Abstract
Astrocytes display context-specific diversity in their functions and respond to noxious stimuli between brain regions. Astrocytic mitochondria have emerged as key players in governing astrocytic functional heterogeneity, given their ability to dynamically adapt their morphology to regional demands on ATP generation and Ca2+ buffering functions. Although there is reciprocal regulation between mitochondrial dynamics and mitochondrial Ca2+ signaling in astrocytes, the extent of this regulation in astrocytes from different brain regions remains unexplored. Brain-wide, experimentally induced mitochondrial DNA (mtDNA) loss in astrocytes showed that mtDNA integrity is critical for astrocyte function, however, possible diverse responses to this noxious stimulus between brain areas were not reported in these experiments. To selectively damage mtDNA in astrocytes in a brain-region-specific manner, we developed a novel adeno-associated virus (AAV)-based tool, Mito-PstI expressing the restriction enzyme PstI, specifically in astrocytic mitochondria. Here, we applied Mito-PstI to two brain regions, the dorsolateral striatum and dentate gyrus, and we show that Mito-PstI induces astrocytic mtDNA loss in vivo, but with remarkable brain-region-dependent differences on mitochondrial dynamics, Ca2+ fluxes, and astrocytic and microglial reactivity. Thus, AAV-Mito-PstI is a novel tool to explore the relationship between astrocytic mitochondrial network dynamics and astrocytic mitochondrial Ca2+ signaling in a brain-region-selective manner.
Similar content being viewed by others
Introduction
Astrocytes have emerged as critical participants in governing neuronal function during health and disease1. Once thought of as a group of rather homogenous support cells, compelling evidence has revealed that protoplasmic astrocytes are heterogenous2,3 and display context-dependent heterogeneity in their functions and in their response to injurious stimuli within and across brain regions4,5.
Astrocytic heterogeneity between brain regions extends to important organelles within these cells, including mitochondria. Indeed, we have recently shown that astrocytic mitochondria in the hippocampus (HPC) and dorsolateral striatum (DLS) are morphologically and functionally distinct6. Astrocytic mitochondria are a particularly relevant organelle in terms of astrocytic heterogeneity because they have robust spontaneous Ca2+ fluxes that respond to neurotransmitter agonists6,7,8, a distinct functional proteomic profile9, and spatial segregation into somata versus processes with regard to Ca2+ fluxes and morphology6,8. In addition, mitochondria are very dynamic organelles, constantly undergoing fission, fusion, and turnover. They specialize in the production of ATP by oxidative phosphorylation (OXPHOS), but are also capable of buffering intracellular Ca2+. Interestingly, there is reciprocal regulation between mitochondrial dynamics and mitochondrial Ca2+ signaling in astrocytes8,10, however, to our knowledge, the extent of this regulation into the rich diversity of astrocytes in different brain regions remains largely unexplored.
In this study, we exploit mitochondrial targeting of bacterial restriction enzymes as an approach to manipulate the mitochondrial genome, giving rise to mitochondrial DNA (mtDNA) loss in a cell-type-specific manner11. Thus, mitochondrially targeted PstI, which cuts at two sites on mouse mtDNA, has been expressed in skeletal muscle fibers and neurons in vivo, primarily resulting in mtDNA depletion, deletion by recombination, and chronic reduction of OXPHOS12,13,14. Brain-wide astrocytic mtDNA depletion was previously achieved by conditional abrogation of Twinkle, a nuclear encoded helicase required for mtDNA replication15. Although this work demonstrated the absolute necessity of mtDNA integrity for proper astrocyte function, few insights into possible diverse responses to this noxious stimulus from astrocytes in different brain areas were reported in these experiments. To selectively damage mtDNA in astrocytes in a brain-region-specific fashion, we have developed a novel adeno-associated virus (AAV)-based tool, Mito-PstI, which expresses PstI, specifically in astrocytic mitochondria. Here, we have applied this tool to the HPC dentate gyrus and the DLS, two well-studied areas of the brain, relevant for Alzheimer’s and Parkinson’s disease, respectively. Our choice of HPC and DLS as brain regions in this study stems from our previous report in which we demonstrate differences in spontaneous mitochondrial Ca2+ influx events in astrocytes from these two brain regions6.
