Abstract
Lung images often have the characteristics of strong noise, uneven grayscale distribution, and complex pathological structures, which makes lung image segmentation a challenging task. To solve this problems, this paper proposes an initial lung mask extraction algorithm that combines threshold and gradient. The gradient used in the algorithm is obtained by the time series feature extraction method based on differential memory (TFDM), which is obtained by the grayscale threshold and image grayscale features. At the same time, we also proposed a lung contour repair algorithm based on the improved convex hull method to solve the contour loss caused by solid nodules and other lesions. Experimental results show that on the COVID-19 CT segmentation dataset, the advanced lung segmentation algorithm proposed in this article achieves better segmentation results and greatly improves the consistency and accuracy of lung segmentation. Our method can obtain more lung information, resulting in ideal segmentation effects with improved accuracy and robustness.
Similar content being viewed by others
Introduction
The lungs are an integral part of the human respiratory system, whose proper functioning is crucial to maintaining human life. However, they are also among the organs with the highest incidence of diseases in humans. Each year, numerous new cases and deaths are caused by lung cancer1. The commonly used imaging detection technologies include X-rays, magnetic resonance imaging (MRI), and computed tomography (CT), etc. Among these, CT images are widely used in auxiliary screening of lung diseases due to their higher sensitivity, accuracy and ease of access. However, due to reasons such as noise, artifacts, and the patient’s own pathological structure, it is difficult for doctors to accurately and quickly obtain lesion characteristics from CT images. Therefore, computer-assisted segmentation is needed so that doctors can clearly observe the shape, density, and structure of lung tissue, thereby helping them diagnose various diseases and improving the accuracy and reliability of the diagnosis.
Lung CT images usually have the characteristics of strong noise, uneven gray distribution and complex pathological structures, which often lead to three problems when performing lung segmentation. Firstly, striking a balance between noise removal and edge detail preservation is a problem2. Due to issues with device imaging, lung CT images usually have strong noise3, which has a great impact on the accurate extraction of lung areas. Although some traditional computer vision filtering algorithms3,4 and deep learning filtering algorithms5 can effectively remove noise, they often excessively blur the edge information and cannot preserve the edge details well. Secondly, the intensity inhomogeneity common in lung CT images makes accurate segmentation challenging6. The intensity inhomogeneity of images may be caused by insufficient scattering compensation, air, alveoli and other factors. Although methods such as region growing7 and random walk8 can automatically grow connected regions according to the similarity of pixels, parameter selection is sensitive, and the seed points and growing conditions need to be set manually. Moreover, it is difficult to segment connected or overlapping lesions accurately. However, deep learning algorithms9 may lead to over-segmentation or under-segmentation due to the inability to identify pathological features in uneven grayscale images. Finally, dealing with pathological tissues with complex structures (such as juxta-pleural nodules)10 is also a challenge. The anatomical structures and pathological tissues Lung CT images show diverse morphology and density characteristics. When the pathological tissue structure is complex, the contour of the segmented lung may be incomplete or broken. For this problem, deep learning methods11,12,13,14,15 can automatically learn through a large amount of training data, to capture the complex features in the image, which makes it achieve better performance in segmentation. However, the deep learning model16,17,18 has poor interpretability, a large number of parameters, complex network models, and requires a long time to train, and usually need the support of high-performance processors. Medical systems often have limited Random-access memory(RAM), computing power, storage, and no GPUs, and can only run smaller deep learning models.19. Traditional computer vision methods are usually based on feature engineering2 and machine learning algorithms, they may have limitations for complex feature extraction in images, which leads to the inability of traditional computer vision methods7,10,20,21 is hard to achieve the accuracy of deep learning when facing the above problems. However, traditional computer vision methods usually have low computational complexity and are suitable for resource-limited environments. At the same time, traditional methods can provide strong interpretability results, making it easier for doctors to understand and verify the segmentation results. Considering the hardware conditions and interpretability, we use the traditional computer vision method for lung segmentation based on the above point of view.
To achieve more accurate and robust lung CT image segmentation, we propose a lung CT image segmentation algorithm based on the combination of threshold gradient and an improved convex hull method. Based on the principle that the gradient of the lung edge is large, this algorithm uses the time series feature extraction method (TFDM) based on differential memory to obtain the gradient information of the image, and combines the threshold of the image to obtain the region of interest(ROI) gradient matrix, and then passes the adaptive gradient threshold to convert the ROI gradient The matrix is binarized to obtain the initial lung mask. Next, based on prior knowledge, the interference information in the initial lung mask is removed. Finally, an improved convex hull method is used to fill in the missing parts of the lung edges to achieve the effect of repairing the consolidated nodules and eliminating shadows, thereby obtaining ideal segmentation results.
