Abstract
Radiomics features (RFs) serve as quantitative metrics to characterize shape, density/intensity, and texture patterns in radiological images. Despite their promise, RFs exhibit reproducibility challenges across acquisition settings, thus limiting implementation into clinical practice. In this investigation, we evaluate the effects of different CT scanners and CT acquisition protocols (KV, mA, field-of-view, and reconstruction kernel settings) on RFs extracted from lumbar vertebrae of a cadaveric trunk. Employing univariate and multivariate Generalized Linear Models (GLM), we evaluated the impact of each acquisition parameter on RFs. Our findings indicate that variations in mA had negligible effects on RFs, while alterations in kV resulted in exponential changes in several RFs, notably First Order (94.4%), GLCM (87.5%), and NGTDM (100%). Moreover, we demonstrated that a tailored GLM model was superior to the ComBat algorithm in harmonizing CT images. GLM achieved R2 > 0.90 in 21 RFs (19.6%), contrasting ComBat's mean R2 above 0.90 in only 1 RF (0.9%). This pioneering study unveils the effects of CT acquisition parameters on bone RFs in cadaveric specimens, highlighting significant variations across parameters and scanner datasets. The proposed GLM model presents a robust solution for mitigating these differences, potentially advancing harmonization efforts in Radiomics-based studies across diverse CT protocols and vendors.
Similar content being viewed by others
Introduction
Utilizing Radiomics Features (RFs) to quantitatively assess clinical or pathological conditions remains challenging for implementation in clinical practice due to issues with reproducibility and generalizability1. Consequently, no RFs-based software has yet gained approval from regulatory bodies such as those in the United States (US) or the European Union (EU).
As a response, international scientific endeavors have focused on enhancing technical stability and achieving better reproducibility across various clinical scenarios, exemplified by initiatives like The Image Biomarker Standardization Initiative2,3.
Nonetheless, challenges related to non-reproducibility persist in current studies utilizing RFs derived from CT scans. Primarily, normalization algorithms for pre-processing rely heavily on either acquired images or pre-acquired digital phantoms, which serve as the benchmark for evaluating RFs' stability under different CT scanner parameters, thus minimizing patient exposure to X-rays. However, studies have demonstrated that not all materials used in digital phantoms remain stable when CT acquisition protocols are altered, potentially failing to accurately represent the intricate structures of the human body4,5,6. Cadaveric studies offer a more precise depiction of organ textural structures, yet research in this area is limited, particularly regarding the impact of CT protocols on cadaveric specimens acquired externally from the donor's body.
This study aims to assess the reproducibility and behavior of RFs derived from CT images of vertebrae obtained from a cadaveric thoraco-abdominal trunk7. CT scans from the same cadaver were conducted using varied acquisition parameters across three CT scanners from different manufacturers and with different detector counts. A test–retest procedure was also executed on a single CT scanner. We conduct a detailed evaluation of the influence of each CT acquisition parameter on individual RFs and provide a quantitative comparison of RFs' harmonization accuracy between a novel Generalized Linear Model (GLM) and the ComBat algorithm. Our analysis exhibits the GLM as a more precise method for standardizing data in Radiomics CT analyses. We make both the entire image dataset and the GLM model accessible for further analyses, whether for customizing analyses to different body organs, exploring alternative Radiomics libraries, or developing optimized standardization algorithms.
Results
The results are organized as follows: each paragraph reports a summary of the specific effects of each protocol parameter on the RFs both in terms of univariate and multivariate (GLM) statistical analyses, whereas all the complete details are reported in exhaustive tables accompanied by a description of all the possible comparisons in the Supplementary Material.
Effects of KV variation on features reproducibility
The statistical analysis of CT acquisitions at fixed mA (300 mA) and variable kV showed significant differences on texture RFs, with variable impact according to the considered scanners. No effects on shape features were found on all scanners. Multivariate analyses through GLM showed a general agreement with univariate testing in terms of voltage effects. Of note, specific voltage effects were also found for each scanner on texture features.
The complete statistical description is available in the supplementary material Sects. 1.1, 1.5 and Supplementary Table 1. The post-hoc analyses of ANOVA statistical tests are reported in Fig. 1.
Post-Hoc test of Radiomics features. The Heatmap shows the number of statistically significant (p < 0.05) pair-wise comparison using paired T-test or Mann–Whitney according to normal distribution. Statistical comparisons were performed for kV variation protocol and mA variation protocol. Benjamini-Hochberg’s correction was performed for multiple comparison.
