Abstract
Banana (Musa spp.) is the most widely consumed fruit globally. Fusarium wilt, caused by Fusarium oxysporum f. sp. cubense (Foc), is a highly threatening disease to banana production. Resistance genes to Foc exist in wild Musa genotypes such as Musa acuminata subsp. burmannicoides var. Calcutta 4. Whilst real-time PCR (RT-qPCR) is appropriate for accurate analysis of gene expression in pathways involved in host defence responses, reference genes with stable expression under specific biotic stress conditions and host tissue types are necessary for normalization of sample variation. In this context, the stability in potential host reference genes ACT1, APT, EF1α, GAPDH, αTUB, RAN, UBIQ1, UBIQ2, βTUB1, βTUB3, L2 and ACTA1 was evaluated in total RNA samples from root tissues in Calcutta 4 (resistant) and Musa sp. cultivar Prata-anã (susceptible) extracted during interaction with Foc subtropical race 4 (STR4). Expression stability was calculated using the algorithms geNorm, NormFinder and BestKeeper. βTUB3 and L2 were identified as the most stable in Calcutta 4, with ACTA1 and GAPDH the most stable in Prata-anã. These reference genes for analysis of gene expression modulation in the Musa-Foc STR4 pathosystem are fundamental for advancing understanding of host defence responses to this important pathogen.
Similar content being viewed by others
Introduction
Banana (Musa spp.) is one of the most widely traded and consumed fruits globally and, as a basic carbohydrate component of the diet in numerous developing countries, is highly relevant for global food security1,2. With cultivated edible bananas and plantains derived from the fertile diploid progenitor species of Musa acuminata (AA) and Musa balbisiana (BB), many of today’s commercial cultivars such as 'Cavendish' (AAA), ‘Pome’ (AAB), and ’Silk’’ (AAB) are sterile, with seedless fruits developing via parthenocarpy3. As genetic diversity is low across such important cultivars, resistance to evolving pathogens is limited4.
Fusarium wilt is currently the most devastating and widely disseminated fungal disease of banana globally2,5,6. Vascular wilt due to pathogen advance in xylem vessels restricts water movement, resulting in leaf yellowing, pseudostem splitting and eventual plant death7. Fusarium wilt is caused by the soilborne fungus Fusarium oxysporum f. sp. cubense (Foc)6,8, with three physiological races of Foc capable of infecting banana, namely race 1, race 2, and race 4, with the latter subdivided into tropical race 4 (TR4) and subtropical race 4 (STR4)8. Cultivars of the 'Gros Michel' group (AAA), 'Pome' (AAB), and 'Silk' (AAB) are susceptible to all races of Foc, while cultivars of the 'Cavendish' subgroup (AAA) are typically resistant to race 1 of Foc but susceptible to STR4 and TR46. Breakdown of resistance to Foc race 1 (VCG01249, 0125 and 0122010) in Cavendish cultivars has, however, been reported in certain growing regions11,12,13,14.
The fungal pathogen Foc can survive in soils in the absence of the host plant, with chlamydospores, which are thick-walled survival spores, able to maintain viability for periods in excess of 20 years15. Given such pathogen persistence, the employment of resistant varieties is essential for disease management. In contrast to susceptible cultivars, fertile resistant wild diploid Musa genotypes, that have coevolved with pathogens at the centre of origin of the genus, represent a source of defence genes and alleles that are appropriate for genetic improvement of cultivars via genetic engineering or gene modification.
For candidate gene discovery, RNAseq-based high-throughput sequencing approaches for investigation of host transcriptome responses in Musa materials contrasting in resistance are advancing understanding of the cellular-level defence response involved in resistance to Foc16,17,18,19. Following such investigations, in silico-derived candidate genes in the defence response require lab-based validation via the reverse transcription real-time polymerase chain reaction (RT-qPCR), which is widely employed to accurately quantify gene expression levels in cDNA derived from RNA20,21,22. The guidelines of the Minimum Information for Publication of Real-Time Quantitative PCR Experiments (MIQE)23 recommend that for accurate measurement of target gene expression, normalization is necessary to that of a reference gene which displays stable expression for any particular tissue type or experimental treatment24. As reference genes are subject to correction of the effect of cDNA quality25,26, normalization of target gene expression to reference genes also corrects for variation between sample replicates that may occur during cDNA synthesis.
