Abstract
Using a new red blood cell (RBC) metabolite extraction protocol, we performed a metabolomic analysis on RBCs in rheumatoid arthritis (RA) patients treated or not with methotrexate (MTX), with the two following objectives: to compare the RBC metabolic profiles of MTX-naïve RA patients and healthy controls (HC), and to investigate whether RBC profiles before and after MTX treatment in RA differed between responders and non-responders. Plasma analysis was performed in parallel. Metabolites were extracted and identified in RBCs and plasma by liquid chromatography-mass spectrometry. We compared the metabolomic fingerprints of 31 DMARD-naïve RA patients and 39 HCs. We also compared the RBC and plasma metabolomes of 25 RA patients who responded or not to MTX therapy before (M0) and after a 3-month treatment period (M3). Significance was determined by Storey’s false discovery rate (FDR) q-values to correct for multiple testing. RA patients and HCs differed in the metabolomic signature of RBCs. The signature mainly contained amino acids (AA). Eleven metabolites, including 4 metabolites belonging to the carbohydrate subclass and 2 amino acids (creatine and valine) showed accumulation in RBCs from RA patients. Conversely, citrulline (fold change = 0.83; q = 0.025), histidine (fold change = 0.86; q = 0.014) and ergothioneine (EGT) (fold change = 0.66; q = 0.024), were lower in RBC of RA patients. Five plasma metabolites, including succinic acid and hydroxyproline, were higher in RA patients, and 7 metabolites, including DHEA sulfate, alanine, threonine and ornithine, were lower. Among RA patients undergoing MTX treatment pre-treatment (M0), EGT values were significantly lower in non-responders. In conclusion, low RBC levels of EGT, a food-derived AA barely detectable in plasma, characterize DMARD naïve RA patients and lack of response to MTX treatment.
Similar content being viewed by others
Introduction
Omics sciences are increasingly used to study complex multifactorial diseases. Among these omics, metabolomics study the whole set of metabolites in a biological system that define its metabolome. Given that metabolites are the final products of cellular regulatory pathways, the metabolome integrates and reflects all the interactions between biochemical entities in cells or tissues and their environment. Thus, metabolomics enable the qualitative and quantitative description of a biological system’s ultimate response to environmental changes1. With regard to rheumatoid arthritis (RA), metabolomics can identify specific signatures of the disease (e.g. RA vs. healthy controls (HC)), trace putative metabolicpathways relevant for disease pathogenesis and treatment, and provide additional information on how patients with RA respond to the environment, including diet and exposure to xenobiotic substances. Several metabolomics studies have been published in the field of RA and have identified a large number of metabolites as putative markers of RA2, response to treatment3, and disease activity4. Most of them are involved in glycolysis (lactic acid, glucose), citrate cycle (citric acid), amino acid (leucine, isoleucine, valine, threonine, serine, proline, alanine, histidine) and lipid (cholesterol, glycerol) metabolisms5. All the RA metabolomic studies were performed on serum, plasma, urine and synovial fluid, but none of these studies examined red blood cells (RBC) as an alternative matrix.
RBCs play a major role in interorgan transport of metabolites during their life span, thereby reflecting exogenous metabolic processes. A case in point is the measure of glycated hemoglobin A1c, which reflects mean glycemia over the past 3 months. RBCs are basic cells devoid of nucleus, mitochondria and ribosome. They therefore have an elementary metabolism.
In this proof-of-concept study, we developed a metabolite extraction protocol to perform metabolomic analysis on RBCs with three objectives. The first was to characterize any differences in the RBC metabolic profiles of Disease-Modifying antirheumatic drugs (DMARD)-naïve RA patients and HCs. The second was to determine whether RBC profiles before and after methotrexate (MTX) treatment in RA patients differed between responders and non-responders. The third was to identify relevant RBC discriminant metabolites. In parallel, we performed metabolomic analysis of the plasma to demonstrate the diagnostic value of RBC metabolome and to study any association between RBC and plasma metabolomes.