Effects of mA variation on features reproducibility
The analysis of CT acquisitions at fixed kV (120 kV) and variable mA did not show statistically significant differences on all RFs for Scanner 1 and 2, whereas GLCM, GLDM, and GLRLM showed a significant difference in Scanner 3. These results are confirmed by the multivariate analysis, which showed no significant effect of current alone (see Fig. 2). The observed differences in univariate testing for Scanner 3 were also observed as a significant interaction effect between Scanner 3 and current in the GLM analysis.
Generalized Linear Model results on RFs. The table reports the significance of each effect considered in the model (columns) in respect to each RFs (rows). Red cells represent a significative (p-value < 0.05) and positive contribution (Coefficient > = 0), whereas blue cells represent a significative (p-value < 0.05) and negative contribution (Coefficient < 0).
The complete statistical description is available in the supplementary material Sects. 1.2, 1.5 and Supplementary Table 2. Post-hoc analyses of ANOVA statistical tests are reported in Fig. 1.
Field-of-view
Several RFs (First Order, GLRLM and NGTDM) resulted significantly different when varying the FOV (Abdomen = 500 mm, Spine = 320 mm). Similar results were also observed after multivariate testing where almost all RFs showed significant effect of FOV covariate, mainly with a reduction when moving from Spine to Abdomen.
The complete statistical description is available in the supplementary material Sects. 1.2, 1.5 and Supplementary Table 3.
Reconstruction kernel
The choice of the Reconstruction Kernel (Standard/Bone) showed no statistically significant difference on Shape features acquired on all scanners, whereas most of the texture features showed a significant change due to different kernels. The GLM analyses showed similar results on Shape features and a general reduction in RFs when switching to "Bone” reconstruction Kernel. Of note, with respect to Scanner 1, the interactions between Scanner 2 and Scanner 3 with the reconstruction kernel showed opposite impact on the RFs (Fig. 2).
The complete statistical description is available in the supplementary material Sects. 1.2, 1.5 and Supplementary Table 4.
Test—retest
Intra-class correlation analysis of Test–Retest sequences showed an overall high agreement between test and retest protocol, with 71 (76.3%) RFs with ICC higher than 0.90, 10 (10.8%) RFs with ICC between 0.8 and 0.9, 9 (9.7%) RFs with ICC between 0.8 and 0.7 and only 3 (3.2%) RFs with ICC less than 0.70. Complete results are reported in Supplementary Table 5.
Parameter effects: scanner comparison
The statistical analysis through GLM showed minimal difference on shape features on images acquired on both Scanner 2 and Scanner 3 with respect to images acquired on Scanner 1, a low-to-moderate effect on texture features on images acquired on Scanner 2 with respect to Scanner 1 (same vendor), and a strong difference on texture features on images acquired on Scanner 3 (different vendor) compared to those obtained from images acquired on Scanner 1. The complete statistical description is available in the supplementary material Sects. 1.2, 1.5 and Supplementary Fig. 1.
Normalization algorithm classification: GLM vs ComBat
We applied both ComBat and the developed GLM algorithm to the above-mentioned data/techniques. We evaluated the performance in predicting each RF by iteratively masking a random subset of recordings, which will be used as unseen test sets in a tenfold cross-validation procedure. The results below report the mean prediction metric for each fold.
ComBat normalization algorithm obtained a mean R2 across 10-folds cross-validation higher than 0.90 in 1 RFs (0.9%), whereas GLM normalization algorithm obtained high R2 in 21 RFs (19.6%). When comparing accuracy metrics between normalization algorithm, GLM algorithm was statistically superior in 39 RFs with respect to ComBat (which was superior in 16 RFs) in terms of R2. Moreover, GLM algorithm obtained lower values of MSE in 44 RFs in respect to ComBat (which had lower values in 14 RFs). All results concerning R2 comparison are reported in Supplementary Table 6.
Discussion
This study represents the first attempt in describing the effects of different CT acquisition parameters and different CT scanners (below referred to as “protocol parameters”) on RFs obtained from a cadaveric donor. Given the relatively low post-mortem changes of the bone8, we focused our study on Radiomics of lumbar vertebrae. Vertebral RFs showed important differences according to the protocol parameters employed. More in detail, within each RFs class, the number of RFs that were significantly different and the extent of modification was varying for each of the considered protocol parameters (mA, kV, FOV, reconstruction kernel, CT scanner manufacturer and model). This should be considered when planning to perform or when analyzing retrospective CT data in both single- and multi-center studies.