Several studies have been conducted in recent years for the development of stable reference genes for different genotypes and tissue types in Musa sp.27,28,29,30, with data highlighting that genes involved in basal cell activities, such as ACT, UBQ, GAPDH, EF, L2, TUB, GAPDH, RAN, and APT, amongst others28, may not necessarily show a stable constitutive expression under particular experimental conditions. As such, the identification of specific stable reference genes is required for each plant genotype, tissue type and experimental treatment.
Several mathematical algorithms are available for assessing the stability of potential reference genes, with three programs widely employed, namely geNorm25, NormFinder26 and BestKeeper31. Each offers distinct statistical parameters for the prediction of gene stability and the optimal number of reference genes for accurate normalization of RT-qPCR data.
Here, 12 candidate reference genes were compared for gene expression normalization in root tissues in Musa genotypes contrasting in resistance to the vascular wilt pathogen F. oxysporum f. sp. cubense STR4, namely Musa acuminata subsp. burmannicoides var. Calcutta 4 (C4) (resistant) and Musa sp. cultivar Prata-anã (PA) (susceptible). The stability of the candidate reference genes was validated through examination of the relative expression of a Thaumatin-like protein 1 (TL1) target gene, encoding a Pathogenesis-related protein 5 (PR5). The reference gene sets developed in this study will enable accurate analysis of expression of target genes involved in defence responses to this important pathogen.
Results
Analysis of expression levels and Cq values of candidate reference genes
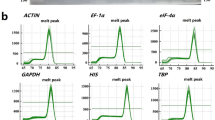
Information regarding primer sequences and PCR amplification conditions for the 12 candidate reference genes is summarized in Table 1. Dissociation curve analysis confirmed amplification specificity for all the tested primers, with single peaks observed in melting curves with cDNA material from inoculated and non-inoculated treatments in C4 and PA (Fig. 1). Variations in cycle quantification (Cq) values across treatments were depicted in a Box-plot representation (Fig. 2), with intervals, means, medians, and outliers presented for the tested reference genes across the four treatments. Overall, Cq values were generally lower in C4 when compared to PA, indicating greater expression. The lowest Cq values were observed for the genes ACTA1 and bTUB3 in C4, and for GAPDH and bTUB3 in PA. Highest Cq values, by contrast, were observed for the genes APT, RAN and Ubiq1 in both genotypes, indicating lowest expression levels. In the case of C4, greater variation in outlier Cq values was observed in the genes bTUB1 and Ubiq1. No such variation was observed in the case of PA.
Representative boxplot of quantification cycle (Cq) values for 12 candidate reference genes tested in root material in Musa acuminata subsp. burmannicoides Calcutta 4 (C4) and Musa sp. cultivar Prata-anã (PA), both inoculated (inoc) with Fusarium oxysporum f. sp. cubense STR4 and non-inoculated (mock).
Determination of candidate reference genes stability
Stability analysis and rankings for the candidate reference genes were conducted utilizing the mathematical algorithms geNorm, NormFinder and BestKeeper. Data was analysed separately for both genotypes, with gene expression data originating from root tissue samples collected at 1, 2, and 4 days after inoculation (DAI) and similarly from mock-inoculated controls. The combined analysis of gene expression stability was based on the total datasets generated for the two genotypes, combining data for biotic stress treatments and non-stressed controls.
When considering all C4 samples, L2 and BTUB3 were ranked as the most stable genes according to analyses conducted with geNorm and NormFinder, exhibiting the lowest M (average expression stability) and SV (stability value). Based on BestKeeper, UBIQ2 and UIBIQ1 genes were the most highly ranked in terms of stability, with the lowest SD (standard deviation) observed in expression values (Table 2). In the case of all PA samples, GAPDH and ACTIN 1 were identified as the genes with the lowest M values according to analysis with geNorm. By contrast, NormFinder identified TUBa and APT as the most stable genes based on SV values, with BestKeeper indicating the greatest stability in bTUB3 and L2, with these genes displaying the lowest SD. For both genotypes, following the M ≤ 0.5 cutoff set by geNorm, the first 7 genes would qualify as stable and suitable for use (Table 2).
In analysis of the Global datasets, geNorm identified only TUBa and ACTIN1 as stable genes, with M values ≤ 0.5. NormFinder identified TUBa and bTUB3, based on SV values, while BestKeeper indicated L2 and bTUB3 as the most stable genes, based on SD values (Table 2).