Results
Clinical parameters
RA patients were predominantly female (71.0%), with a mean age of 50.0 years, and mostly ACPA (92.3%) and RF (95.2%) positive. Disease activity before MTX initiation was moderate to high (mean DAS28-CRP: 4.7 ± 1.4) (Table 1). Of the 31 initial RA patients, 25 were treated with MTX for at least 3 months. After a 3-month MTX treatment period, 12 patients were classified as responders and 13 as non-responders. The two groups before treatment did not significantly differ in baseline DAS28-CRP (4.6 ± 1.1 vs. 3.8 ± 1.5, p = 0.17 for responders and non-responders, respectively) or in tender and swollen joint counts (Supplementary Table S1).
RBC metabolites associated with RA (vs. healthy controls)
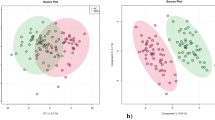
Under our conditions, 33 metabolite features from the C18( +) and 74 from the HILIC(-) analysis matched the accurate masses and retention times of the metabolites included in our chemical database, yielding 105 unique annotated metabolites (See supplementary file). Univariate analysis of the differences in mean expression between the two groups showed a significant difference in HILIC(−) analysis (p-value < 0.05) for 14 metabolites (Fig. 1A, Table 2) and all of them remained significant after correction for multiple testing (FDR q-value < 0.05) (Supplementary Figure S1). Three of them – histidine, ergothioneine (EGT), and citrulline—were found to be less expressed in RBCs from RA patients (RA/HC ratio: 0.86, 0.66 and 0.83, respectively, with all three FDR qvalue < 0.02). Meanwhile, eleven—including 4 metabolites in the carbohydrate subclass and 2 amino acids (creatine and valine)—showed accumulation. Although the results obtained under C18( +) conditions were statistically less robust (Fig. 1B, Supplementary Table S2), the same trend was observed for EGT (RA/HC ratio: 0.70, p = 0.040) and citrulline (RA/HC ratio: 0.83, p = 0.027), showing lower concentrations in the RA group compared to HCs. These differences did not reach FDR-adjusted significance. In this dataset, 5 metabolites involved in the arginine and proline pathway showed accumulation in RA RBCs compared to HCs (aminovaleric acid, creatine, spermidine, spermine and stachydrine). Metabolites were also studied in plasma samples from RA patients and HCs (See supplementary Data, tables S3 and S4). We found lower concentrations of several amino acids in the plasma of RA patients compared to HCs (DHEA, alanine, threonine, ornithine, theophylline) and higher hydroxyproline.
Red blood cell metabolite levels in RA patients compared to HCs Volcano plots showing the results of comparisons of RBC metabolite levels (using the HILIC(−) dataset) in RA patients relative to HCs. The vertical dashed lines indicate the threshold for the 0.2Log2 fold abundance difference. The horizontal dashed line indicates the p = 0.05 threshold. Between-group comparisons as described in statistical methods. Figure 1A shows the results obtained with the HILIC(−) dataset. Figure 1B shows the results obtained with the C18( +) dataset.
High level of RBC ergothioneine is associated with favorable methotrexate (MTX) response
Since RBC analysis identified EGT as a differentiating metabolite between RA patients and HCs, we investigated whether the concentration of EGT differed between responders and non-responders before (M0) and/or after (M3) MTX initiation. Of the 31 included patients, 25 completed the follow-up and were available for analysis. Among the 6 patients that did not complete the study, 4 missed their 3 month follow-up visit and 2 did not tolerate MTX. Those 6 patients, had similar values of DAS28-CRP (p = 0.47), mean corticosteroid dose (p = 0.21) and EGT levels (p = 0.10) as those that completed their follow-up.
A total of 12 patients responded to MTX, while 13 were classified as non-responders. We found significantly lower EGT values in non-responders at M0 (p = 0.036) when using the HILIC(−) dataset (Fig. 2). A similar trend was observed using C18( +) (p = 0.07, Wilcoxon test). Responders and non-responders did not differ in baseline corticosteroid dose (p = 0.84) or in baseline disease activity expressed by DAS28CRP values (p = 0.17) (supplementary table 1). Higher that the median EGT was the only variable associated with clinical response to methotrexate (OR 6.00; 95% CI 1.02 to 35.38) in multivariable logistics regression model taking into account these two confounders.
Discussion
This study is the first RBC metabolomics characterization in RA. This proof-of-concept study identified EGT—an amino acid barely detectable in plasma—as a key metabolite that is reduced in untreated RA and particularly in future non-responders to MTX. In alignment with several RA metabolomic studies, our RBC and plasma analysis shows a predominantly amino acidic RA signature. Since the RBC metabolome is not limited to pathways essential for RBC functions, but also includes metabolites from external sources such as plasma and other cells, the observed variations in AA concentration may reflect impaired systemic metabolism.