Related work
Lung segmentation is a classic topic in computer vision and medical image processing, and has received widespread attention in the fields of image segmentation and medical image processing in recent years. In the field of traditional computer vision lung segmentation, it mainly includes two tasks: lung tissue extraction and lung contour repair. However, lesions will cause the patient’s lung structure to be complex, so it is necessary to remove interference terms to remove non-pulmonary tissue to improve segmentation accuracy. Therefore, we will discuss it in three parts: extracting lung tissue, completing lung contours, and removing interference items.
Extraction of lung tissues
In the work of extracting lung tissue, researchers usually improve the algorithm from two perspectives, namely improving threshold selection and finding more accurate boundaries. The selection of threshold is crucial for lung tissue extraction, which determines whether we can obtain more lung information from the original lung CT images. Threshold is the most commonly used method20. However, selection of threshold is easily affected by the complexity and gray uniformity of lung tissue, resulting in the inability to obtain the correct threshold. Therefore, thresholding is often combined with other methods to improve the accuracy and consistency of lung tissue extraction.
Samy22 combines thresholding with gray-level co-occurrence matrix to improve segmentation consistency and reliability. Rao23 incorporates attention mechanisms into the segmentation algorithm to achieve lung segmentation in cases of non-uniformity in grayscale distribution. These methods solve the problem of selecting appropriate thresholds. However, their edges are sharp and lack smoothness, which is quite different from the actual situation. To find more accurate boundaries, Li24 uses the gradients to correct threshold segmentation results and obtain precise lung boundaries, while Chen25 uses the gradients to refine lung boundaries. Rajeswari26 combines global threshold with gradient features to extract lung boundary information. However, when the similarity in gray levels between the lesion area and the surrounding tissue is high, the gradient at the boundary between the lungs and other human tissues does not change much, which directly results in the inability to obtain accurate boundaries, leading to blurred boundaries and reduced segmentation accuracy. Based on the existing problems in the above research, we proposed an algorithm that combines thresholding and gradients. The TFDM algorithm is used to extract the gradient. It can also play a filtering role while obtaining the gradient. It is then combined with the threshold to obtain the ROI gradient matrix. The algorithm can effectively solve the problem of fuzzy boundaries and difficult-to-determine boundaries.
Repairing of lung contour
In the work on contour repair methods, the convex hull method27 is often used to repair the contour of the lung. Recent studies show that, Tavakoli28 integrated the convex hull method and curvature features to address pulmonary nodule repair within the lung parenchyma. Wang29 combined the rolling ball method with convex hull approaches to rectify lung contour irregularities. Shi30 proposed combining the convex hull algorithm and derivative frequency analysis of discrete points for lesion area restoration. However, the aforementioned repair methods are only suitable for cases with mild lung tissue lesions. When the lung tissue lesions exhibit calcification or large areas of cavities, the existing convex hull method and rolling ball method cannot cope with such symptoms, causing a large amount of lung tissue loss and making it difficult to repair the defect contour. In order to solve such problems, we propose an improved convex hull method, which uses the size and center of the connected domain to determine whether this part of the contour needs to be repaired, so as to solve the problem that the contour cannot be repaired when the tissue lesions are severe.
Interference terms removal
The interference terms31 refer to specific tissues or phenomena observed in CT images that can potentially affect lung segmentation. Common interference terms include large blood vessels, noise. Raj32 used morphological methods to remove interference terms, which can effectively remove interference from the heart and blood vessels, but the effect is not good for effusion lesions and calcified lesions. Zhou33 utilized attention mechanisms to remove interference. Zheng34 used receptive fields to remove interference terms. But the method easily mistakes calcified lungs as interference items, affecting the accuracy and consistency of segmentation. Therefore, according to medical knowledge, we divide the images into three categories based on the location and area of the connected area center. By considering the characteristics of the human body structure and lung lesions35, corresponding structural parameters are used for different types of images to effectively eliminate interference terms and improve the consistency and reliability of lung segmentation.