Our results show that Mito-PstI can induce astrocytic mtDNA loss in vivo, but with remarkable brain-region-dependent effects on mitochondrial dynamics, spontaneous Ca2+ fluxes and astrocytic and microglial reactivity. Thus, AAV-Mito-PstI is a novel tool to explore the relationship between astrocytic mitochondria network dynamics and astrocytic mitochondria Ca2+ signaling in a brain-region-selective manner.
Results
Mito-PstI targeted to astrocytes depletes and deletes mtDNA in vivo
Prior work showed that targeting of PstI to mitochondria in murine cells led to mtDNA depletion and a deletion of mtDNA between the two PstI restriction sites that are specifically present in murine mtDNA12,13,14. Based on this, we designed an AAV in which the well-established, GfaABC1D astrocyte-specific promoter16 drives expression of PstI, specifically targeted to astrocytic mitochondria via a mitochondria signal sequence. In addition, we utilized previously published AAV reporter constructs to specifically drive expression of either GCaMP6f (Mito-GcaMP6f) or GFP (Mito-GFP) in astrocytic mitochondria (Fig. 1A)6. The expression pattern of Mito-GFP in the DLS and the HPC dentate gyrus (Fig. 1B) demonstrated the highly selective astrocyte specificity allowed by these vectors. As a first step, we determined whether our Mito-PstI construct would lead to depletion of mtDNA in astrocytes in vivo. To do this, we introduced 1 × 1010 gc of our AAV via stereotaxic surgery unilaterally into the DLS of 2- to 3-month-old mice, and after ~ 3 wk, microdissected the injected DLS and isolated total DNA from this tissue and from DLS of age-matched, uninjected mice that served as control. Mitochondrial DNA content in the samples were quantified as the mtDNA/nuclear DNA ratio by sybr green quantitative RT-PCR17 using primers to Cytochrome b (Mt-cyb, white arrows in Fig. 1C), a mitochondrial gene outside the PstI sites, and Erk2 (Mapk1), as nuclear gene. As seen in Fig. 1D, Mito-PstI-infected tissue had on average 60.1 ± 4.6% mtDNA content relative to the control (n = 3; p = 0.0044, t-test). ~ 40% depletion in mtDNA content in vivo is consistent with selective targeting of astrocytic mtDNA, and not of mtDNA of all cell types in the DLS. Since mitochondria-targeted PstI has also been shown to induce mtDNA deletions by recombination in neurons and muscle12,14, we used two pairs of nested primers flanking the two PstI sites in mitochondria (black and gray arrows in Fig. 1C) to amplify the region between them by standard PCR. While the expected 4512 bp fragment from wildtype mtDNA is too large to be detected with our amplification conditions (see “Materials and methods”), the much smaller DNA bands generated by recombination following PstI digestion would be amplified using our protocol. In agreement with this rationale, we detected a ~ 700 bp fragment only in samples from mice treated with Mito-PstI in vivo (Fig. 1E). The size of this fragment was similar to that reported by the Moraes group using primers that also flanked the PstI sites in experiments in which PstI was expressed in dopaminergic neurons12. The bands were excised, extracted, and sent for sequencing, which confirmed that recombination indeed took place between the two mtDNA PstI sites (Supplementary Material Fig. 1). Thus, Mito-PstI targeted to astrocytes in vivo causes significant depletion of mtDNA and recombination events consistent with prior results in neurons12.
