Everris NA Inc.; Dublin, OH, USA) and covered with a transparent plastic cap. The plants were then placed in a growth chamber maintained at 22 °C with a 12-h photoperiod for two weeks before being transferred into six-inch pots containing Sunshine #3 soilless mix (Sun Gro Horticulture Canada Ltd.; Seba Beach, AB) (Fig. S1).
To restore seed fertility in the F1 hybrids, the roots of each F1 plant were submerged in a 3.4 g/L solution of colchicine for 1.5 h, then thoroughly washed with water and transplanted into the soil to double the chromosome numbers, following a previously described method66. In cases where F1 plants failed to produce fertile pollen, a 0.05% colchicine solution was applied to the leaf axils of hybrid plants, as described by Chen et al.34. Amphidiploids (AACC) were identified by the presence of fertile pollen and the development of fully formed stamens. Seed fertility (seeds/pod) in the early generations of the resynthesized B. napus lines was low due to self-incompatibility. To obtain self-pollinated seeds, plants with open flowers and unopened buds were enclosed in an airtight plastic bag, which was then filled with CO2 to raise the internal concentration to optimize the conditions for flower development and pollen production, which can ultimately lead to improved seed production. After 3–4 h, the bag was removed and replaced with a pollinating bag. Seeds were increased for further experiments. The resynthesized F1 plants were self-pollinated to produce subsequent progenies, from F2 to F6. As it was challenging to produce self-pollinated seeds in some resynthesized lines from T19 × ECD11; selected F2 plants were crossed with the doubled haploid (DH) B. napus canola line DH16516, developed by Dr. Séguin-Swartz at the SRDC, AAFC, to obtain semi-resynthesized F1 progeny (Fig. 1).
Evaluation of newly resynthesized and semi-resynthesized B. napus lines for clubroot resistance
Strains of P. brassicae collected from canola fields in Alberta, Saskatchewan, and Manitoba were provided by Dr. S. E. Strelkov at the University of Alberta, Drs. A. Akhavan and B. Ziesman at the Government of Saskatchewan, and Dr. L. A. Murphy and Mr. X. Guo at Pest Surveillance Initiative, Canada (Table S1). The strains were characterized with five B. napus lines (unpublished data) carrying single clubroot resistance genes Rcr1, Rcr3, Rcr8, Rcr9 and Rcr10, respectively10,11,16,17. These strains represent eight races of P. brassicae in the canola fields in Western Canada (Table S1). In addition, the strains from Alberta were characterized with the Canadian Clubroot Differentials8.
The newly resynthesized and semi-resynthesized B. napus, the parental lines, and control plants were tested for clubroot resistance with the P. brassicae strains in a greenhouse at the SRDC, AAFC. The inoculum preparation method used in this study was as described by Karim and Yu29. Seedlings of the susceptible canola line DH16516 or B. napus line NRC11-24 (Nutrien Ag. Solutions, Saskatoon, Saskatchewan) with a known resistance gene Rcr1 were inoculated and maintained under controlled conditions to increase clubroot galls. Clubbed roots were harvested from infected plants after 5–6 weeks and stored at −20 °C. Fresh inoculum of each pathotype was prepared by softening about 5 g of frozen club in distilled water for 1–2 h, homogenizing it in a blender for 2 min, and straining it through 2–3 layers of nylon mesh cloth. The resulting spore suspension was diluted with deionized water to achieve a final concentration of 1 × 107 resting spores/mL.
The inoculation method used in this study was as described by Karim and Yu29. Seeds of the resynthesized, semi-resynthesized lines, their parental lines T19, ECD11, and JL04, and the two controls DH16516 and NRC11-24 were sown into Sunshine #3 soilless mix (Sun Gro Horticulture Canada Ltd.; Seba Beach, AB) with Osmocote (Everris NA Inc.; Dublin, OH, USA) in 32-pot inserts held by trays (The HC Companies; Twinsburg, OH, USA). Adequate water was added to each tray to soak the soilless mix overnight. Seven days after planting, inoculation was performed by adding 15 ml of inoculum (1 × 107 spores/ml) into each pot with 6–9 seedlings of each line with two replications. The inoculated plants were grown in a growth chamber set at 22/18 °C day/night temperature with a 16-h photoperiod. The pots were kept moist by adding water whenever necessary. Six weeks after inoculation, plant roots were examined for clubroot symptoms. Clubroot severity was evaluated on a 0 to 3 scale67, where 0 = no clubbing, 1 = a few small clubs (small galls on fewer than one-third of the roots), 2 = moderate clubbing (small to medium-sized galls on between one-third and two-thirds of the roots), and 3 = severe clubbing (medium to large-sized galls on over two-thirds of the roots). A disease severity index (DSI) was calculated for each line using the method of Horiuchi and Hori68:
Kompetitive allele specific PCR (KASP) analysis
Young leaves from resynthesized B. napus lines, parental lines, and A- and C-genome susceptible control lines were harvested for DNA extraction. The leaves were freeze-dried in a Freezone 6 dryer (Labconco Corp, Kansas City, MO) for 48 h and ground using the Mixture Mills 300 (Retsch Inc., Newtown, PA). DNA extraction was carried out with the DNeasy 96 Plant Kit (Qiagen, Toronto, ON, Canada) following the DNeasy plant handbook from QIAGEN. The extracted DNA was quantified using a NanoVue Plus spectrophotometer (GE Healthcare, Piscataway, NJ), diluted to 10 ng/μl, and stored at −20 °C until subsequent use for genotyping.
