Abstract
Gaining a comprehensive understanding of the role played by the oral microbiome in moderate to severe plaque psoriasis and its potential implications for disease management and development holds significant importance. With the objective of exploring correlations between the oral microbiota and severe psoriasis, this study involved 72 severe psoriasis patients and 16 healthy individuals, whose clinical manifestations and living habits were carefully recorded. Cutting-edge techniques such as 16S rRNA gene sequencing and bioinformatics analysis were employed to compare the microbial flora, investigating dynamic changes among severe plaque psoriasis patients, psoriatic arthritis patients and healthy individuals. The findings revealed noteworthy patterns including increased levels of Aggregatibacter in the psoriatic arthritis group, accompanied by a decrease in the level of Prevotella. Moreover, the enrichment o Capnocytandophaga (P = 0.009), Campylobacter (P = 0.0022), and Acetobacter (P = 0.0292) was notably more substantial in the psoriasis group compared to the control group, whereas certain bacterial species such as Bacteroides (P = 0.0049), Muribaculaceae (P = 0.0048) demonstrated decreased enrichment. Additionally, the psoriatic arthritis group exhibited significantly higher levels of Ralstonia, Bifidobacterium and Micromonospora. Based on these findings, it can be inferred that individuals with lower levels of Prevotella and higher levels of Corynebacterium may be more susceptible to psoriasis exacerbation.
Similar content being viewed by others
Introduction
Psoriasis is a recurring, chronic inflammatory skin disease with a global incidence of approximately 3–4%, of which 90% comprises psoriasis vulgaris1. The etiology of psoriasis is unknown but it may be linked to factors such as heredity, infection, and immune dysfunction2. The pathogenesis of psoriasis is primarily driven by overactive keratinocytes and dysfunctional immune cells, with T-cell imbalance playing a critical role in disease occurrence3. Among the essential pathways in psoriasis progression, the IL-23IL-17a-Th17 axis is prominent, regulated by various chemokines including IL-17, TNF-α, IL-1β, and IL-64,5.
The oral microbiome is a highly complex ecosystem shaped by teeth structure, gingival sulcus, tongue, buccal mucosa, hard and soft palate, and tonsils, providing a unique microenvironment. It harbors billions of bacteria, fungi, and viruses that establish a complete microbial community6. Microorganisms play a pivotal role in maintaining oral environment stability, where different parasites locally colonize the oral cavity7. Metagenomic and molecular analyses have revealed that the human mouth carries over 700 microbial species belonging to 150 genera, making it the second-largest microecosystem after the gastrointestinal tract8,9.
Disturbance in the oral flora can lead to localized infections like gingivitis and mucosal inflammation, with repercussions on the entire body through the interaction between pathogens and the immune system. Bacterial dysbiosis has been associated with various conditions such as chronic kidney failure, obesity, cancer (e.g., male digestive tract, colorectal, pancreatic and female uterine), mild cognitive impairment, metabolic syndrome, pneumonia, endocarditis, low birth weight infants, arteriosclerosis, rheumatoid arthritis, pancreatic cancer, and hepatitis8,10,11,12. It is also implicated in certain autoimmune skin diseases.
Research has shown that psoriasis patients experience more severe gingivitis, significant loss of alveolar bone, fewer teeth, and more severe dental caries compared to individuals without psoriasis13.The severity of these oral conditions correlates with the severity of psoriasis and the presence of psoriatic arthritis. Periodontitis severity is higher in the group of individuals with psoriatic arthritis compared to the control group14.
Metagenomic and micro genomic analyses have identified Streptococcus as a crucial genus in the oral bacterial community15. Streptococcal infection has been associated with the onset and exacerbation of psoriasis. Salivary levels of neutrophil gelatinases associated with lipocalin and transferrin in psoriasis patients are lower than those in non-psoriatic individuals with an abnormal oral environment16. Lower concentrations and secretory rates of immunoglobulin IgA and lysozyme, biomarkers linked to decreased oral humoral immunity, suggest that psoriasis patients may face a heightened risk of microbial infection, potentially triggering psoriasis exacerbation17.