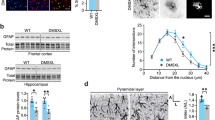
Mito-PstI depletes and deletes astrocytic mtDNA in vivo. (A) Schematic showing the AAV 2/5 vectors for stereotaxic injection into specific brain regions. GfaABC1D: Gfap promoter. Mito-7: COX8A mitochondrial signal sequence. 3XGGS: 12 amino acid linker. (B) Images depicting Mito-GFP fluorescence on NeuN stained HPC and DLS tissue, respectively. GFP expression was highly astrocyte specific. Scale bar = 500 μm. (C) Diagram for mtDNA genome with location of PstI sites (red bars), PCR primers for Cytochrome b (white arrows), and for detection of possible deletions (black and gray arrows). Nucleotide (nt) positions are indicated. (D) Relative mtDNA content quantification showing a ~ 40% reduction in mtDNA content in Mito-PstI-treated tissue. Each data point is midbrain homogenate from a single, male mouse. Errors bars are ± SEM, p-value for the relative mtDNA content quantification is based on a two-sample t-test, n = 3 male mice per condition. (E) Detection of potential deletions. Size markers in the right-most lane; ~ 700 bp fragment, suggestive of mtDNA recombination, only observed in Mito-PstI treated samples (n = 3/condition).
Mito-PstI causes significant morphological changes to astrocytic mitochondria in DLS, but not in HPC
We recently demonstrated robust morphological and functional differences in astrocytic mitochondria between the hippocampus and striatum6. Therefore, in this study, we sought to utilize AAV-Mito-Pst1 as a tool to determine if damaging astrocytic mtDNA within the HPC and DLS would result in divergent effects on astrocytic mitochondria in these two brain regions. Thus, we unilaterally co-injected a 1:1 mixture of AAV-Mito-PstI and AAV-Mito-GFP (1 × 1010 gc each AAV, 2 × 1010 gc total) either into the DLS or the dentate gyrus of the hippocampus (HPC) of 2–3-month-old mice. As a control, we injected mice with 2 × 1010 vg of AAV-Mito-GFP in either DLS or HPC. This AAV vector utilized the same GfaABC1D promoter and mitochondrial signal sequence that was used to target PstI (Fig. 1A), which allowed clear visualization of astrocytic mitochondria (Fig. 2A–D). After ~ 3 wk, brains were dissected, sectioned in a cryostat and these were imaged in a confocal microscope. Images of individual Mito-GFP-expressing astrocytes were taken (Fig. 2A–D) and analyzed using ImageJ. We first quantified the number, size, and combined area of mitochondrial particles in individual astrocytes within each of the two brain regions (Fig. 2E–G). Results revealed that baseline mitochondrial number and size were significantly different between DLS and HPC as Mito-GFP control mitochondrial particles in the DLS were ~ twofold more abundant (Fig. 2E) and ~ fivefold larger (Fig. 2F) than those in HPC (Table 1). Under control Mito-GFP injected conditions, no significant differences were observed in whole network area of mitochondria between the HPC and DLS (Fig. 2G, Table 1). Astrocytic PstI expression did not significantly alter the number, size and combined area of mitochondrial particles in the HPC, however, by contrast, we observed significant changes in the DLS. Thus, in the DLS, Mito-PstI-treated astrocytes had more mitochondria (Fig. 2E, Mito-GFP + PstI: 849 ± 49; Mito-GFP: 540 ± 23; p < 0.0001, Mann Whitney test), that were smaller (Fig. 2F, Mito-GFP + PstI: 4.4 ± 0.39 µm2; Mito-GFP: 8.5 ± 0.83 µm2; p < 0.0001, Mann Whitney test) than mito-GFP expressing control astrocytes. Additionally, Mito-PstI-expressing astrocytes in the DLS showed a significantly larger area of mitochondrial coverage per astrocyte (609 ± 69 µm2) than the Mito-GFP control group (331 ± 40 µm2; p = 8.46E-4, Mann Whitney test). Thus, these baseline morphological parameters suggest that astrocytic mitochondria in DLS and HPC show heterogeneity at baseline in mitochondrial morphology, as well as diverging structural changes in mitochondria following astrocytic mtDNA damage. Of note, DLS mitochondria, in particular, appeared more fragmented with Mito-PstI expression.