The B. rapa parental line T19 carried race-specific resistance genes Rcr4, Rcr8, and Rcr911. Two QTLs, Rcr_C03-1 and Rcr_C08-1, conferring race non-specific resistance to P. brassicae races in the cabbage cultivar ECD11, were recently identified29. SNP markers linked to these genes/QTLs were developed. The presence of Rcr4, Rcr8, and Rcr9 from B. rapa T19 and the QTLs, Rcr_C03-1 and Rcr_C08-1 from B. oleracea ECD11 in the newly resynthesized B. napus lines were confirmed using SNP markers genotyped with the KASP method (http://www.lgcgroup.com/), following the manufacturer’s instructions. PCR was performed in a StepOne Plus Real-Time PCR System (Applied Biosystem, Mississauga, ON).
Statements
B. rapa breeding line, T19, was provided from the Saskatoon Research and Development Centre, Agriculture and Agri-Food Canada (SRDC, AAFC), in Saskatoon, Saskatchewan, Canada. The seeds of the resistant B. rapa line (T19), crossed with two B. oleracea lines, ECD11 and JL04, were provided by Dr. G. R. Dixon (The University of Warwick, Wellesbourne, Warwick, UK), and Dr. Zhen Huang (Northwest A&F University, Yangling, Shaanxi, China), respectively. Voucher specimen of these developed material from the current research has not been deposited yet in a publicly available herbarium. We have permission to collect the plants used in this study. All methods were performed in accordance with the Saskatoon Research and Development Centre, Agriculture and Agri-Food Canada (SRDC, AAFC), Saskatchewan, Canada guidelines/regulations/legislation
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Woronin, M. S. Plasmodiophora brassicae, der Organismus, der die unter dem Namen Hernie bekannte Krankheit der Kohlpflanzen verursacht. Arbeiten Sankt Petersburger Naturforschender Gesellschaft. 8, 169–201 (1877).
Hollman, K. B., Hwang, S. F., Manolii, V. P. & Strelkov, S. E. Pathotypes of Plasmodiophora brassicae collected from clubroot resistant canola (Brassica napus L.) cultivars in western Canada in 2017–2018. Can. J. Plant Pathol. 43, 622–630 (2021).
Canola Council of Canada. https://www.canolacouncil.org/about-canola/industry/#:~:text=Canola%20is%20grown%20by%2043%2C000,of%20all%20farm%20crop%20receipts (2021).
Froese, R. D., Derksen, H., Guo, X. & McLaren, D. L. Monitoring and occurrence of clubroot in Manitoba in 2018. Can. Plant Dis. Surv. 99, 1–179 (2019).
Dixon, G. R. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 28, 194–202 (2009).
Williams, P. H. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 56, 624–626 (1966).
Strelkov, S. E., Manolii, V. P., Cao, T., Xue, S. & Hwang, S. F. Pathotype classification of Plasmodiophora brassicae and its occurrence in Brassica napus in Alberta, Canada. J. Phytopathol. 155, 706–712 (2007).
Strelkov, S. E. et al. Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada. Can. J. Plant Pathol. 40, 284–298 (2018).
Chu, M. et al. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genom. 15, 1166. https://doi.org/10.1186/1471-2164-15-1166 (2014).
Yu, F. et al. Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene Rcr1 through bulked segregant RNA sequencing. PLoS ONE 11(4), e0153218. https://doi.org/10.1371/journal.pone.0153218 (2016).
Yu, F. et al. Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 7, 4516. https://doi.org/10.1038/s41598-017-04903-2 (2017).
Huang, Z. et al. Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front. Plant Sci. 8, 1448. https://doi.org/10.3389/fpls.2017.01448 (2017).