Methods
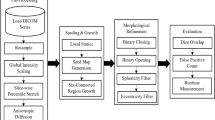
This section describes the specific methods used in the study. We divide the algorithm into two stages: initial lung mask extraction and contour repair. Specifically, in the first stage, we combine thresholding with gradient information to obtain the ROI gradient matrix. Then, the ROI gradient matrix is converted into a lung initial mask using adaptive gradient threshold. In the second stage, we utilize a modified convex hull method for lung contour repair. The specific steps of the entire process are illustrated in Fig. 1. In the following chapters, we will explain the specific processes and principles of these two stages in detail.
Lung initial mask extraction
In the initial stage, the image gray threshold is first obtained, and the adaptive gradient threshold is obtained from the gray threshold and lung gray distribution information. Then, the adaptive gradient threshold is used to binarize the ROI gradient matrix to generate an initial lung mask. In this section, we divide the algorithm into three parts to illustrate its application: using the TFDM algorithm to obtain the gradient, combining the threshold with the TFDM to extract the ROI gradient matrix, and obtaining the adaptive gradient threshold.
Time series feature extraction based on differential memory
Differential time memory sequence technology can effectively remove trend components while suppressing systematic noise . It can better retain the details of the image, improve the clarity of the image, and reduce the impact of uneven grayscale distribution, artifacts and other phenomena on lung segmentation. We introduce memory factors and forgetting factors as adjustable parameters in TFDM to adjust the impact of the model on observations at different time points. By adjusting these parameters, the TFDM method can effectively balance long-term trends and short-term fluctuations in the feature extraction process. This design enables our method to mange noise in the lung segmentation task more robustly, thereby improving the accuracy and reliability of the segmentation algorithm.
Assuming that the input sequence L represents a time series, this method can be divided into three steps.
-
1.
Establish the memory model by introducing user-adjustable parameters \({f_1}\) and \({f_2}\) to balance the degrees of forgetting and remembering. The forgetting factor \({F_1}\) for A is calculated as \({f_1-f_2}\), while the memory factor M is calculated as \({1-F_1}\). The forgetting factor \({F_2}\) for B equals to \({f_1}\), while the memory factor \({M_2}\) is calculated as \({1-F_2}\). The forgetfulness factor \({F_3}\) for C is determined by calculating \({f_1+f_2}\), while the memory factor \({M_3}\) is obtained as \({1-F_3}\).
-
2.
Traverse through a length k of the time series list and calculate both current information’s degree of memorization (based on past data) and its degree of forgetting using the formulas below to generate sequences \({L_j}\) representing memories. Three memory sequences \({(L_1, L_2, L_3)}\) are generated corresponding to models A, B, and C respectively. The formula of \({L_j}\) is as follows:
$$\begin{aligned} {{\textrm{L}}_{\textrm{j}}}[i] = {M_j}{L_j}[i - 1] + {F_j}L[i], \end{aligned}$$(1) -
3.
Differentiate the memory sequence to obtain the final difference result.
$$\begin{aligned} result = {L_2} - {L_1} - ({L_3} - {L_2}) = 2{L_2} - {L_1} - {L_3}, \end{aligned}$$(2)
To illusrate the TFDM algorithm better, we present in Fig. 2 the process of using the TFDM algorithm. When the input is a grayscale image (two-dimensional matrix), the direction is horizontal and vertical, and the sequence length is 5, the process of using the TFDM algorithm: Select a pixel point, obtain the sequence of neighborhood pixel values in the horizontal and vertical directions of this point, use the TFDM algorithm to process this sequence to obtain a gradient sequence with a length of 5; use the gradient with the largest absolute value in the sequence as the gradient of this sequence Gradient,take the sum of the squares of the gradients in the horizontal and vertical directions as the final result, representing this pixel‘s gradient.
The threshold was combined with the TFDM algorithm to obtain the initial lung mask
Lung CT images are grayscale images, so we expect to obtain the gradient of the entire image, not just the gradient of a single pixel. Therefore, we need to combine the threshold with the TFDM algorithm to obtain the ROI gradient matrix of the image. Next, we will describe the process of getting the ROI gradient matrix:
The input image is the initial CT image M, and the size of the image is \({m*n}\).
-
1.