Mito-PstI causes significant morphological changes to astrocytic mitochondria in DLS, but not in HPC. Representative confocal images (A–D), MitoGraph surface reconstructions (Aʹ–Dʹ), and MitoGraph mitochondrial network skeletons (A”-D”) of Mito-GFP expression in HPC (A,B) and DLS (C,D) astrocytic mitochondria. Scale bars = 10 μm. Mitochondrial particle count (E), size (F), and network area (G) as quantified using ImageJ. Mitochondrial network connectivity parameters assessed using MitoGraph (H–P). For image analyses, each data point is a single astrocyte. Data expressed as mean ± SEM, p-values are based on a two-sample t-test or Mann Whitney test, depending on normality. For (E–G), n = 21 Mito-GFP only astrocytes and 17 Mito-GFP + PstI astrocytes in the HPC, n = 25 astrocytes per condition in the DLS. For (H-P), n = 30 astrocytes per condition in the HPC, n = 93 Mito-GFP-only astrocytes and n = 43 Mito-GFP + Mito-PstI astrocytes in the DLS. Brain slices were taken from 3–5 mice, using both males and females, per condition per brain region.
To further study the morphological effects of Mito-PstI on astrocytic mitochondria, we next used MitoGraph, a software package specifically designed to quantitatively analyze mitochondrial networks18. The confocal images were processed using MitoGraph to reconstruct the mitochondrial network surfaces (Fig. 2Aʹ–Dʹ) and skeletons (Fig. 2A″–D″). A number of network connectivity parameters (Fig. 2H–P) resulted from the skeletonizing of the segmented mitochondrial network into a set of edges (i.e., mitochondrial tubules), and nodes (i.e., endpoints and branch points). Edges are the connection between nodes and individual mitochondrial particles are referred to as connected components. In the HPC, node (Fig. 2H, Mito-GFP: 1504.8 ± 78.87, Mito-GFP + PstI: 1835 ± 133.1, p = 0.06173, Mann Whitney test) and connected component counts (Fig. 2J, Mito-GFP: 632 ± 40, Mito-GFP + PstI: 797 ± 63, p = 0.03107, t-test) were elevated in the Mito-GFP + Mito-PstI group compared to Mito-GFP controls, however, the overall connectivity scores were not different from the controls (Fig. 2P). This suggests that Mito-PstI expression failed to significantly change morphological characteristics of the mitochondrial network within astrocytes in the HPC. In stark contrast, the mitochondrial network in DLS astrocytes for the Mito-GFP + Mito-PstI group had a significantly higher number of nodes (Fig. 2H, Mito-GFP: 883 ± 49, Mito-GFP + PstI: 1234 ± 74; p < 0.0001, Mann Whitney test), connected components (Fig. 2J, Mito-GFP: 270 ± 15, Mito-GFP + PstI: 474 ± 29; p < 0.0001, Mann Whitney test) and free ends (Fig. 2K, Mito-GFP: 0.76 ± 0.01, Mito-GFP + PstI: 0.85 ± 0.012; p < 0.0001, Mann Whitney test) than control. In addition, Mito-Pst1 expressing DLS astrocytes had significantly fewer 4-way junctions (Fig. 2L, Mito-GFP: 0.0391 ± 0.0022, Mito-PstI: 0.022 ± 0.0023; p < 0.0001, Mann Whitney test), lower Average Degree (Fig. 2M, Mito-GFP: 1.44 ± 0.018, Mito-GFP + PstI: 1.28 ± 0.020, p < 0.0001, Mann Whitney test), Average Edge Length (Fig. 2N, Mito-GFP: 0.925 ± 0.0117 µm, Mito-GFP + PstI: 0.754 ± 0.0195 µm, p < 0.0001, Mann Whitney test), PHI (Fig. 2O, Mito-GFP: 0.29 ± 0.02, Mito-GFP + PstI: 0.102 ± 0.013; p < 0.0001, Mann Whitney test ) and connectivity score (Fig. 2P, Mito-GFP: 0.96 ± 0.033, Mito-GFP + PstI: 0.56 ± 0.036; p < 0.0001, Mann Whitney test). Importantly, the connectivity score is a single measure, intended to capture the diverse nature of the mitochondrial network, calculated by using as numerator the sum of parameters elevated in highly fused networks (e.g., PHI: relative size of the largest connected mitochondrial component compared to the total mitochondrial size, average edge length, and average degree), and as denominator the sum of parameters elevated in highly fragmented networks (e.g., nodes, edges, and connected components). A reduction in connectivity score tightly correlates with increased mitochondria fission18. Thus, these data suggest that Mito-PstI specifically causes increased fission, or decreased fusion, of mitochondrial networks within astrocytes in the DLS.