Dakouri, A. et al. Analysis of genome-wide variants through bulked segregant RNA sequencing reveals a major gene for resistance to Plasmodiophora brassicae in Brassica oleracea. Sci. Rep. 8, 17657. https://doi.org/10.1038/s41598-018-36187-5 (2018).
Chang, A. et al. Clubroot resistance gene Rcr6 in Brassica nigra resides in a genomic region homologous to chromosome A08 in B. rapa. BMC Plant Biol. 19, 224. https://doi.org/10.1186/s12870-019-1844-5 (2019).
Huang, Z., Peng, G., Gossen, B. D. & Yu, F. Fine mapping of a clubroot resistance gene from turnip using SNP markers identified from bulked segregant RNA-Seq. Mol. Breed. 39, 1–10 (2019).
Karim, M. M. et al. Two clubroot-resistance genes, Rcr3 and Rcr9wa, mapped in Brassica rapa using bulk segregant RNA sequencing. Int. J. Mol. Sic. 21(14), 5033. https://doi.org/10.3390/ijms21145033 (2020).
Yu, et al. Identification of two major QTLs in Brasica napus lines with introgressed clubroot resistance from turnip cultivar ECD01. Front. Plant Sci. 12, 785989. https://doi.org/10.3389/fpls.2021.785989 (2021).
Karim, M. M. & Yu, F. A promising Brassica napus source to combat clubroot: Identification of QTLs for resistance to 12 pathotypes of Plasmodiophora brassicae in B. napus cultivar ECD10 through genotyping-by-sequencing. In ASA, CSSA, SSSA International Annual Meeting, St. Louis, Missouri, USA. https://scisoc.confex.com/scisoc/2023am/meetingapp.cgi/Paper/153159 (2023).
Gao, F. et al. Fine mapping a clubroot resistance locus in chinese cabbage. J. Am. Soc. Hort. Sci. 139, 247–252 (2014).
Hirani, A. H. et al. Combinations of independent dominant loci conferring clubroot resistance in all four turnip accessions (Brassica rapa) from the European clubroot differential set. Front. Plant Sci. 9, 1628. https://doi.org/10.3389/fpls.2018.01628 (2018).
Zhang, H. et al. Mapping of clubroot (Plasmodiophora brassicae) resistance in canola (Brassica napus). Plant Pathol. 65, 435–440 (2015).
Hasan, M. J. & Rahman, H. Genetics and molecular mapping of resistance to Plasmodiophora brassicae pathotypes 2, 3, 5, 6, and 8 in rutabaga (Brassica napus var. napobrassica). Genome 59, 805–815 (2016).
Fredua-Agyeman, R. & Rahman, H. Mapping of the clubroot disease resistance in spring Brassica napus canola introgressed from European winter canola cv. ‘Mendel’. Euphytica 211, 201–213 (2016).
Fredua-Agyeman, R., Jiang, J., Hwang, S.-F. & Strelkov, S. E. QTL mapping and inheritance of clubroot resistance genes derived from Brassica rapa subsp. Rapifera (ECD02) reveals resistance loci and distorted segregation ratios in two F2 populations of different crosses. Front. Plant Sci. 11, 899. https://doi.org/10.3389/fpls.2020.00899 (2020).
Fredua-Agyeman, R. et al. Clubroot resistance derived from the European Brassica napus cv. ‘Tosca’ is not effective against virulent Plasmodiophora brassicae isolates from Alberta, Canada. Sci. Rep. 11, 14472. https://doi.org/10.1038/s41598-021-93327-0 (2021).
Hasan, M. J., Shaikh, R., Basu, U. & Rahman, H. Mapping clubroot resistance of Brassica rapa introgressed into Brassica napus and development of molecular markers for the resistance. Crop Sci. 61, 4112–4127 (2021).
Wang, Z., Megha, S., Kebede, B., Kav, N. N. V. & Rahman, H. Genetic and molecular analysis reveals that two major loci and their interaction confer clubroot resistance in canola introgressed from rutabaga. Plant Genome 15, e20241. https://doi.org/10.1002/tpg2.20241 (2022).
Piao, Z., Ramchiary, N. & Lim, Y. P. Genetics of clubroot resistance in Brassica species. J. Plant Growth Regul. 28, 252–264 (2009).
Karim, M. M. & Yu, F. Identification of QTLs for resistance to 10 pathotypes of Plasmodiophora brassicae in Brassica oleracea cultivar ECD11 through genotyping-by-sequencing. Theor. Appl. Genet. 136, 249. https://doi.org/10.1007/s00122-023-04483-y (2023).