Studies have revealed an association between nail involvement and psoriatic arthritis18. Additionally, age at onset, disease duration, and specific nail conditions including erythematous lunula, onychorrhexis, oil drop, and subungual hyperkeratosis, have been correlated with a higher risk of PSA19. Nail destruction serves as a significant risk factoring the progression from plaque psoriasis to psoriatic arthritis.
Materials and methods
Ethical approval and informed consent
The study complied with the Helsinki Declaration and was approved by the Medical Ethics Committee at the Jiangsu Province Hospital and the first affiliated hospital of Nanjing Medical University (ethics approval No. 2022-SR-170).
Sample collection
All saliva samples were collected at the Department of Dermatology, Jiangsu Province Hospital. Written informed consent was obtained from all participants. Unstimulated whole saliva was harvested from patients and healthy controls according to our standardized protocol for sample collection7.
Inclusion and exclusion criteria
Saliva samples were collected from moderate to severe psoriasis patients before initiating treatment with any biological agent. These patients did not have a family history of psoriasis and had more than 20 teeth. The inclusion criteria for these patients included having 10% or more of their body surface area affected by psoriasis and a Psoriasis Area and Severity Index score (PASI) of ten or greater20.
Individuals who exhibited certain pathological conditions or systemic autoimmune diseases, including malignant tumors, diabetes, cardiovascular diseases, immune dysfunction, herpes infections, systemic autoimmune diseases, rheumatoid arthritis or acute oral diseases such as herpes simplex and oral mucosal ulcers, local oral trauma, or had undergone surgery within the past three months were considered as exclusion criteria. Additionally, Subjects with a smoking history or who had consumed alcohol within the preceding three months were also excluded from the study.
DNA extraction and polymerase chain reaction (PCR) amplification
A Qiagen Gel Extraction Kit was used to extract total genomic DNA (Qiagen, Germany). A total amount of 1.0 μg DNA per sample was used as input material for the DNA sample preparations. Sequencing libraries were generated using NEBNext® DNA Library Prep Kit following manufacturer's recommendations and indices were added to each sample. The genomic DNA is randomly fragmented to a size of 350 bp by shearing, then DNA fragments were end polished, A-tailed, and ligated with the NEBNext adapter for Illumina sequencing, and further PCR enriched by P5 and indexed P7 oligos. The PCR products were purified (AMPure XP system) and resulted libraries were analyzed for size distribution by Agilent 2100 Bioanalyzer and quantified using real-time PCR. Sequencing was performed by using Illumina HiSeq 2000 with the V3 and V4 data of 16S rRNAs were sequenced.
Bioinformatics and statistical analysis
Paired-end reads were assigned to samples based on their unique barcodes and were truncated by cutting off the barcodes and primer sequences. FLASH (Version 1.2.11) was used to merge paired-end reads. Quality filtering of the raw tags was performed using fastp (Version 0.20.0) software to obtain high-quality clean tags. The clean tags were compared with the Silva database, using Vsearch (Version 2.15.0) to detect the chimera sequences, and then the chimera sequences were removed to obtain the effective tags. For the effective tags obtained previously, denoising was performed with DADA2 in QIIME2 software (Version QIIME2-202,006) to obtain initial amplicon sequence variants (ASVs). Alpha diversity was calculated in QIIME2. Beta diversity was calculated based on unweighted UniFrac distances in QIIME2.
Institutional review board statement
The study complied with the Helsinki Declaration and was approved by the Medical Ethics Committee at the Jiangsu Province Hospital and the first affiliated hospital of Nanjing Medical University (ethics approval No. 2022-SR-170).
Results
Clinicopathological characteristics of the enrolled population
Based on the presence or absence of nail destruction, psoriasis patients were categorized into two groups: PSO1 for those without nail destruction, and POS2 for those with nail psoriasis (Fig. 1). The psoriatic arthritis group consisted of patients who fulfilled the CASPAR criteria21 and had nail psoriasis (NP).
The clinical characteristics of these four groups were summarized at baseline. The age, gender, BMI, onset, disease duration, psoriasis area, severity index, BSA and variety of gingival indexes (such as dental plaque, bleeding on probing, Periodontitis) were comparable among the subjects.