Mito-PstI causes a striking divergence in the kinetics of spontaneous mitochondrial Ca2+ influx events between astrocytes in the HPC versus DLS
We previously reported that astrocytic mitochondria display robust, spontaneous Ca2+ influx events in situ with baseline inter-regional differences between astrocytes in the DLS and HPC6. Thus, to evaluate the effects of PstI on spontaneous mitochondrial Ca2+ events, we unilaterally co-injected the DLS or the HPC dentate gyrus of 2–3-month-old mice with a mixture of AAV-Mito-PstI (1 × 1010 gc) and AAV-Mito-GCaMP6f (2 × 109 gc). As control, we injected other mice with 2 × 109 vg of AAV-Mito-GCaMP6f in either DLS or HPC. This virus delivered the GCaMP6f Ca2+ indicator19 selectively to astrocytic mitochondria in the two brain regions, (Fig. 1A6). After 3–4 wk, Ca2+ imaging of individual astrocytes was performed in live brain slices as detailed in “Materials and methods”, and recordings were analyzed with the AQuA software package20. In preliminary analysis of the data, we discovered we could bin the observations into two distinct types of Ca2+ influx events, based on their propagation distance. Considering the size of mitochondria (~ 1 μm in length), and movement in the x–y axis due to mitochondrial and optical jitter, we defined non-propagating events as those with a traveling distance of 0.99 μm or below, while propagating events were those with a traveling distance of 1 μm or higher. Figure 3, and Supplementary Movies 1–4, show representative examples of these types of events. Using these parameters to discriminate non-propagating from propagating events, we examined whether the introduction of Mito-PstI would cause changes in mitochondrial Ca2+ influx kinetics dependent on this segregation. Overall, we found that non-propagating events vastly outnumbered propagating events by about 10:1 in frequency, regardless of the brain region examined (Fig. 4). At baseline, we observed significant differences in kinetic parameters for non-propagating and propagating events between the HPC and DLS. Specifically for non-propagating events, the DLS displayed a significant ~ 20% greater amplitude in mitochondrial calcium events, while duration of events in the DLS were ~ threefold greater than the HPC (Table 2). We also observed significant baseline changes between the HPC and DLS for propagating events. Specifically, the DLS showed ~ fourfold greater duration of events, along with ~ 40% reduction in propagation distance (Table 2). As for PstI effects, non-propagating events in the DLS showed a significant increase in frequency (Mito-GCaMP: 147.70 ± 36.481, Mito-GCaMP + PstI: 336.27 ± 278.53, p = 0.012, t-test), while amplitude (Mito-GCaMP: 1.23 ± 0.044, Mito-GCaMP + PstI: 1.11 ± 0.036, p = 0.036) and duration (Mito-GCaMP: 2.88 ± 0.104, Mito-GCaMP + PstI: 2.608 ± 0.083, p = 0.048, t-test) were reduced. By contrast, in the HPC (Fig. 4A), non-propagating event frequency and duration were unaffected by PstI, while amplitude was significantly increased (Mito-GCaMP: 0.997 ± 0.070, Mito-GCaMP + PstI: 1.188 ± 0.062, p = 0.037, Mann–Whitney test).