The original image M is transformed into a new two-dimensional image, and each element of the two-dimensional image is the grayscale sequence obtained in a fixed direction at the corresponding pixel position. Here, the default is horizontal and vertical directions, so the two matrices are respectively \({M_h}\) and \({M_v}\), and the size of the two matrices is \({m*n}\), the length of the grayscale list \({L=l}\), the gray interval of the sequence is l, in which each element of the horizontal grayscale matrix is \({M_{h_{i,j}}=list[{M_{i - l/2,j}},{M_{i - l/2 + 1,j}}, \ldots \ldots {{\textrm{M}}_{{\mathrm{i + l}}/2 - 2,j}},{M_{i + l/2 - 1,j}}]}\), and each element of the vertical grayscale matrix is \({M_{v_{i,j}} = list[{M_{i,j - l/2}},{M_{i,j - l/2 + 1}}, \ldots \ldots {{\textrm{M}}_{i,j + l/2 - 2}},{M_{i,j + l/2 - 1}}]}\)
-
2.
Determine whether to use the TFDM algorithm according to the pixel gray level. If \({M_{i,j}} < {T_g}\), the sequences at the corresponding positions in \({M_h}\) and \({M_v}\) are input into the TFDM algorithm to obtain the gradient sequences at H and V matrices’ (i, j) positions, Otherwise, the sequence [0, 0, ..., 0, 0] with length L is used to fill the corresponding position, and finally the matrix H and V of the horizontal and vertical gradients are obtained.
-
3.
Get the maximum gradient value. Each element of the two matrices H and V is traversed. The maximum absolute value in the list is taken as the gradient value of the corresponding direction at this position, and the gradient matrices H and V are updated. The formula is \({H_{i,j}} = Max(abs({H_{i,j}}))\),\({V_{i,j}} = Max(abs({V_{i,j}}))\).
-
4.
Get the ROI gradient matrix G. The gradient values of the two matrices \({H_{i,j}}\) and \({V_{i,j}}\) at the same position were squared to obtain the ROI gradient matrix.
$$\begin{aligned} {{\textrm{G}}_{{\textrm{i,j}}}} = \sqrt{H_{i,j}^2 + V_{i,j}^2}, \end{aligned}$$(3)
Adaptive gradient threshold algorithm
After obtaining the ROI gradient matrix, it must be converted into a binary image using an adaptive gradient threshold. During this conversion process, if the grayscale image itself has low brightness, it may result in an overall decrease in the image’s grayscale and subsequently reduce its overall gradient. Therefore, adjusting the size of the gradient threshold based on the grayscale characteristics of the image, it becomes necessary. Determining an adaptive gradient threshold involves parameters such as the average value of thoracic cavity grayscale and OTSU thresholds. We can use the following formula to derive an adaptive gradient threshold based on these parameters.
Where the pixels inside the thoracic cavity are represented by \({n_i}\), the total number of pixels is N, the grayscale value is \({g_i}\), the average grayscale value is \({\mu }\), the maximum gradient value in the grayscale image is \({g_m}\), and the OTSU threshold is h. With the calculated threshold \({g_t}\), we can compare the element values of the region of interest with the threshold. Pixels greater than the threshold are set to white, while pixels smaller than the threshold are set to black. In this way, the gradient image is converted into a binary image using an adaptive gradient threshold. Then, by performing operations to remove interference, we can obtain binary images of the lungs that can be used for contour repair.
Interference terms removal
Lung images are often interfered with by many factors, such as noise, artifacts, and other human tissues. These factors will cause the segmentation results to contain a large number of tissues that are inconsistent with the actual lungs. In order to prevent these tissues from being mistakenly identified as lung tissues, interference items need to be removed.We first remove tissues with an area of less than 20 in the image, and then determine whether the area of the largest tissue in the image is greater than 1500, and divide the binary images of the lungs into two categories accordingly. Then it will be judged according to certain conditions. The specific judgment indicators are shown in the figure Table 1.
The improved convex hull repair algorithm
In the second stage, we proposed a contour repairing algorithm based on the convex hull method to address incomplete or fragmented lung edges caused by pathological tissues in images.
-
1.
Choose the structural parameters of the closing operation. Choose the appropriate structure element parameters based on the number of connected domains and the area of the largest connected domain. The structural parameters can be adjusted according to the actual situation. When the image has more lesions and the gray distribution is extremely uneven, the parameters of this formula can be referred to. The structural parameters of the closed operation are as follows:
$$\begin{aligned} {\textrm{structure}} = \left\{ \begin{array}{l} 3 \times 3,{A_{\max }}< = 2000,{A_n}< = 3\\ 5 \times 5,{A_{\max }}< = 2000,{A_n}> 3\\ 15 \times 15,{A_{\max }} > 2000,{A_n} < = 3\\ 30 \times 30,else \end{array} \right. \end{aligned}$$(7)\({A_{max}}\) is the maximum connected area of the binary image of the lung, and \({A_n}\) is the number of connected domains.