Kindly refer to the Table 1, there was no significant difference in age of presentation, age of onset, clinical course, PASI score and BSA score between groups. The results of inter-group comparison showed that the BMI of the study groups was significant different (P = 0.039). The chart also implied no significant difference in Indicators of gingivitis severity such as dental plaque, bleeding on probing and biologic width dental between study groups.
Differences of oral microflora between psoriasis patients and healthy people
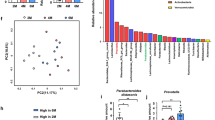
The ASV Venn diagram showed that the psoriasis patients (PSO) and healthy controls (Ctrl) had 1244 common ASVs, while the PSO group (n = 4864) had more individual ASVs than the Ctrl group (n = 981) (Fig. 2A). We performed PCoA analysis based on the unweighted UniFrac distance and selected the combination of principal coordinates with the highest contribution rate for graphic display. PCoA diagram (Fig. 2B) showed that the two groups contributed relative and distinguishable clusters in two-dimensional spatial distribution. Analysis of similarities (ANOSIM) (R = 0.167, P = 0.015) showed a more significant difference between the PSO and Ctrl groups than the intragroup comparison.
Comparison of bacterial diversity between psoriasis patients and healthy controls. (A)The ASVs between the PSO group(blue) and the control group(green). (B) PCoA based on unweighted UniFrac distance illustrated the patients’ grouping patterns (red) and healthy control group (blue). (C) LEfSe compared bacterial community structure between controls (red) and psoriasis patients (blue). Only taxa meeting an LDA significant threshold > 2 were shown. (D) Patient-enriched taxa were indicated with a positive LDA score (green), and taxa enriched in the Ctrl group had a negative score (red). Interaction graph of network for species correlation analysis in psoriasis patients(E) and heathy people(F), green line showed positive correlation and red line showed negative correlation between nodes (ρ > 0.6, p < 0.001, top 50 ASVs).
LEfSe (LDA effect size) revealed significant differences in 168 species of bacteria between the two groups at the genus level (Fig. 2C and Fig. 2D). In contrast, the proportions of 15 genera were significantly different between the cohorts at the genus level. Compared with the normal population, enrichment increased in seven bacterial species, including Capnocytandophaga (P = 0.009), Campylobacter (P = 0.0022) and Acetobacter (P = 0.0292), while enrichment of eight bacterial species decreased, including Bacteroides (P = 0.0049), Muribaculaceae (P = 0.0048), and Lactobacillus (P = 1.72E−6).
The horizontal correlation network diagram of genera level abundance of the top 20 showed that the synergism between Capnocytophage, Prevotella, and Streptococcus is essential in maintaining oral microbiota's structural and functional stability in psoriasis patients (Fig. 2E). While Patescibacteria, Actinobacteriota and Bacteroidota is essential in maintaining oral microbiota's structural in healthy people (Fig. 2F).
Differences of oral microflora between PSO and PSA patients
The ASV Venn diagram showed that the people in the PSO and PSA groups had 1732 common ASVs, while the PSO group had more individual ASVs than the PSA group (4376 vs. 1238) (Fig. 3A). PCoA diagram showed there was no significant difference between two groups in contributed relative and distinguishable clusters (Fig. 3B).
Comparison of bacterial diversity between psoriasis patients (PSO) and psoriatic arthritis patients (PSA). (A) The ASVs between the PSO group (blue) and the PSA group (green). (B) PCoA based on unweighted_unifrac distance illustrated the PSA patients’ grouping patterns (red) and PSO patients’ grouping patterns (green). (C) LEfSe identified the most differential abundant taxon between the groups. (D)Taxa enriched in PSO and PSA groups were indicated with green and red LDA scores. Co-occurrence patterns of cropping sensitive ASVs. Co-occurrence networks visualizing significant correlations (ρ > 0.6, p < 0.001, top 50 ASVs) between bacteria and fungi ASVs in PSO (E) and PSA patients (F).