RFs obtained from CT scanners produced by the same vendor were statistically more similar than those obtained from datasets from different CT vendors, even if equipped with different numbers of detectors. This result is in line with phantom-based studies which demonstrated that same vendors CT scanners were clustered together9, and multi-vendors dual-energy CT studies10 demonstrated lower reproducibility.
The effects of mA and KV variations on the reproducibility of RFs were different. In fact, modification of mA was not associated with alteration of RFs values 11,12, whereas kV variations were associated with exponential variation of several RFs (especially First Order and GLCM)13. As this result might have strong implications in multicenter studies, standardization of kV in CT protocols among different centers is strongly suggested to obtain homogenous data. We can speculate that current and voltage might have different effects on the reproducibility of RFs because of the Photoelectric and Compton effects. Specifically, the high-density vertebral trabecular structure could be better characterized by high-energy photons (higher kV) that could better describe biological related characteristics of the patients’ bony tissues14.
CT acquisitions with different FOV showed a significant modification of the shape features, which is likely due to the different voxel size. However, also other classes of RFs were significantly modified. This is in line with results previously obtained on phantom data6, that showed low reproducibility when varying FOV and reconstruction Kernel. When compared to the standard image kernel, reconstruction kernel applied to the original raw data was associated with significant differences especially in First Order, GLRLM, GLSZM and NGTDM features. The results obtained analyzing the interactions between reconstruction algorithm and different scanners suggested a stronger agreement between RFs reconstructed from scanners of the same vendor compared to RFs obtained from scanners of different vendors. This outcome is probably due to the diverse proprietary kernel algorithms used by different vendors, suggesting caution when processing images from different vendors particularly when proprietary filters are applied.
From the GLM analysis, we observed that the Intercept is almost always significantly associated with the target RF. This information suggests that these features contain information independent from protocol parameters which might be related to patho/physiological subject’s characteristics.
We compared the GLM to a well-established harmonization algorithm (ComBat) when predicting unseen acquisitions. We found that the GLM approach provides better performances than the ComBat algorithm in predicting RFs in terms of both R2 and MSE. IBSI guidelines and several research studies recommend the use of ComBat to counteract batch effects15. The proposed GLM algorithm could represent a reference for future research of Radiomics and could be applied to normalize CT acquisitions performed on different CT scanners.
Overall, we showed that RFs standardization can be significantly improved if data are mapped on a reference machine with a model previously calibrated using a specific CT protocol. For this purpose, the collection and sharing of CT images recorded with the presented protocol with additional vendors and machine models would allow for a broader and more accurate standardization of RFs with the aim of improving generalizability and repeatability of Radiomics studies. Further, we propose this GLM algorithm as a more accurate method for data harmonization in Radiomics CT studies. Both the dataset and the GLM code are made available for further analyses, either tailored to other body organs, or to different Radiomics libraries/features, or to the development of further optimized standardization algorithms. Also, by sharing our data we aim at encouraging worldwide researchers to provide additional data by including other CT scanners and body districts.
There are several limitations in our study. Unfortunately, we could not gather a complete test–retest for all CT scanners. Second, as the GLM approach is very robust according to the available sample size, a larger data collection would allow for exploring the potential of other machine-learning (e.g., Support Vector Machines, Random Forest) and deep-learning (e.g., CNN) approaches, which would possibly improve the presented results. Third, this study mainly focused on the analysis of the complete segmentation of lumbar vertebrae, whereas the analysis of each vertebral structure would allow for the development of a vertebra-specific standardization model for RFs.
Despite these limitations, we believe this benchmarking work will guide Radiomics-based studies towards a much more accurate and standardized approach thus encouraging worldwide research in creating a collaborative CT image datasets which would define the references for RFs standardization across different scanners and body districts, possibly derived from cadaveric bodies instead of phantoms or animals.
In conclusion, we evaluated the effects of several CT acquisition parameters (mA, kV, FOV, Reconstruction Kernel, CT Scanner) on RFs of lumbar vertebrae in a cadaveric trunk. All the considered effects were included in a multivariate model (GLM) to standardize RFs. This model was found to be more accurate than the ComBat algorithm.
The complete dataset is publicly available to be applied for future research in the RFs field, and to be considered as the starting reference point for the creation of a collaborative open CT image database to increase the sample size, the range of available scanners, and the available body districts.