-
2.
The length of convex hull line segments was calculated. Calculate the size \({A_a}\) of each connected domain in the binary image after the closing operation, and calculate its corresponding convex hull to obtain the convex hull line segments set of each connected domain. For each convex hull line segment set, calculate the Euclidean length of each line segment in this set. The length calculation formula is \({L = sqrt({({s_1} - {e_1})^2} + {({s_2} - {e_2})^2})}\). The starting point and ending point of each convex hull segment are \({S (s_1, s_2)}\) and \({E(e_1, e_2)}\) .
-
3.
Contour patching. According to the area of the connected domain and the length of the convex hull line segment, determine whether to keep the convex hull line segment. The judgment condition is as follows: when \({A_a<2000}\), delete the two longest convex hull line segments; when \({2000<A_a<5000}\), delete the two longest convex hull segments and the convex hull segments whose length is greater than 70. Otherwise, delete the longest convex hull line segment and the convex hull line segment with length greater than 100.
Two sets of contour correction results are shown in Fig. 3.
Result
The public dataset used in this study is the COVID-19 CT segmentation dataset from https://medicalsegmentation.com/covid19/. There are 2112 CT images of patients with new coronavirus pneumonia. The size of the image is 512*512 pixels. We compared the results obtained by various segmentation algorithms with the results of manual segmentation performed by three experienced radiologists, using comprehensive lung area as the evaluation criterion. During the experiment, we used manually segmented images in medical records as the final gold standard to evaluate and verify the accuracy and reliability of various segmentation algorithms. Such a design can provide a comprehensive comparison and important reference and guidance for further improving and optimizing the lung image segmentation algorithm.
This paper verifies the effectiveness of the proposed method by calculating the Dice similarity coefficient (DSC) and sensitivity of the image segmentation. By measuring the overlap between the segmentation results and the expert manual segmentation results, the Dice similarity coefficient can be obtained, which is used to measure the performance of the model in lung segmentation; by measuring the ratio of the segmentation results of the predicted method to the expert manual segmentation results, we can get sensitivity, used to determine the degree of correct segmentation of the lungs, that is:
Among them, urepresents the segmentation result of this article, and v represents the doctor’s gold standard segmentation result, and the accuracy index IOU is used to evaluate the experimental results.
Among them, \({N_{11}}\) represents the correctly segmented lung parenchyma region. \({N_{10}}\) represents the correctly segmented background area. \({N_{01}}\) represents an incorrectly segmented lung parenchyma area, and \({N_{00}}\) represents an incorrectly segmented background area. The greater the accuracy of IOU, the more accurate the image segmentation is and the closer it is to standard segmentation.
Experimental results
To verify the accuracy of the segmentation algorithm, we compared the algorithm with 8 advanced methods, including Inf-Net36, Chain code-SVM37, CNN-Clustering38, MLT39, GAN-Unet40, U-Net++41,TDRMFA42, PACO43 to lung CT images were segmented as a comparison experiment. Our method achieves the best DSC and IOU, which are 0.9599 and 0.9911 respectively. The sensitivity is slightly lower than TDRMFA, but still higher than other methods. This fully proves the superiority of our method. The specific comparative experimental results are displayed in Table 2.