Determination of the optimal number of reference genes for normalizing the expression of target genes was assessed using the algorithm geNorm (Fig. 3). Based on a V ≤ 0.15 cutoff, two genes were considered sufficient for normalization in each of C4 and PA, with V values of 0.141 and 0.092, respectively. However, when considering the global sample, two genes would be insufficient (V = 0.175), with three genes required for data normalization (V = 0.134).
Pairwise variation values obtained using geNorm to determine the optimal number of reference genes for accurate RT-qPCR normalization, for both genotypes Musa acuminata subsp. burmannicoides Calcutta 4 (C4) and Musa sp. cultivar Prata-anã (PA), conditions, and the global analysis. When values are below the cutoff of 0.15, the inclusion of an additional reference gene is not necessary.
Following analysis of stability values of gene pairs with the lowest SV values generated by NormFinder (Fig. 4), L2-RAN, bTUB3-EF1a, and L2-EF1a were identified as pairs with SV values ≤ 0.15 in the case of C4, and APT-TUBa and GAPDH-TUBa in the case of PA. No pairs were observed with SV ≤ 0.15 in the case of the global dataset.
Expression analysis of the TL1 gene (PR5) using selected reference genes
To verify the stability of any potential reference gene, analysis of normalization of expression of a target marker gene for a specific experimental condition is required. Here, the relative expression of a TL1 gene, which is a PR5 gene involved in plant host resistance to various pathogens, including Foc32,33, was evaluated in cDNA samples for the Foc STR4-resistant genotype C4, inoculated at 1, 2, and 4 DAI, as well as for corresponding non-inoculated controls. Evaluations were also conducted in the susceptible Musa genotype PA, under the same conditions.
The four most stable genes for relative expression normalization, based on rankings through geNorm and NormFinder analyses, were tested for each Musa genotype. In the case of C4, the most stable genes employed comprised L2, bTUB3, EF1a, and TUBa, while for PA, these comprised GAPDH, ACTIN1, APT, and TUBa. Ubiq1 and bTUB1 were also included in relative expression normalization analysis of the target gene TL1 to represent the least stable genes for employment in normalization for both genotypes.
In C4, differential expression of the TL1 gene between inoculated and non-inoculated controls was observed when normalizing using the two pairs formed by the four most stable genes. This contrasted with the relative expression data normalized using the least stable genes, where no differential expression of the TL1 gene between inoculated and non-inoculated controls was observed. In PA, where there was no differential expression of TL1 between inoculated and non-inoculated controls, expression levels following normalization using the most or least stable genes did significantly differ, with the same expression trends observed (Fig. 5).
Relative expression levels of Thaumatin-like protein 1 (TL1) in Musa acuminata subsp. burmannicoides var. Calcutta 4 (C4) and Musa sp. cultivar Prata-anã (PA), under infection with Fusarium oxysporum f. sp. cubense STR4 isolate 218A 1, 2 and 4 days after inoculation. Normalization was examined using the two most stable gene pairs for C4 (L2 and bTUB3; EF1a and TUBa), and for PA (GAPDH and ACTIN1; APT and TUBa), with each compared with the least stable pair for C4 and PA (UBIQ1 and bTUB1). **** indicated significance at p < 0.0001, ** indicated significance at p < 0.001 and * indicated significance at p < 0.5.
Discussion
The interaction between different Musa genotypes and Foc has been the subject of several transcriptome analyses, with a focus on host gene characterization during both the compatible and incompatible response28. To verify in silico-derived gene expression data, RT-qPCR is widely employed to quantify the expression of a specific gene in cDNA samples originating from different treatments17,27,29,30,34,35,36,37. Accurate analysis of gene expression, however, requires normalization to reference genes that display stable expression under the investigated experimental conditions. Such normalization will then correct for variation inherent between cDNA samples. Screening of potential reference genes for normalization thus requires consideration of specific experimental samples30, whether originating from different organisms, tissue types, biotic or abiotic stresses or cDNA sample.