Materials and methods
Dataset description
The analysis comprises the dataset described in a previously published paper and freely accessible at https://zenodo.org/records/10053317. Briefly, the dataset comprises the acquisition of a human cadaver belonged to an 80-year-old Caucasian man. The cause of death was septic shock due to a pseudomonas infection which first compromised the urinary tract and the lungs. The man was 183 cm high and weighted 104 kg with a BMI of 31.19 kg/m2. The cadaver was imaged without equipment and/or clothing, at room temperature.
The dataset comprises multiple Computed Tomography (CT) acquisitions of the cadaveric trunk performed on 3 different CT scanners: (a) Revolution CT (GE HealthCare, 256 slices, defined as Scanner 1); (b) Revolution EVO (GE HealthCare, 64 slices, defined as Scanner 2); (c) Ingenuity CT (Philips Healthcare, 64 slices, defined as Scanner 3). Test–Retest protocol was performed on a single scanner (Scanner 1).
The complete acquisition protocol comprises 2 main parts:
-
KV variable: the acquisitions were performed at 300 mA, changing the kV parameter from 80 to 140 kV with 20 kV steps.
-
mA variable: the acquisitions were performed at 120 kV, changing the mA parameter from 250 to 400 mA with 50 mA steps.
Each acquisition includes also:
-
two fields of view (FOV): Abdomen (500 mm) and Spine (320 mm);
-
two reconstruction algorithms: Standard Soft Tissue Kernel and the Bone Kernel;
Therefore, a total of 112 acquisitions were included in the analysis.
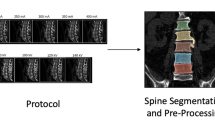
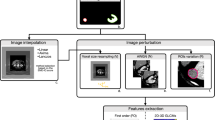
Each images is matched with the relative segmentation of lumbar vertebrae (from L1 to L5), obtained by training a convolutional neural network (CNN) within nnU-Net framework. The complete workflow is reported in Fig. 3 and segmentation results are reported in Fig. 4
Radiomics feature extraction
RFs were extracted using the pyradiomics library (version 3.0.1), a software adhering to the Image Biomarker Standardization Initiative (IBSI) protocol3. Features were extracted from a composite volume of interest (VOI) formed by the union of all the lumbar vertebrae VOIs. Pre-processing steps included only setting the bin width to 15 HU, to preserve the effects due to the single acquisition parameters. We included a total of 107 RFs on each VOI, divided into 14 shape features, 18 first order features, 24 grey level co-occurrence matrix (GLCM) features, 16 grey level run length matrix (GLRLM) features, 16 grey level size zone matrix (GLSZM) features, 14 grey level dependence matrix (GLDM), and 5 neighboring gray-tone difference matrix (NGTDM).
Statistical analysis
Each RF was tested for normal distribution using the Shapiro–Wilk test.
Intra- and Inter-scanner analyses were assessed using either Analysis of Variance Repeated Measurements (ANOVARM, parametric test) or Friedman’s Test (non-parametric test) according to feature distribution for kV and mA fixed sequences. Post-Hoc analyses were performed using pairwise T-Test (parametric test) or Mann–Whitney (non-parametric test) using Benjamini–Hochberg correction for multiple comparison. Intra- and Inter-scanner assessment of FOV and Kernel effects, as well as Test–Retest analyses were assessed using paired T-test (parametric test) or paired Wilcoxon (non-parametric test). Test–Retest analyses were also assessed for correlation with Intra-class Correlation (ICC).
Each RF was tested using a generalized linear model (GLM) to assess the effects of the following CT acquisition parameters: scanner (Scanner 1, Scanner 2, Scanner 3), field of view (“Abdomen” or “Spine”), reconstruction kernel (“Standard” or “Bone”), current, voltage, and the interaction of CT scanners with current, voltage and reconstruction kernel.
The Bayesian Information Criteria (BIC) was used to select between a linear (Normal/Gaussian link function) or nonlinear (Gamma link function) baseline model for each RF. Since the Gamma function is designed for strictly positive dependent variables, each RF was increased by the absolute value of the minimum.
Statistical significance was set to p < 0.05.
Comparison between standardization algorithms
The proposed GLM model was successively employed to standardize RFs across different acquisitions, which include different scanners and CT parameters.
We compared the obtained results with respect to the ComBat algorithm, the current state of the art for RFs harmonization.
Specifically, we applied a 10-folds cross-validation procedure for both GLM and ComBat algorithm and we computed for each testing fold the R2 and Mean Squared Error (MSE). Eventually, T-test was applied to statistically compare the distribution of R2 and MSE obtained from GLM and ComBat algorithm.