The LEfSe revealed significant differences among the two groups at the genus level (Fig. 3C and Fig. 3D). Ralstonia, Bifidobacterium, and Micromonospora levels were significantly higher in the PSA group, while Saccharimonadales was significantly higher in the PSO group.
Co-occurrence networks of Actinobacteriota, Proteobacteria, Firmicutes is essential pays an important role in maintaining oral microbiota's structural and functional stability in psoriasis patients (Fig. 3E). While microorganism such as Cyanobacteria, Poteobacteria, Bcteroidota and Spirochaetotais was essential in maintaining oral microbiota's structural in healthy people (Fig. 3F).
Differences of oral microflora between PSO patients with or without NP
The ASV Venn diagram showed that people in the PSO1 and PSO2 groups had 1605 common ASVs, the PSO1 group had 2751 individual ASVs, and the PSO2 group had 1752(Fig. 4A). The PCoA analysis showed that the oral microbiota of the PSO1 group appeared to be more tightly clustered than the PSO2 groups, suggesting that there were more similar bacterial communities among patients in the PSO1 group (Fig. 4B).
Comparison of bacterial diversity between psoriasis patients without nail psoriasis (PSO1) or with nail psoriasis (PSO2). (A)The ASVs between the PSO1 (blue) and PSO2 (green) groups. (B) PCoA based on unweighted_unifrac distance illustrated the PSO1 patients’ grouping patterns (red) and PSO2 patients’ grouping patterns (blue). (C) Tphylogenetic trees in PSO1 and PSO2 groups at genus level. (D) LEfSe identified the most differential abundant taxon between the groups. Co-occurrence networks visualizing significant correlations (ρ > 0.6, p < 0.001, top 50 ASVs) between bacteria and fungi ASVs in PSO1 (E) and PSO2 patients (F).
In order to study the phylogenetic relationships of horizontal species at genus level, the representative sequences of Top100 genera were obtained by multiple sequence alignment, the species abundance of PSO1 and PSO2 group were listed (Fig. 4C).
LEfSe (LDA effect size) revealed significant differences between the groups (Fig. 4D). Streptococcus (P = 0.022) and Alloprevotella (P = 0.042) were significantly higher in the PSO1 group, while the levels of 19 taxa were significantly higher in the PSO2 group, including Desulfuromonas (P = 0.0062) and Anaerococcus (P = 0.0272).
The horizontal correlation network diagram of genera level abundance of the top 50 showed that the synergism between Cyanobacteria, Patescibacteria, Actinobacteriota and Proteobacteria was essential in maintaining oral microbiota's structural and functional stability in PSO1 group (Fig. 4E). While co-occurrence networks of Actinobacteriota, Proteobacteria and Spirochate essential played an important role in maintaining oral microbiota's structural and functional stability in PSO2 group (Fig. 4F).
Species abundance distribution of three groups at the genus level
The top ten genera were selected according to all genus level samples' species annotation and abundance information. A heat map was drawn for the species and samples clustered in each group (Fig. 5) to calculate the species concentrations in the samples.
Species and samples clustered in Ctrl, PSO, and PSA groups. (A) Validation of selective oral bacteria, the level of Prevotella decreased from the Ctrl, PSO to PSA groups, while the level of Aggregatibacter acting in the opposite direction. (B). Relative abundance of bacteria in each group at the genus level.
The absolute abundance of Prevotella in Ctrl, PSO and PSA group is 0.073737, 0.061876 and 0.053603, and the relative abundance of Prevotella in the three groups is 0.073913, 0.06186 and 0.053497. The absolute abundance of Aggregatibacter in Ctrl, PSO and PSA group is 0.012015,0.018627and 0.02636, and the relative abundance of which in the three group is 0.012034, 0.018636 and 0.026347. The level of Prevotella decrsased as the deterioration of diseases, contrary to the abundance of Aggregatibacter.