In order to prove the effectiveness of the contour repair algorithm, we take the binary image after removing interference items as the input image, and compare the results of our contour repair algorithm with the contour repair algorithms in GCH44 and TCH45. Experimental results show that the improved convex hull algorithm we proposed is better than the existing convex hull algorithm, especially achieving an increase of 0.0407 in IOU. The specific experimental results are displayed in Table 3:
Component validation
In order to prove the effectiveness of each part of our experiment, with the interference items removed by default, we selected the following algorithms for comparison: Adaptive threshold combined with Sobel gradient and improved convex hull repair method (TSR), adaptive threshold combined with Sobel gradient algorithm (TS), adaptive threshold and improved convex hull repair algorithm (TR), adaptive threshold and gradient combined algorithm (TO). Among them, the TO and TR algorithms are part of our algorithm. TO is the method that only uses lung initial mask extraction in Sect. "Lung initial mask extraction" to test the effectiveness of our repair algorithm; TR is an ablation experiment that does not use our proposed ROI gradient calculation method to test the effectiveness of our gradient matrix method; TSR only changes the TFDM algorithm we proposed into the Soble operator, which is used to test the effectiveness of the gradient matrix calculation method we proposed; The difference between TS and TSR is that TS does not use the repair algorithm, which is used to prove the effectiveness of the repair algorithm. The specific comparison experiment is reflected in Table 4:
By comparison, we can find that the ROI matrix extraction method we proposed is better than the adaptive threshold compared with the Soble operator. All indicators of TO are higher than TS, and our indicators are better than those of TSR. It is proved that the performance of our ROI gradient matrix extraction algorithm is better than the traditional gradient algorithm; through the comparison of TS and TSR, TO and ours, except for the Sensitivity index, the effect of the method using the patching algorithm is better than the effect of not using the patching algorithm. The effectiveness of the patching algorithm is fully proved. The specific comparison results are shown in Figs. 4 and 5: Among them, Fig. 4 is a relatively complete image of the lung parenchyma, and Fig. 5 is an image with uneven grayscale distribution, large noise, and complex structure. It can be seen from the comparison between TR and other images that the use of gradients can significantly remove large-area interference items and avoid mistakenly identifying other lung tissues as lungs. Through the comparison between TO and ours and TS and TSR, it can be seen that the use of repair algorithm can effectively repair the lung parenchyma, eliminate holes caused by better than threshold segmentation, and make the lung parenchyma more complete. The effectiveness of each part of the algorithm is fully proved.
Discussion
Our algorithm employs relatively simple and easily implementable techniques that yield favorable outcomes. However, we face challenges in accurately segmenting certain images with severe grayscale unevenness and complex lung structures due to the low internal density and irregular shape of the lungs. To address this issue, we will further optimize the selection strategy for thresholds and gradients to adapt to more intricate lung segmentation tasks. Although our TFDM algorithm inherently possesses smoothing effects, there is a possibility of erroneously segmenting interference tissues into the lungs when interference tissues are present or when the lungs are close to each other, as depicted in Fig. 6. This is an undesirable situation that we aim to avoid. At the same time, the line segments we use when using the improved convex hull method for contour repair are straight lines, which affects the segmentation results to a certain extent. We will work hard to find a new line segment fitting method that can be fitted into a curve and better match the actual situation. In future work, we will strive to solve the above two types of situations.
Conclusions
Lung segmentation plays a vital role in detecting and diagnosing lung diseases using CAD systems and is critical to achieving accurate and efficient detection and diagnosis. However, due to the unique properties of lung images, such as strong noise, uneven grayscale distribution, complex lung structures, etc, traditional lung segmentation algorithms perform poorly in handling these problems.
To effectively overcome these challenges, we propose a lung extraction algorithm that combines threshold with TFDM and a contour repair method that improves the convex hull method. First, our algorithm generates the ROI gradient matrix by combining thresholds with TFDM gradients. This matrix is then binarized using an adaptive gradient threshold to accurately separate lung regions from background regions. Finally, we enhance the contour repair algorithm of the convex hull method to repair the missing contours caused by disease. We compare the results of the proposed algorithm with other commonly used methods to verify the effectiveness of the algorithm. By comparing the results of analysis and ablation experiments, we can conclude that the lung segmentation method we proposed is significantly ahead of the current methods. It can accurately segment the lung parenchyma, thus reducing the time for doctors to outline lesions and achieving the goal of lung parenchyma. The purpose of fully automatic segmentation.
Data availability
Data will be made available on request.The code and related materials can be accessed at https://github.com/zdyihong/LungCTseg.
References
Bray, F. et al. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424 (2018).
Jung, S., Liao, C., Wu, Y., Yuan, S. & Sun, C. Efficiently classifying lung sounds through depthwise separable cnn models with fused stft and mfcc features. Diagnostics 11(4), 732 (2021).
Liu, C., Xie, W., Zhao, R. & Pang, M. Segmenting lung parenchyma from ct images with gray correlation-based clustering. IET Image Proc. 17(6), 1658–1667 (2023).
Ben Gharsallah, M. & Seddik, H. Phase congruency-based filtering approach combined with a convolutional network for lung ct image analysis. Imaging Sci. J. 69(5–8), 275–287 (2021).
Ilhan, A., Alpan, K., Sekeroglu, B. & Abiyev, R. Covid-19 lung ct image segmentation using localization and enhancement methods with u-net. Procedia Comput. Sci. 218, 1660–1667 (2023).
Yu, H. et al. Design of lung nodules segmentation and recognition algorithm based on deep learning. BMC Bioinform. 22, 1–21 (2021).