Rocherieux, J. et al. Isolate-specific and broad spectrum QTLs are involved in the control in Brassica oleracea. Theor. Appl. Genet. 108, 1555–1563 (2004).
Wang, X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1040 (2011).
Chalhoub, B. et al. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014).
Lv, H., Wang, Y. & Han, F. A high-quality reference genome for cabbage obtained with SMRT reveals novel genomic features and evolutionary characteristics. Sci. Rep. 10, 12394. https://doi.org/10.1038/s41598-020-69389-x (2020).
Chen, B. Y., Heneen, W. K. & Jonsson, R. Resynthesis of Brassica napus L. through interspecific hybridization between B. aloboglabra Bailey and B. campestris L. with special emphasis on seed colour. Plant Breed. 101, 52–59 (1988).
Inomata, N. Embryo rescue techniques for wide hybridization. In Breeding Oilseed Brassicas (Labana, K.S., Banga, S.S., Banga, S.K. eds.) Monogr. Theor. Appl. Genet. 19, 94–107 (1993).
Olsson, G. Species cross within the genus Brassica. II. Artificial Brassica napus L.. Hereditas 46, 351–386 (1960).
Takeshita, M., Kato, M. & Tokumasu, S. Application of ovule culture to the production of intergeneric or interspecific hybrids in Brassica and Raphanus. Jpn. J. Genet. 55, 373–387 (1980).
Inomata, N. Production of interspecific hybrids between Brassica campestris and Brassica oleracea by culture in vitro of excised ovaries. I. Effects of yeast extract and casein hydrolysate on the development of excised ovaries. Jpn. J. Breed. 27, 295–304 (1977).
Inomata, N. Production of interspecific hybrids between Brassica campestris and Brassica oleracea by culture in vitro of excised ovaries. II. Effects of coconut milk and casein hydrolysate on the development of excised ovaries. Jpn. J. Genet. 53, 1–11 (1978).
Song, K., Tang, K. & Osborn, T. C. Development of synthetic Brassica amphidiploids by reciprocal hybridization and comparison to natural amphidiploids. Theor. Appl. Genet. 86, 811–821 (1993).
Karim, M. M. et al. Production of high yield short duration Brassica napus by interspecific hybridization between B. oleracea and B. rapa. Breed Sci. 63, 495–502 (2014).
Katche, E. I. & Mason, A. S. Resynthesized rapeseed (Brassica napus): Breeding and genomics. Crit. Rev. Plant Sci. 42, 65–92 (2023).
Kräling, K. Utilization of genetic variability of resynthesized rapeseed. Plant Breed. 99, 209–217 (1987).
Seyis, F., Friedt, W. & Luhs, W. Yield of Brassica napus L. hybrids developed using resynthesised rapeseed material. Anadolu J. Agric. Sci. 25, 159–167 (2010).
Girke, A., Schierholt, A. & Becker, H. C. Extending the rapeseed gene pool with resynthesized Brassica napus II: Heterosis. Theor. Appl. Genet. 124, 1017–1026 (2012).
Katche, E. I., Schierholt, A., Becker, H. C., Batley, J. & Mason, A. S. Fertility, genome stability, and homozygosity in a diverse set of resynthesized rapeseed lines. Crop J. 11, 468–477 (2023).
Li, Y. et al. Breakdown of self-incompatibility due to genetic interaction between a specific S-allele and an unlinked modifier. Nat. Commun. 14, 3420. https://doi.org/10.1038/s41467-023-38802-0 (2023).
Srivastava, A. et al. Resynthesis of Brassica juncea through interspecific crosses between B. rapa and B. nigra. Plant Breed. 123, 204–206 (2004).
Hasan, M. J. & Rahman, H. Resynthesis of Brassica juncea for resistance to Plasmodiophora brassicae pathotype 3. Breed. Sci. 68, 385–391 (2018).
Szadkowski, E. E. F. et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186, 102–112 (2010).
Xiong, Z., Gaeta, R. T. & Pires, J. C. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. 108, 7908–7913 (2011).
Pires, J. C. et al. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. 82, 675–688 (2004).
Rahman, M. H. Resynthesis of Brassica napus L. for self incompatibility: Self-incompatibility reaction, inheritance and breeding potential. Plant Breed. 124, 13–19 (2005).
Diederich, E. & Sacristan, M. Disease response of resynthesized Brassica napus L. lines carrying different combinations of resistance to Plasmodiophora brassicae Wor.. Plant Breed. 115, 5–10 (1996).