Previous reference genes developed for Musa have included those encoding essential and constitutive proteins in plants. These have included the ribosomal protein 2 (RPS2), ribosomal protein L2 (L2), and actin (Actin), which constitutes an essential component of the cell cytoskeleton. Proteins responsible for translation and formation of other proteins, such as the elongation factor 1α (EF1α) and ubiquitin (UBIQ1 and UBIQ2), have also been employed, as well as proteins responsible for cellular metabolism, such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH), among others27,28,29,30. In all these cases, however, gene expression, even when considered basal, is often variable, such that different genotypes, tissues, developmental stages, and responses to biotic and abiotic stresses must all be considered in the examination of gene expression stability.
Previously, Chen and colleagues27 evaluated expression stability of 20 candidate genes in M. acuminata Cavendish subgroup fruit tissues, during different developmental stages, including post-harvest, different abiotic stresses (chilling and high temperature), biotic stress (Colletotrichum musae), and during ethylene hormone treatment. Stability ranking varied among all treatments, with only the APT gene being less stable in four out of the six tested conditions. Rego and coworkers29 also characterized the expression of eight candidate genes in leaves of M. acuminata Cavendish Grand Naine and Calcutta 4 under biotic stress (Pseudocercospora musae). Data revealed APT and UBQ2 as the most stable genes for normalization in Calcutta 4, whilst RAN and α-TUB were more stable in Cavendish Grand Naine. Podevin and coworkers30 similarly investigated the expression of six candidate reference genes in various tissues of Musa genotypes from the AAA, AA, and ABB groups, and under various experimental treatments that included physiological growth conditions, biotic (Pseudocercospora fijiensis) and abiotic (osmotic) stress conditions. Data demonstrated that for each experimental condition, an investigation of appropriate reference genes is necessary.
In our study on the stability of 12 candidate reference genes in Musa C4 and PA during the interaction with Foc STR4, data based on analyses of stability using geNorm were supported by NormFinder-based analysis. BestKeeper-derived rankings were not considered for determining genetic stability, with different stability rankings to the former algorithms based on high coefficient of variation values. We demonstrate that the employment of a set of distinct most stable genes is necessary for each genotype: L2 and bTUB3 for C4 and GAPDH and ACTIN1 for PA. The least stable reference genes comprised UBIQ1 and bTUB1 for both C4 and PA. Rankings were also corroborated based on Cq value outlier numbers, as observed in Fig. 1. Whilst more outliers were apparent for UBIQ1 and bTUB1, the opposite was observed for the most stable genes, with a low amplitude of Cq for bTUB3 and L2 between the inoculated and mock treatments in C4, and similarly for GAPDH and ACTIN1 in the inoculated and mock treatments in PA. Previously, Zhang and colleagues28 reported candidate reference genes for normalization of target gene expression in the Musa genotypes Guangfen No.1 (ABB) and Cavendish Brazilian (AAA) following inoculation with Foc TR4 and Foc R1, with L2 and TUB most stable in Guangfen No. 1, and ACT1 and TUB most stable in Brazilian. Whilst L2 and TUB were also found to be the most stable genes for C4 in our study, indicating that certain genotypes from different groups (AA and ABB, respectively) may share stable reference genes appropriate for studies with Foc, comparisons also revealed differences between specific genotypes. Such data highlights that for each experimental condition and genotype examined, specific reference genes are essential and that disregarding biological differences is not suitable for correct normalization, potentially interfering with accurate analysis of target gene expression via RT-qPCR.
Validation of the stability of the reference genes was conducted through analysis of the relative expression of the TL1 gene during the interaction of each genotype with Foc STR4. This gene encodes a thaumatin-like protein 1, which is a PR-5 protein involved in the defence response to biotic stress. Previously, Mahdavi and coworkers33 demonstrated that insertion of a thaumatin-like protein (TLP) gene in susceptible Pisang Nangka bananas (AAB) conferred resistance to Foc TR4. Here, expression analysis of the TL1 gene in C4 when performed with the four most highly ranked stable reference genes based on geNorm (Table 2) demonstrated that all four genes were appropriate for normalization of target gene expression when there was differential expression of the gene in response to Foc STR4. As expected, the TL1 gene did not show differential expression in response to infection by Foc STR4 in the susceptible genotype PA, with low relative expression even after infection. No difference in the normalization pattern was observed between the most and least stable reference genes in this genotype.