Avinash, S., Manjunath, K., & Kumar, S. An improved image processing analysis for the detection of lung cancer using gabor filters and watershed segmentation technique. In 2016 International Conference on Inventive Computation Technologies (ICICT), vol 3, pp 1–6 (IEEE, 2016).
Chen, C. et al. Pathological lung segmentation in chest ct images based on improved random walker. Comput. Methods Progr. Biomed. 200, 105864 (2021).
Das, A. Adaptive unet-based lung segmentation and ensemble learning with cnn-based deep features for automated covid-19 diagnosis. Multimed. Tools Appl. 81(4), 5407–5441 (2022).
Suji, R., Godfrey, W. & Dhar, J. Border to border distance based lung parenchyma segmentation including juxta-pleural nodules. Multimed. Tools Appl. 82(7), 10421–10443 (2023).
Tuncer, S., Çınar, A., Tuncer, T. & Çolak, F. Determining and measuring the amount of region having covid-19 on lung images. Appl. Comput. Syst. 26(2), 183–193 (2021).
Asnawi, M., Pravitasari, A., Darmawan, G., Hendrawati, T., Yulita, I., Suprijadi, J., & Nugraha, F.: Lung and infection ct-scan-based segmentation with 3d unet architecture and its modification. In Healthcare, vol 11, pp 213 (MDPI, 2023).
Zhang, G., Yang, Z. & Jiang, S. Automatic lung tumor segmentation from ct images using improved 3d densely connected unet. Med. Biol. Eng. Comput. 60(11), 3311–3323 (2022).
Singh, A. et al. Deep lf-net: Semantic lung segmentation from Indian chest radiographs including severely unhealthy images. Biomed. Signal Process. Control 68, 102666 (2021).
Bruzadin, A., Boaventura, M., Colnago, M., Negri, R. & Casaca, W. Learning label diffusion maps for semi-automatic segmentation of lung ct images with covid-19. Neurocomputing 522, 24–38 (2023).
Yan, X., Wu, Y. & Tan, W. Segmentation of pulmonary parenchyma from pulmonary ct based on resu-net++ model. J. Med. Imaging Health Inform. 11(3), 760–766 (2021).
Jalali, Y., Fateh, M., Rezvani, M., Abolghasemi, V. & Anisi, M. Resbcdu-net: A deep learning framework for lung ct image segmentation. Sensors 21(1), 268 (2021).
Sun, W. et al. Covid-19 ct image segmentation method based on swin transformer. Front. Physiol. 13, 981463 (2022).
Hooda, R., Mittal, A. & Sofat, S. Segmentation of lung fields from chest radiographs-a radiomic feature-based approach. Biomed. Eng. Lett. 9, 109–117 (2019).
Sudha, V. & Jayashree, P. Lung nodule detection in ct images using thresholding and morphological operations. Int. J. Emerg. Sci. Eng. 1(2), 17–21 (2012).
Ban, Z., Liu, J. & Cao, L. Superpixel segmentation using gaussian mixture model. IEEE Trans. Image Process. 27(8), 4105–4117 (2018).
Bakheet, S. & Al-Hamadi, A. Automatic detection of covid-19 using pruned glcm-based texture features and ldcrf classification. Comput. Biol. Med. 137, 104781 (2021).
Rao, Y. et al. Covid-19 ct ground-glass opacity segmentation based on attention mechanism threshold. Biomed. Signal Process. Control 81, 104486 (2023).
Li, X., Feng, B., Qiao, S., Wei, H. & Feng, C. Sift-gvf-based lung edge correction method for correcting the lung region in ct images. PLoS One 18(2), 0282107 (2023).
Chen, G. et al. Automatic pathological lung segmentation in low-dose ct image using eigenspace sparse shape composition. IEEE Trans. Med. Imaging 38(7), 1736–1749 (2019).
Rajeswari, J., Raja, J. & Jayashri, S. Gradient contouring and texture modelling based cad system for improved tb classification. Autom. Softw. Eng. 29, 1–12 (2022).
Xiao, X. et al. An automated segmentation method for lung parenchyma image sequences based on fractal geometry and convex hull algorithm. Appl. Sci. 8(5), 832 (2018).
Tavakoli, M., Orooji, M., Teimouri, M. & Shahabifar, R. Segmentation of the pulmonary nodule and the attached vessels in the ct scan of the chest using morphological features and topological skeleton of the nodule. IET Image Proc. 14(8), 1520–1528 (2020).