Gaeta, R. T., Pires, J. C., Iniguez-Luy, F., Leon, E. & Osborn, T. C. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19, 3403–3417 (2007).
Gaeta, R. T. & Pires, J. C. Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol. 186, 18–28 (2010).
Udall, J. A., Quijada, P. A. & Osborn, T. C. Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L.. Genetics 169, 967–979 (2005).
Wit, F. Inheritance of reaction to clubroot in turnips. Hortic. Res. 5, 47–49 (1965).
Diederichsen, E., Frauen, M., Linders, E. G. A., Hatakeyama, K. & Hirai, M. Status and perspectives of clubroot resistance breeding in crucifer crops. J. Plant Growth Regul. 28, 265–281 (2009).
Fredua-Agyeman, R., Hwang, S. F., Strelkov, S. E., Zhou, Q. & Feindel, D. Potential loss of clubroot resistance genes from donor parent Brassica rapa subsp. rapifera (ECD04) during doubled haploid production. Plant Pathol. 67, 892–901 (2018).
Rahaman, M. M. Identification and genetic mapping of resistance genes against Canadian pathotypes of Plasmodiophora brassicae in Brassica rapa and Brassica napus. A thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Plant Science, Department of Agricultural, Food and Nutritional Science, University of Alberta. https://era.library.ualberta.ca/items/71656066-b33d-41b3-bf4e-f34c0afa7360/download/acc1512d-c0a0-4ae3-a3d4-83aba3a419ee (2022).
Bradshaw, J. E., Gemmell, D. J. & Wilson, R. N. Transfer of resistance to clubroot (Plasmodiophora brassicae) to swedes (Brassica napus L. var. napobrassica Peterm) from B. rapa. Ann. Appl. Biol. 130, 337–348 (1997).
Kawasaki, M., Ohara, T., Ishida, M., Takahata, Y. & Hatakeyama, K. Development of novel clubroot resistant rapeseed lines (Brassica napus L.) effective against Japanese field isolates by marker assisted selection. Breed. Sci. 71, 528–537 (2021).
Hu, H., Zhang, Y. & Yu, F. A CRISPR/Cas9-based vector system enables the fast breeding of selection-marker-free canola with Rcr1-rendered clubroot resistance. J. Exp. Bot. 75, 1347–1363 (2024).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15, 473–497 (1962).
Coventry, J. Manual for Microspore Culture Technique for Brassica napus. Department of Crop Science Technical Bulletin, OAC Publication No. 0489, University of Guelph, Guelph, Ontario, Canada. https://books.google.ca/books?id=kZzLMQAACAAJ (1988).
Kuginuki, Y., Hiroaki, Y. & Hirai, M. Variation in virulence of Plasmodiophora brassicae in Japan tested with clubroot-resistant cultivars of Chinese cabbage (Brassica rapa L. spp. pekinensis). Eur. J. Plant Pathol. 105, 327–332 (1999).
Horiuchi, S. & Hori, M. A. Simple greenhouse technique for obtaining high levels of clubroot incidence. Bull. Chugoku Natl. Agric. Exp. Station E 17, 33–55 (1980).
Acknowledgements
We would like to thank Drs. Stephen Strelkov, Alireza Akhavan, Barbara Ziesman, Lee Anne Murphy and Xiaowei Guo for providing clubroot strains, Drs. Kevin Falk and Gary Peng (AAFC, Saskatoon) for B. rapa T19, Dr. Dr. G. R. Dixon (The University of Warwick, Wellesbourne, Warwick, UK) for B. oleracea ECD11, Dr. Zhen Huang (Northwest A&F University, Yangling, Shaanxi, China) for B. oleracea JL04 and Alberta Innovates, Alberta Canola Producers Commission & Alberta Government for funding the project. We would also like to thank Ms. Melissa Kehler for her assistance in conducting some of the experiments.
Funding
This study was funded by Alberta Innovates, Alberta Canola Producers Commission and Alberta Agriculture and Forestry.
Author information
Authors and Affiliations
Contributions
MK conducted all the experiments for the project; performed crosses, embryo rescue, colchicine treatment, seed increasing, designed the KASP primers, performed KASP analysis, evaluated plants for clubroot resistance and wrote the manuscript. FY collected materials including the parental lines, strains of P. brassicae, prepared the research proposal, designed the KASP primers, revised manuscript and managed the overall project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Masud Karim, M., Yu, F. Resynthesizing Brassica napus with race specific resistance genes and race non-specific QTLs to multiple races of Plasmodiophora brassicae. Sci Rep 14, 14627 (2024). https://doi.org/10.1038/s41598-024-64795-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-64795-x