Discussion
Psoriasis is a chronic and recurring inflammatory skin disease. While genetic susceptibility has traditionally been considered the primary cause of inflammatory disease, mounting evidence suggesting a connection between psoriasis and the microbial communities in the intestines and skin. The diversity of intestinal flora is deemed to be related to the pathogenesis of psoriatic arthritis, the application of fecal bacteria transplantation in psoriatic arthritis has achieved good results23. According to Alekseyenko’s finding, patients with plaque psoriasis exhibited an increased abundance of Corynebacterium, Cutibacterium, Staphylococcus, and Streptococcus in the affected skin areas21. Another study suggests that the dry microenvironment in psoriatic lesions may not be suitable for Cutibacterium colonization24. However, research on the correlation between psoriasis and oral microflora is still limited24,25,26 Oral flora diversity between plaque psoriasis and the normal population is now receiving increasing attention27. Differences in oral flora between psoriatic patients with and without a geographical tongue has been discovered recently28.
The oral microbiome is a highly complex ecosystem consisting of various microorganisms Periodontitis, a condition characterized by inflammation of the gums, can potentially worsen or advance psoriasis and psoriatic arthritis due to the immune-modifying effects of oral dysbiosis. Periodontitis has been identified as a potential comorbidity in psoriasis and psoriatic arthritis29. Bacteria in the oral cavity can enter the bloodstream through gingival pockets, and microbiota-derived molecules can circulate systemically, stimulating specific cytokines and potentially influencing arthritis development. A study observed that reduced levels of Helicobacter, Flexispira, Clostridium, and Dehalobacterium suppressed arthritis in mice with interleukin-1 receptor antagonist deficiences30. In the current study, significantly high levels of Ralstonia, Bifidobacterium, and Micromonospora were found in the PSA group, while the level of Saccharimonadales was significantly elevated in the PSO group.
Analyzing the deep sequencing data in this study revealed a decrease in the abundance of Prevotella species from the control (Ctrl) and PSO groups to the PSA group. Recent research on gut microbiota has shown a decreased abundance of Prevotella in patients with psoriasis compared to control populations31,32. Prevotella plays an essential role in maintaining the integrity of the colonic mucosa and be involved in gut permeability, systemic exposure to microbial antigens, and systemic inflammation32,33. Prevotella can stimulate dendritic cells to release interleukin IL-1β, IL-6, and IL-23 through Toll-like receptor 2, which in turn activate neutrophils producing IL-17. Changes in the oral microbiome can increase the number of anaerobic bacteria in periodontitis and local inflammatory mediators such as TNF-α, IL-1, IL-2, IL-8, and prostaglandins6. In humans, a specific strain of Prevotella has been shown to modify systemic inflammation through Th1, Th2, and Th17-driven inflammation in the gut33. The effectiveness of oral treatment with Prevotella histicola in mice depends on regulation by CD103 + dendritic cells and the generation of regulatory T cells in the gut, leading to the suppression of pro-inflammatory Th1 and Th17 responses and increased transcription of interleukin-1034. The decline of Prevotella may be one of the potential factors contributing to the aggravation of psoriasis and the occurrence of joint lesions.
Furthermore, the present study found an increase in Aggregatibacter actinomycetemcomitans (Aa) from the control PSO group to the PSA groups. Aa, a pathogenic bacterium associated with periodontitis, has been considered a potential trigger of autoimmunity in rheumatoid arthritis35. However, the correlation between Aa and PSA has not been reported. in vitro studies have shown that Aa might stimulate a Th17/IL-17 response. Previous research demonstrated that stimulation of monocytes with Porphyromonas gingivalis (Pg) or Aa resulted in increased expression of CD40, CD54, and HLA-DR, along with enhanced production of proinflammatory cytokines TNF-α, IL-1β, IL-6, and IL-2336. The present study also discovered significantly higher levels of IgG antibodies against E. coli and Aa in some patients with pustulosis palmariset plantairs compared to the control group37.
Our findings suggest that the transition from severe plaque psoriasis, regardless of nail damage, to psoriatic arthritis does not notably intensify gingivitis symptoms like dental plaque, bleeding on probing, or periodontitis. It appears that alterations in flora may play a more direct role in exacerbating psoriasis, rather than being closely linked to the severity of gingivitis symptoms. Given the limitation in terms of research scope and sample size, further basic studies are necessary to elucidate the specific role of Prevotella and Aggregatibacter actinomycetemcomitans in the exacerbation of psoriasis. It is important to explore the potential of actively intervening with these microorganisms to potentially delay the progression of psoriatic onychomycosis and arthropathic psoriasis. However, to validate these findings, additional comprehensive studies encompassing both fundamental and clinical research are required.