Wang, Y., & Yue, S. Ground glass nodule segmentation based on regional adaptive mrf model. In 2020 39th Chinese Control Conference (CCC), pp 6295–6300 (IEEE, 2020).
Shi, X., Liu, J., Xu, J., & Lu, M. A lung segmentation method based on an improved convex hull algorithm combined with non-uniform rational b-sample. In International Conference on Sensing and Imaging, pp 311–319 (Springer, 2022).
Wang, H. et al. Lung ct image enhancement based on image segmentation and total variational. Chin. J. Lasers 49(20), 8 (2022).
Raj, S., Vinod, D., Mahanand, B. & Murthy, N. J. Intuitionistic fuzzy c means clustering for lung segmentation in diffuse lung diseases. Sens. Imaging 21(1), 37 (2020).
Zhou, C., Song, J., Zhou, S., Zhang, Z. & Xing, J. Covid-19 detection based on image regrouping and resnet-svm using chest x-ray images. IEEE Access 9, 81902–81912 (2021).
Zheng, Y., Jiang, S. & Yang, Z. Deformable registration of chest ct images using a 3d convolutional neural network based on unsupervised learning. J. Appl. Clin. Med. Phys. 22(10), 22–35 (2021).
Chu, Z., Li, W., Fu, B. & Lv, F. Ct characteristics for predicting invasiveness in pulmonary pure ground-glass nodules. Am. J. Roentgenol. 215(2), 351–358 (2020).
Fan, D. et al. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging 39(8), 2626–2637 (2020).
Kumar, S. & Latte, M. Lung parenchyma segmentation: Fully automated and accurate approach for thoracic ct scan images. IETE J. Res. 66(3), 370–383 (2020).
Xu, M. et al. Segmentation of lung parenchyma in ct images using cnn trained with the clustering algorithm generated dataset. Biomed. Eng. Online 18, 1–21 (2019).
Liu, W. et al. Integrating lung parenchyma segmentation and nodule detection with deep multi-task learning. IEEE J. Biomed. Health Inform. 25(8), 3073–3081 (2021).
Yan, W., Wang, Y., Gu, S., Huang, L., Yan, F., Xia, L., & Tao, Q. The domain shift problem of medical image segmentation and vendor-adaptation by unet-gan. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference Shenzhen China October 13–17 2019 Proceedings Part II 22, pp. 623–631 (Springer, 2019).
Ibtehaz, N. & Rahman, M. Multiresunet: Rethinking the u-net architecture for multimodal biomedical image segmentation. Neural Netw. 121, 74–87 (2020).
Wang, Gi., Guo, S., Han, L. & Cekderi, A. Two-dimensional reciprocal cross entropy multi-threshold combined with improved firefly algorithm for lung parenchyma segmentation of covid-19 ct image. Biomed. Signal Process. Control 78, 103933 (2022).
Rodriguez, O., Diomar, E., Mejia, R. & Aldo, R. Semi-supervised covid-19 volumetric pulmonary lesion estimation on ct images using probabilistic active contour and cnn segmentation. Biomed. Signal Process. Control 85, 104905 (2023).
Saha, M., Amin, S., Sharma, A., Kumar, T. & Kalia, R. Ai-driven quantification of ground glass opacities in lungs of covid-19 patients using 3d computed tomography imaging. PLoS One 17(3), 0263916 (2022).
Li, Q., Yuan, J. & Xiao, L. Lung segmentation method based on k-means and convex hull algorithm. Netw. New Media Technol. 11, 42–51 (2022).
Funding
This work is supported by the National Key Research and Development Program of China under Grant No.2022YFF0606200.
Author information
Authors and Affiliations
Contributions
JZ and LW proposed ideas and innovations, JZ was responsible for applying for fund support and making constructive suggestions for the entire experiment, LW conducted experimental verification and wrote the main text, LW and JG was responsible for checking and revising the writing of the paper, and AY was responsible for detailed inspection of the paper.All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, J., Wang, L., Gui, J. et al. Study on lung CT image segmentation algorithm based on threshold-gradient combination and improved convex hull method. Sci Rep 14, 17731 (2024). https://doi.org/10.1038/s41598-024-68409-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-68409-4
Keywords
This article is cited by
-
Three-dimensional reconstruction of lung tumors from computed tomography scans using adversarial and transductive learning
Scientific Reports (2025)
-
Semi-supervised segmentation of lung CT images based on contrastive learning
Signal, Image and Video Processing (2025)