We found a significant decrease in EGT, histidine (EGT precursor), and citrulline in RA patient RBCs vs. HCs. This decrease may be related to the predominance of the ammonia recycling pathway and/or the nitric oxide (NO) pathway involved in RA, and may reflect increased catabolism and resulting muscle loss6. Interestingly, the plasma metabolome also revealed the potential role of the ammonia-recycling pathway, notably through the lower ornithine in RA patients. Furthermore, the fatigue frequently experienced by RA patients has been associated with downregulation of the urea cycle that involves citrulline7. Citrulline concentration was lower in RA patients’ RBC metabolome compared to HCs. Since RBCs do not have any NO synthase activity, the citrulline may be coming from plasma. The presence of endothelial dysfunction in RA patients, even at an early stage, is well established and it has been suggested that patients may exhibit impaired NO reactivity8. Thus, we can hypothesize that the citrulline decrease reflects the endothelial dysfunction observed in untreated RA and may be a consequence of decreased NO synthase activity and/or lack of arginine (NO synthase substrate) intake.
The metabolomic analysis performed on plasma in our study is consistent with the relevant literature. In a meta-analysis of 10 publications reporting metabolomic analyses, Li et al. identified 17 metabolites that could differentiate RA patients and controls5. This study confirmed two of them: threonine and alanine. Of note, these metabolites were significantly reduced in collagen-induced arthritis mice compared to controls9. We also identified DHEA sulfate as the metabolite with the lowest RA/HC ratio, in alignment with the literature10.
The data obtained from the RBC metabolome analysis is therefore partly consistent with the plasma analysis (involvement of the ammonia recycling pathway), but also identified other metabolites and pathways that appear to be dysregulated in RA patients, especially EGT. Our analyses showed a lower EGT concentration in RA RBCs compared to HC RBCs, and a higher EGT concentration in MTX responders compared to non-responders before MTX onset. EGT is a natural AA that derives from histidine. It is strongly associated with a “health conscious food pattern” that was linked to a reduced risk of cardiometabolic disease in a 3,236-participant study11. EGT cannot be synthesized by humans and is provided through diet. It can be found in actinobacteria, cyanobacteria, some fungi (such as porcini), oysters and garlic. EGT is a powerful antioxidant and can induce the expression of SIRT112, which appears lower in RA endothelial and synovial vessels13.
EGT uses OCTN1—a dependent organic cation transporter primarily expressed in blood cells including erythrocytes—to enter cells14. EGT is therefore up to 100 times more concentrated in RBCs than in plasma, where it is barely detectable. Interestingly, a study highlighted a polymorphism of SLC22A4 (coding for OCTN1) strongly associated with RA in East Asian15 but not Caucasian populations16. The functional consequences of this polymorphism are unknown, as is the actual biological role of EGT.
Our result differs from a study showing that the erythrocyte concentration of ergothioneine in RA patients was higher than in HCs17. This discrepancy may be related to the cohorts used, since the patients included in this previous study were nearly all treated with MTX and showed low disease activity (i.e. responders). Conversely, our study focused on DMARD-naïve RA patients. If, as our results suggest, patients with higher levels of EGT have a higher probability of responding to methotrexate treatment, it is conceivable that these patients are also more likely to remain on treatment over the long term. Therefore, by selecting patients in remission on methotrexate treatment, the other study might have focused on RA patients with the highest levels of EGT.
Our work has some limitations, mainly the absence of matching of RA patients and HCs on potential confounders like smoking status and BMI.
Moreover, like most proof-of-concept studies, this work is underpowered to predict the response to methotrexate. Based on our ratio of responders to non-responders, an ideal number of 59 subjects should have been included to achieve 80% power with an alpha error of 0.05. Also, our results were not replicated in a validation cohort. However, identifying a metabolic signature that may predict response to methotrexate treatment in early RA is beyond the scope of this work and would require larger cohorts and adjustments for multiple confounders.