Conclusions
The research paper demonstrates a potential correlation between psoriasis and microbial communities, both in the oral cavity and the skin. The findings highlight the importance of considering the role of oral dysbiosis and periodontitis in the development and progression of psoriasis and psoriatic arthritis. The decrease in Prevotella abundance and the increase in Aggregatibacter actinomycetemcomitans suggest their potential involvement in the inflammatory processes underlying these conditions. Further studies are needed to explore the mechanisms through which these microbial shifts contribute to the pathogenesis of psoriasis and to assess the potential for targeted therapies aimed at modulating the oral microbiome in the management of psoriatic diseases.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Menter, A. et al. Guidelines of care for the management of psoriasis and psoriatic arthritis. J. Am. Acad. Dermatol. 58, 826–850 (2008).
Bissonnette, R. et al. TNF-α antagonist and vascular inflammation in patients with psoriasis vulgaris: A randomized placebo-controlled study. J. Invest. Dermatol. 137, 1638–1645 (2017).
Lowes, M. A., Suárez-Fariñas, M. & Krueger, J. G. Immunology of psoriasis. Annu. Rev. Immunol. 32, 227–255 (2014).
Cai, Y. et al. Differential roles of the mTOR-STAT3 signaling in dermal γδ T cell effector function in skin inflammation. Cell Rep. 27, 3034-3048.e5 (2019).
Chen, C. et al. Metabolomic profiling reveals amino acid and carnitine alterations as metabolic signatures in psoriasis. Theranostics 11, 754–767 (2021).
Samaranayake, L. & Matsubara, V. H. Normal oral flora and the oral ecosystem. Dent Clin North Am 61, 199–215 (2017).
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I. & Dewhirst, F. E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732 (2005).
Gao, L. et al. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell 9, 488–500 (2018).
Diaz, P. I., Hoare, A. & Hong, B.-Y. Subgingival microbiome shifts and community dynamics in periodontal diseases. J Calif Dent Assoc 44, 421–435 (2016).
Mysak, J. et al. Porphyromonas gingivalis : Major periodontopathic pathogen overview. J. Immunol. Res. 2014, 1–8 (2014).
Linden, G. J., Lyons, A. & Scannapieco, F. A. Periodontal systemic associations: Review of the evidence. J. Clin. Periodontol. 40, 8–19 (2013).
Li, X., Kolltveit, K. M., Tronstad, L. & Olsen, I. Systemic diseases caused by oral infection. Clin. Microbiol. Rev. 13, 547–558 (2000).
Qiao, P. et al. Psoriasis patients suffer from worse periodontal status—A meta-analysis. Front Med (Lausanne) 6, 212–222 (2019).
Egeberg, A., Mallbris, L., Gislason, G., Hansen, P. R. & Mrowietz, U. Risk of periodontitis in patients with psoriasis and psoriatic arthritis. J. Eur. Acad. Dermatol. Venereol. 31, 288–293 (2017).
Komatsu, T. et al. E-selectin mediates porphyromonas gingivalis adherence to human endothelial cells. Infect. Immun. 80, 2570–2576 (2012).
MacKlis, P. et al. The association between oral health and skin disease. J. Clin. Aesthetic Dermatol. 13, 48–53 (2020).
Belstrøm, D. et al. Salivary microbiota and inflammation-related proteins in patients with psoriasis. Oral Dis. 26(3), 677–687 (2020).
Scher, J. U., Ogdie, A., Merola, J. F. & Ritchlin, C. Preventing psoriatic arthritis: Focusing on patients with psoriasis at increased risk of transition. Nat. Rev. Rheumatol. 15, 153–166 (2019).
Liu, P. et al. Predicting the risk of psoriatic arthritis in plaque psoriasis patients: Development and assessment of a new predictive nomogram. Front. Immunol. 12, 740968 (2022).