In summary, given the importance of reference genes for accurate normalization of target gene expression in the Musa-Foc SRT4 pathosystem, this study developed reference gene sets for application in candidate gene discovery in genotypes contrasting in resistance to this important pathogen. Based on algorithm ranking, L2 and bTUB3 should be prioritized for use in C4 root materials both inoculated with Foc STR4 or non-inoculated, and similarly with GAPDH and ACTIN1 in PA. These reference genes will contribute to advancing understanding of the host defence responses to this important pathogen.
Methods
Plant material
In vitro propagated plantlets of the susceptible Musa sp. cultivar 'Prata-anã' (subgroup 'Pome' AAB) and the resistant wild genotype 'Calcutta 4' (Musa acuminata subsp. burmannicoides, subgroup AA) were provided by Embrapa Cassava and Tropical Fruits. Plantlets were acclimatized and subsequently grown in pots containing coconut fibre substrate. Eighteen plants of each genotype were maintained in a greenhouse at an average temperature of 24 °C, with watering at 48-h intervals for 60 days prior to inoculation.
Root inoculation with Foc STR4
Plants of each genotype were inoculated with F. oxysporum f. sp. cubense STR4 (isolate CNPMF 218A from Embrapa Cassava and Tropical Fruits). Inoculation was conducted according to Rocha and colleagues38. Inoculum comprised 50 g of rice, previously infested with the pathogen at a concentration of 106 CFU/g of rice, added directly to the substrate. Controls comprised non-infested rice, with application to the substrate as above. Following a complete randomized design, with three distinct biological replicates per treatment, root samples were collected from Calcutta 4 (C4) and Prata-Anã (PA) at 1, 2, and 4 DAI and similarly from mock-inoculated controls. Samples were immediately flash-frozen in liquid nitrogen and stored at—800 C.
RNA extraction and cDNA synthesis
Total RNA was extracted from frozen 1 g root samples using the Plant RNA Reagent (Invitrogen ThermoFisher Scientific, MA, USA) and purified using the Directzol Plant RNA Purification kit (Zymo Research, CA, USA) following the manufacturer's protocol. RNA quality was verified using 1% agarose gels and quantified using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, MA, USA). A 1 µg aliquot of RNA from each treatment and replicate was used for cDNA (complementary DNA) synthesis via the Super Script IV Reverse Transcriptase kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer's protocol.
Selection of candidate reference genes
A total of 12 candidate genes were selected for testing as potential reference genes based on recent literature for genes with stable expression in Musa spp. and involvement in basal cell activities. Actin-3 (ACTIN1), APT, EF1α, GAPDH, α-TUB, RAN, β-TUB (bTUB1), Ubiq1, and Ubiq2 were selected from Rego et al. (2019), L2 and β-TUB (bTUB3) from Zhang and coworkers28, and Actin-2 (ACTA1) from Castañeda and coworkers39 (Table 1).
Real-time quantitative PCR
Analysis of gene expression was conducted using the iTaq™ Universal SYBR® Green kit (Bio-Rad, Hercules, CA, USA). Each qPCR reaction was conducted in a final volume of 10 µL, containing 2 µL of a 1:20 dilution of each cDNA stock, 0.2 µM of each primer, and 5 µL of the iTaq™ Universal SYBR® Green kit. PCR amplifications were carried out on an ABI StepOne™ Real-Time PCR thermocycler (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) according to the following parameters: an initial two-step phase of 50 °C for 2 min and 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 15 s, and primer annealing and extension at 60 °C for 60 s. Three biological and three technical replicates were included for each gene. Melting temperatures (Tm) were determined using the StepOne Software v2.3 (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA).
Determination of candidate reference gene stability
Expression analysis of each of the 12 candidate genes was measured for all C4 and PA replicate cDNA samples based on quantification cycle (Cq) values. Gene expression stability was determined using the statistical algorithms geNorm25, NormFinder26 and BestKeeper31, according to default parameters. Analyses were conducted for expression data from each individual genotype, as well as based on a global analysis of data for both genotypes, both inoculated and non-inoculated. In the case of geNorm, the lower the average M value, below a threshold of ≤ 0.5, the greater the stability for a candidate reference gene. This algorithm also estimates a V value (Vn/n + 1) to determine the optimal number of reference genes for use in normalization. NormFinder calculates expression stability based on the stability value (SV), where lower values indicate higher gene stability. This algorithm also identifies the most appropriate combinations of pairs of stable genes. In the case of BestKeeper, lower coefficient of variation (CV) and standard deviation (SD) values indicate greater expression stability. R software40 was employed for analysis of data generated with all algorithms and for preparation of figures.