Another limitation is the fact that the biological role of EGT and its transporter in the pathogenesis of RA is still unknown. The observation that RA patients have lower levels of EGT compared to healthy controls, and that those with the lowest levels are also less likely to respond to methotrexate treatment, is intriguing. This finding warrants mechanistic studies to understand the biological role of this metabolite. Whether EGT may be a marker of refractory RA, a potential adjuvant to methotrexate treatment, or merely a random bystander cannot be established by this work.
However, this completely novel method in rheumatology preliminarily identifies EGT, a food-derived AA that has antioxidant properties and accumulates in tissues, as a potential metabolite of interest in untreated RA and MTX response. Larger studies are warranted to bolster these original data.
Methods
Patients
The study group consisted of 31 prospectively recruited consecutive patients that met the 2010 ACR-EULAR Classification Criteria for RA and were DMARD-naïve. All patients were recruited from the Avicenne Rheumatology Department (Bobigny, France). Following an overnight fast, RA patients underwent blood sampling and clinical assessment before (M0) and 3 months after (M3) MTX initiation. At M3, all patients with a good or moderate DAS28-CRP-based EULAR response or in DAS28-CRP-based remission (i.e. DAS28-CRP < 2.6)) were classified as MTX responders. Only patients who had completed 3 months of MTX treatment and completed their 3-month follow-up visit were included in the longitudinal analysis (i.e. responders vs. non-responders). Blood samples of age- and sex-matched blood donors (Etablissement Français du Sang) were used as healthy controls (HC). This study was approved by the local review board of Avicenne Hospital (NI-2016-11-01) and performed in accordance with the Declaration of Helsinki and the institutional guidelines. Informed consent was obtained from all subjects.
Preparation of RBC lysates and RBC and plasma metabolite extraction
Preparation of RBC lysates and metabolite extraction were performed as previously described (see supplementary methods).
Untargeted metabolomics by ultra-high performance liquid chromatography (UHPLC) coupled with high-resolution mass spectrometry (HRMS) and data processing
The ultra-high performance liquid chromatographic (UHPLC) separation was performed on a Hypersil GOLD C18 1.9 µm, 2.1 mm × 150 mm column (RP) at 30 °C (Thermo Fisher Scientific, les Ulis, France), and HPLC chromatographic separations were performed on a Sequant ZICpHILIC 5 µm, 2.1 × 150 mm (HILIC) at 15 °C (Merck, Darmstadt, Germany). All chromatographic systems were equipped with an on-line prefilter (Thermo Fisher Scientific, Courtaboeuf, France). The data processing is described elsewhere18 and reported in the supplementary methods. LC–MS analyses were performed using a U3000 liquid chromatography system coupled to an Exactive mass spectrometer from Thermo Fisher Scientific (Courtaboeuf, France) fitted with an electrospray source. The software interface was Xcalibur (version 2.1) (Thermo Fisher Scientific, Courtaboeuf, France) (see supplementary methods).
Data processing and metabolite annotation procedures are described elsewhere (see supplementary methods).
Statistical analysis
Metabolite concentrations expressed as chromatographic peak areas (HILIC (-) or C18 ( +)) were analyzed using R software (version 4.2.1)19. For a given metabolite, the normality of the data distributions for each of the two groups—RA and HC—was evaluated by the Shapiro–Wilk test and visual examination of the qqplot graphs. The equality of variance was tested using the var.test function (R stats library). If the hypothesis of normality of at least one of the two distributions was rejected, the comparison of the values of the two groups was done by the row_Wilcoxon_two-sample test (R MatrixTest library). Otherwise, this comparison was done by Welch t test (unequal variances) or Student t test. Significant changes in metabolite levels were tested using the t-test, and False Discovery Rate (FDR) were determined according to Storey and Tibshirani procedure to control false discovery rate in multiple statistical tests. For the comparison between responders and non-responders, baseline corticosteroid doses and DAS28CRP values were compared with independent sample T test after checking for normality. A multivariable logistic regression model was used to check the independent association between EGT baseline values and 3-month clinical response to methotrexate adjusting on baseline corticosteroids and DAS28CRP values. To check for a potential attrition bias, DAS28-CRP, mean corticosteroid dose and EGT level were compared between patients lost-to-follow-up and those who completed the study by Mann–Whitney test to the sample size.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Semerano, L., Roméo, P.-H. & Boissier, M.-C. Metabolomics for rheumatic diseases: Has the time come?. Ann. Rheum. Dis. 74, 1325–1326 (2015).