Menter, A. et al. Adalimumab therapy for moderate to severe psoriasis: A randomized, controlled phase III trial. J. Am. Acad. Dermatol. 58, 106–115 (2008).
Taylor, W. et al. Classification criteria for psoriatic arthritis: Development of new criteria from a large international study. Arthritis. Rheum. 54, 2665–2673 (2006).
Landzberg, M., Doering, H., Aboodi, G. M., Tenenbaum, H. C. & Glogauer, M. Quantifying oral inflammatory load: Oral neutrophil counts in periodontal health and disease. J. Periodontal. Res. 50, 330–336 (2015).
Kragsnaes, M. S. et al. Safety and efficacy of faecal microbiota transplantation for active peripheral psoriatic arthritis: an exploratory randomised placebo-controlled trial. Ann. Rheum. Dis. 80, 1158–1167 (2021).
Quan, C. et al. Psoriatic lesions are characterized by higher bacterial load and imbalance between Cutibacterium and Corynebacterium. J. Am. Acad. Dermatol. 82, 955–961 (2020).
Alekseyenko, A. V. et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome 1, 31 (2013).
Rogier, R. et al. Aberrant intestinal microbiota due to IL-1 receptor antagonist deficiency promotes IL-17- and TLR4-dependent arthritis. Microbiome 5, 63 (2017).
Belstrøm, D. et al. Salivary microbiota and inflammation-related proteins in patients with psoriasis. Oral Dis. 26, 677–687 (2020).
Horiuchi, Y. Geographic tongue: What is this disease?. J. German Soc. Dermatol. 21, 1465–1467. https://doi.org/10.1111/ddg.15226 (2023).
Dalmády, S., Kemény, L., Antal, M. & Gyulai, R. Periodontitis: a newly identified comorbidity in psoriasis and psoriatic arthritis. Expert Rev. Clin. Immunol. 16, 101–108 (2020).
Drago, L. et al. Oral-gut microbiota and arthritis: Is there an evidence-based axis?. J. Clin. Med. 8, 1753 (2019).
Todberg, T. et al. Characterization of oral and gut microbiota in patients with psoriatic diseases: A systematic review. Acta Dermato Venereologica 101(7), 150 (2021).
Shapiro, J. et al. Psoriatic patients have a distinct structural and functional fecal microbiota compared with controls. J. Dermatol. 46, 595–603 (2019).
Itano, A. et al. Clinical translation of anti-inflammatory effects of Prevotella histicola in Th1, Th2, and Th17 inflammation. Front Med (Lausanne) 10, 1070433 (2023).
Marietta, E. et al. Administration of human derived upper gut commensal Prevotella histicola delays the onset of type 1 diabetes in NOD mice. BMC Microbiol 22, 8 (2022).
Díaz-Zúñiga, J. et al. T-lymphocyte phenotype and function triggered by Aggregatibacter actinomycetemcomitans is serotype-dependent. J. Periodontal. Res. 50, 824–835 (2015).
Cheng, W. et al. Periodontitis-associated pathogens P. gingivalis and A. actinomycetemcomitans activate human CD14 + monocytes leading to enhanced Th17/IL-17 responses. Eur J Immunol 46, 2211–2221 (2016).
Kosugi, M., Ishihara, K. & Okuda, K. Implication of responses to bacterial heat shock proteins, chronic microbial infections, and dental metal allergy in patients with pustulosis palmaris et plantaris. Bull Tokyo Dent. Coll. 44, 149–158 (2003).
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
Writing—original draft preparation Fan, W.; writing—review and editing, Lu, Y. and Lei, N.; saliva collection and clinical data registration, Zheng, Y.J., Lei, Na., Fan, W. and Su, Z.L .;gene 16 s-RNA sequencing supervision, Liu, J. and Su, T.; figures preparation, Zheng, Y.J.; statistics , Fan, W. and Cao, X.C., project administration, Lu, Y.; All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fan, W., Lei, N., Zheng, Y. et al. Oral microbiota diversity in moderate to severe plaque psoriasis, nail psoriasis and psoriatic arthritis. Sci Rep 14, 18402 (2024). https://doi.org/10.1038/s41598-024-69132-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-69132-w