Data availability
The Dataset used for image analysis is freely available at the following link: https://zenodo.org/records/10,053,317.
Code availability
The GLM code and the relative weights associated to each features are available at https://github.com/rikhy967/CadAIver.
References
Huisman, M. & Akinci D’Antonoli, T. What a radiologist needs to know about radiomics, standardization, and reproducibility. Radiology. https://doi.org/10.1148/radiol.232459 (2024).
Whybra, P. et al. The image biomarker standardization initiative: Standardized convolutional filters for reproducible radiomics and enhanced clinical insights. Radiology https://doi.org/10.1148/radiol.231319 (2024).
Zwanenburg, A. et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295(2), 328–338. https://doi.org/10.1148/radiol.2020191145 (2020).
Kalendralis, P. et al. Multicenter CT phantoms public dataset for radiomics reproducibility tests. Med. Phys. 46(3), 1512–1518. https://doi.org/10.1002/mp.13385 (2019).
Varghese, B. A. et al. Reliability of CT-based texture features: Phantom study. J. Appl. Clin. Med. Phys. 20(8), 155–163. https://doi.org/10.1002/acm2.12666 (2019).
Li, Y. et al. The impact of phantom design and material-dependence on repeatability and reproducibility of CT-based radiomics features. Med. Phys. 49(3), 1648–1659. https://doi.org/10.1002/mp.15491 (2022).
Levi, R. et al. CT Cadaveric dataset for radiomics features stability assessment in lumbar vertebrae. Sci. Data 11(1), 366. https://doi.org/10.1038/s41597-024-03191-6 (2024).
Kemp, W. L. Postmortem change and its effect on evaluation of fractures. Acad. Forensic Pathol. 6(1), 28–44. https://doi.org/10.23907/2016.004 (2016).
Mackin, D. et al. Measuring computed tomography scanner variability of radiomics features. Invest. Radiol. 50(11), 757–765. https://doi.org/10.1097/RLI.0000000000000180 (2015).
Lennartz, S. et al. Robustness of dual-energy CT-derived radiomic features across three different scanner types. Eur. Radiol. 32(3), 1959–1970. https://doi.org/10.1007/s00330-021-08249-2 (2022).
Mackin, D. et al. Effect of tube current on computed tomography radiomic features. Sci Rep. 8(1), 2354. https://doi.org/10.1038/s41598-018-20713-6 (2018).
Midya, A., Chakraborty, J., Gönen, M., Do, R. K. G. & Simpson, A. L. Influence of CT acquisition and reconstruction parameters on radiomic feature reproducibility. J. Med. Imaging. 5(01), 1. https://doi.org/10.1117/1.JMI.5.1.011020 (2018).
Foy, J. J. et al. Harmonization of radiomic feature variability resulting from differences in CT image acquisition and reconstruction: Assessment in a cadaveric liver. Phys. Med. Biol. 65(20), 205008. https://doi.org/10.1088/1361-6560/abb172 (2020).
Levi, R. et al. CT-based radiomics can identify physiological modifications of bone structure related to subjects’ age and sex. Radiol. Med. https://doi.org/10.1007/s11547-023-01641-6 (2023).
Orlhac, F. et al. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur. Radiol. 31(4), 2272–2280. https://doi.org/10.1007/s00330-020-07284-9 (2021).
Acknowledgements
RF-2021-12374134, Italian Ministry of Health. We would like to thank Leonardo Zingoni, Gianluca Solitro and Domenico Viccica for their assistance in the management of the cadaver donor.
Author information
Authors and Affiliations
Contributions
R.L. : Conceptualization, Methodology, Software, Formal analysis, Writing—Original Draft, M.M. : Conceptualization, Methodology, Software, Formal analysis, Writing—Original Draft, G.S. : Methodology, Formal analysis, F.G. : Validation, Formal analysis, Data Curation, M.B. : Validation, Investigation, Data Curation, A.A. : Validation, Data Curation, L.A.C. : Validation, Data Curation, Writing—Review & Editing, S.S. : Investigation, Data Curation, M.G. : Validation, Investigation, R.B. : Conceptualization, Methodology, Writing—Review & Editing, Supervision L.S.P.: Conceptualization, Validation, Investigation, Writing—Review & Editing, Supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Levi, R., Mollura, M., Savini, G. et al. A reference framework for standardization and harmonization of CT radiomics features on cadaveric sample. Sci Rep 14, 19259 (2024). https://doi.org/10.1038/s41598-024-68158-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-68158-4