Chu, S. H. et al. Circulating plasma metabolites and risk of rheumatoid arthritis in the Nurses’ Health Study. Rheumatology (Oxford) 59, 3369–3379 (2020).
Priori, R. et al. 1H-NMR-based metabolomic study for identifying serum profiles associated with the response to etanercept in patients with rheumatoid arthritis. PLoS One 10, e0138537 (2015).
Hur, B. et al. Plasma metabolomic profiling in patients with rheumatoid arthritis identifies biochemical features predictive of quantitative disease activity. Arthritis Res. Ther. 23, 164 (2021).
Li, C. et al. Metabolomics in the development and progression of rheumatoid arthritis: A systematic review. Joint Bone Spine 87, 425–430 (2020).
Farrow, M. et al. Muscle deterioration due to rheumatoid arthritis: Assessment by quantitative MRI and strength testing. Rheumatology (Oxford) 60, 1216–1225 (2021).
Surowiec, I. et al. Metabolomics study of fatigue in patients with rheumatoid arthritis naïve to biological treatment. Rheumatol. Int. 36, 703–711 (2016).
Bergholm, R. et al. Impaired responsiveness to NO in newly diagnosed patients with rheumatoid arthritis. Arterioscler. Thromb. Vasc. Biol. 22, 1637–1641 (2002).
He, M. et al. Spontaneous ultra-weak photon emission in correlation to inflammatory metabolism and oxidative stress in a mouse model of collagen-induced arthritis. J. Photochem. Photobiol. B 168, 98–106 (2017).
Lee, Y.-J. et al. A discovery of screening markers for rheumatoid arthritis by liquid chromatography mass spectrometry: A metabolomic approach. Int. J. Rheum. Dis. 23, 1353–1362 (2020).
Smith, E. et al. Ergothioneine is associated with reduced mortality and decreased risk of cardiovascular disease. Heart 106, 691–697 (2020).
D’Onofrio, N. et al. Ergothioneine oxidation in the protection against high-glucose induced endothelial senescence: Involvement of SIRT1 and SIRT6. Free Radic. Biol. Med. 96, 211–222 (2016).
Leblond, A. et al. Implication of the deacetylase sirtuin-1 on synovial angiogenesis and persistence of experimental arthritis. Ann. Rheum. Dis. 79, 891–900 (2020).
Gründemann, D. et al. Discovery of the ergothioneine transporter. Proc. Natl. Acad. Sci. U.S.A. 102, 5256–5261 (2005).
Tokuhiro, S. et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet. 35, 341–348 (2003).
Lee, Y. H. et al. Meta-analysis of SLC22A4 and RUNX1 polymorphisms : Associations with rheumatoid arthritis susceptibility. Z. Rheumatol. 74, 351–358 (2015).
Taubert, D. et al. Association of rheumatoid arthritis with ergothioneine levels in red blood cells: A case control study. J. Rheumatol. 33, 2139–2145 (2006).
Darghouth, D. et al. Pathophysiology of sickle cell disease is mirrored by the red blood cell metabolome. Blood 117, e57-66 (2011).
R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Funding
This work was supported by the French Rheumatology Society (Societé Française de Rhumatologie, SFR) and Entrepreneurs&Go.
Author information
Authors and Affiliations
Contributions
PHR, MCB and LS conceptualized the study JS, PHR and LS created the methodology for the study JS, MP, MB, FC, FF carried out the data collection JS and LS did the data analysis and verified the data JS and CJ supervised the data collection JS and LS wrote the original draft All authors reviewed and edited the manuscript All authors had full access to all of the data and the final responsibility to submit for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sigaux, J., Junot, C., Boissier, MC. et al. Red blood cell metabolomics identify ergothioneine as a key metabolite in DMARD-naïve rheumatoid arthritis and response to methotrexate. Sci Rep 14, 21005 (2024). https://doi.org/10.1038/s41598-024-68477-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-68477-6
Keywords
This article is cited by
-
Development and multi-center validation of machine learning models based on targeted metabolomics for rheumatoid arthritis
Journal of Translational Medicine (2025)
-
Evaluating treatment responsiveness in rheumatoid arthritis through predictive metabolomic profiling: A systematic review of studies examining methotrexate, TNF, and IL-6 inhibitors as therapeutic interventions
Clinical Rheumatology (2025)