Expression analysis of a PR-5-encoding gene
Combinations of the most stable genes in C4 (L2 and bTUB3; EF1a and TUBa) and PA (GAPDH and ACTIN1; APT and TUBa), together with the least stable genes in both C4 and PA (Ubiq1 and bTUB1), were all compared in terms of efficiency in normalization of expression of a gene in C4 encoding a Thaumatin-like protein 1 (TLP1) (Macma4_05_g17130), which is member of the Pathogenesis-related-5 protein family (PR-5). In a previous transcriptome study by the group (data not shown), the Log2 fold-change (LogFC) expression of this gene in C4 in inoculated root materials compared to non-inoculated controls was 3.86, 3.46 and 2.37 for 1, 2 and 4 DAI, respectively. Specific primers designed for this gene comprised TL1-F (GATGCGACGCTGATGAAA) and TL1-R (AGACCGGCCATAAGATACA). RNA extraction, cDNA synthesis and RT-qPCR were conducted as described above.
Data availability
All data generated and analysed during the study is included in the published article.
References
FAO. Banana Statistical Compendium 2021. (FAO, 2022).
Kema, G. H. J. et al. Editorial: Fusarium wilt of banana, a recurring threat to global banana production. Front. Plant Sci. 11, 628888 (2021).
Amorim, E. P., Santos-Serejo, J. A., Amorim, V. B. O. & Silva, S. O. Melhoramento genético. In O agronegócio da banana (eds Ferreira, C. F. et al.) 173–200 (Embrapa, 2016).
Laliberté, B. Global strategy for the conservation and use of Musa genetic resources: A consultative document prepared by the Global Musa Genetic Resources Network (MusaNet). (Biodiversity International, 2016).
Ploetz, R. C. Fusarium wilt of banana. Phytopathology 105, 1512–1521 (2015).
Pegg, K. G., Coates, L. M., O’Neill, W. T. & Turner, D. W. The Epidemiology of Fusarium Wilt of Banana. Front. Plant Sci. 10, 1–19 (2019).
Dita, M., Barquero, M., Heck, D., Mizubuti, E. S. G. & Staver, C. P. Fusarium wilt of banana: Current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. https://doi.org/10.3389/fpls.2018.01468 (2018).
Jamil, F. N., Tang, C.-N., Saidi, N. B., Lai, K.-S. & Baharum, N. A. Fusarium wilt in banana: Epidemics and management strategies. In Horticultural Crops (eds Baimey, K. et al.) (IntechOpen, 2020). https://doi.org/10.5772/intechopen.89469.
Thangavelu, R. & Mustaffa, M. M. First report on the occurrence of a virulent strain of fusarium wilt pathogen (Race-1) infecting cavendish (AAA) group of bananas in India. Plant Dis. 94, 1379–1379 (2010).
Thangavelu, R. et al. First Report of Fusarium oxysporum f. sp. cubense VCG 0125 and VCG 01220 of race 1 infecting Cavendish bananas (Musa sp. AAA) in India. Plant Dis. 105, 1215–1215 (2021).
ICAR–NCRB. Annual Report 2022. (ICAR - National Research Centre for Banana (2023)
Raman, T. et al. Comparative whole-genome sequence analyses of fusarium wilt pathogen (Foc R1, STR4 and TR4) infecting Cavendish (AAA) bananas in India, with a special emphasis on pathogenicity mechanisms. J. Fungi 7, 717 (2021).
Thangavelu, R., Raj, E. E., Pushpakanth, P., Loganathan, M. & Uma, S. Draft genome of Fusarium oxysporum f. sp. cubense strain tropical race-4 infecting Cavendish (AAA) Group of Banana in India. Plant Dis. 105, 481–483 (2021).
Thangavelu, R. et al. Identification of sources resistant to a virulent Fusarium wilt strain (VCG 0124) infecting Cavendish bananas. Sci. Rep. 11, 3183 (2021).
Buddenhagen, I. W. Understanding strain diversity in Fusarium oxysporum f. sp. cubense and history of introduction of tropical race 4 to better manage banana production. Acta Hortic. 828, 193–204 (2009).
Kotari, P., Ajitha, R. & Ravishankar, K. V. Molecular investigations of gene expression analysis in two contrasting genotypes of banana during fusarium wilt (Foc1) infection. Indian Phytopathol. 71, 445–452 (2018).
Zhang, L. et al. Transcriptomic analysis of resistant and susceptible banana corms in response to infection by Fusarium oxysporum f. sp. cubense tropical race 4. Sci. Rep. 9, 1–14 (2019).
Li, W. M. et al. Deep RNA-seq analysis reveals key responding aspects of wild banana relative resistance to Fusarium oxysporum f. sp. cubense tropical race 4. Funct. Integr. Genomics 20, 551–562 (2020).
Kaushal, M., Mahuku, G. & Swennen, R. Comparative transcriptome and expression profiling of resistant and susceptible banana cultivars during infection by Fusarium oxysporum. Int. J. Mol. Sci. 22, 1–29 (2021).
Kubista, M. et al. The real-time polymerase chain reaction. Mol. Aspects Med. 27, 95–125 (2006).
VanGuilder, H. D., Vrana, K. E. & Freeman, W. M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44, 619–626 (2008).
Gutierrez, L. et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618 (2008).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Radonić, A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862 (2004).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Chen, L. et al. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234, 377–390 (2011).
Zhang, L. et al. Identification and validation of reference genes for RT-qPCR analysis in banana (Musa spp) under Fusarium wilt resistance induction conditions. J. Phytopathol. 165, 746–754 (2017).
Rego, E. C. S. et al. Stable reference genes for RT-qPCR analysis of gene expression in the Musa acuminata-Pseudocercospora musae interaction. Sci. Rep. 9, 1–11 (2019).
Podevin, N., Krauss, A., Henry, I., Swennen, R. & Remy, S. Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol. Breed. 30, 1237–1252 (2012).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
de Jesús-Pires, C. et al. Plant thaumatin-like proteins: Function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 21, 36–51 (2020).
Mahdavi, F., Sariah, M. & Maziah, M. Expression of rice Thaumatin-like protein gene in transgenic banana plants enhances resistance to fusarium wilt. Appl. Biochem. Biotechnol. 166, 1008–1019 (2012).
Nolan, T., Hands, R. E. & Bustin, S. A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582 (2006).
Munusamy, U. RT-qPCR profiling of pathogenesis related genes in Musa acuminata cv ‘Berangan’ seedling challenged with Fusarium oxysporum f.sp. cubense tropical race 4. Pakistan J. Agric. Sci. 56, 37–42 (2019).
Li, C. et al. Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense. BMC Genomics 14, 1–16 (2013).
Pinheiro, T. D. M. et al. Transcriptome Profiling of the Resistance Response of Musa acuminata subsp. burmannicoides, var Calcutta 4 to Pseudocercospora musae. Int. J. Mol. Sci. 23, 1–27 (2022).
Rocha, A. D. J. et al. Molecular, histological and histochemical responses of banana cultivars challenged with Fusarium oxysporum f. sp. cubense with different levels of virulence. Plants 11, 2339 (2022).
Castañeda, N. E. N. et al. Gene expression analysis in Musa acuminata during compatible interactions with Meloidogyne incognita. Ann. Bot. 119, 915–930 (2017).
R Core Team. R: A language and environment for statistical computing. at https://www.r-project.org/ (2021).
Acknowledgements
This work was funded by Brazilian funding bodies FAPDF (Grant 00193. 00000778/2021-03), Instituto Nacional de Ciência e Tecnologia Plant Stress Biotech (Grant 465480/2014-4), and CAPES (Finance Code 001). RNGM was supported by a fellowship from CNPq (Grant 308165/2021-7).
Author information
Authors and Affiliations
Contributions
R.N.G.M. conceived and designed the study. E.C.C. conducted the biotic assays and RT-qPCR analysis. E.C.C., L.S.B. and T.G.G. analysed the data. R.N.G.M., E.C.C. and T.G.G. provided intellectual input. E.C.C. and R.N.G.M. wrote and revised the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial and/or non-financial interests in relation to the work described.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
de Castro Costa, É., Bastos, L.S., Gomes, T.G. et al. Reference genes for RT-qPCR analysis in Musa acuminata genotypes contrasting in resistance to Fusarium oxysporum f. sp. cubense subtropical race 4. Sci Rep 14, 16578 (2024). https://doi.org/10.1038/s41598-024-67538-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-67538-0