Representative images of Ca2+ influx events separated by type. (A) Confocal (left) and AQuA (right) representation of Non-Propagating astrocytic mitochondrial Ca2+ influx in live HPC (left) and DLS (right) brain slices. (B) Confocal (left) and AQuA (right) representation of Propagating astrocytic mitochondrial Ca2+ influx with a 5 s interval in the HPC (top) and DLS (bottom) live brain slices. White arrows depict initiation of events, while blue arrows indicate ending-point of propagating events. Propagating events are color-coded red, whereas Non-Propagating events are color-coded green by AQuA. Scale bars = 20 μm.
Mito-PstI causes a striking divergence in the kinetics of spontaneous mitochondrial Ca2+ influx events between astrocytes in the HPC versus DLS. Graphs depicting differences in frequency, amplitude, duration and propagation distance between Mito-GCaMP and Mito-GcaMP + PstI conditions for the two types of Ca2+ events: Non-propagating (A) and Propagating (B). Ca2+ influx from live HPC and DLS brain slices was analyzed utilizing the AQuA software plug-in for ImageJ. For image analyses, each data point is a single astrocyte. Data expressed as mean ± SEM, p-values are based on two-sample t-test or Mann–Whitney test. For HPC, n = 3 mice, 9 cells (Mito-GCaMP) and n = 10 mice, 18 cells (Mito-GCaMP + PstI). For DLS, n = 7 mice, 17 cells (Mito-GCaMP) and n = 8 mice, 30 cells (Mito-GCaMP + PstI).
For propagating events, we found that propagation distance of mitochondrial Ca2+ influx events significantly decreased only in the DLS (Mito-GCaMP: 12.094 ± 1.427 µm, Mito-GCaMP + PstI: 8.983 ± 1.284 µm, p = 0.044, Mann Whitney test), but remained unchanged in the HPC. Although propagating event frequency and duration remained unchanged in both brain regions after PstI treatment (Fig. 4B), the amplitude of propagating events showed a small but significant increase only in the HPC of PstI-treated cells (Mito-GCaMP: 0.770 ± 0.054, Mito-GCaMP + PstI: 1.06 ± 0.055, p = 0.002, t-test). Thus, following mtDNA damage induced by mitochondrial PstI expression, the kinetics of spontaneous mitochondrial Ca2+ influx events in astrocytes showed striking brain region-specific differences between the HPC and DLS. Importantly, significant differences were observed for non-propagating events in the DLS but not in the HPC.
Mito-PstI causes astrocytes to become reactive in the HPC and DLS
Astrocytes adopt a reactive state, known as astrogliosis, in response to infection, injury or chronic disease, which involve variable morphological, functional, and molecular changes21. Since astrocytic mtDNA depletion has been shown to trigger astrogliosis15, and glial fibrillary acidic protein (GFAP) gene and protein induction are classical markers of astrogliosis22, GFAP content was assessed via immunostaining in both the HPC and DLS (Fig. 5A–D). In the HPC, both GFAP staining area (Fig. 5E, Mito-GFP: 319 ± 28 µm2, Mito-GFP + PstI: 523 ± 72 µm2, p = 0.0137, Mann Whitney test) and intensity (Fig. 5F, Mito-GFP: 295 ± 11, Mito-GFP + PstI: 384 ± 29, p = 0.00747, t-test ) increased in the Mito-GFP + Mito-PstI mice. The staining area also increased in the PstI-expressing DLS samples (Fig. 5E, Mito-GFP: 607 ± 43 µm2, Mito-GFP + PstI: 770 ± 46 µm2, p = 0.0122, t-test), however, the intensity decreased (Fig. 5F, Mito-GFP: 330 ± 86, Mito-GFP + PstI: 265 ± 82, p = 0.00832, t-test). Thus, overall these data suggest that astrocytes in both HPC and DLS become reactive in response to Mito-PstI expression.
The HPC and DLS display heterogenous differences in astrocytic and microglial reactivity to Mito-PstI. Representative confocal images of GFAP staining in HPC (A,B) and DLS (C,D) astrocytes expressing either Mito-GFP (A,C) or Mito-GFP + PstI (B,D). ImageJ quantification of GFAP area (E) and intensity (F) per field of view (FOV). Scale bars = 10 μm. Representative confocal images of Iba1 staining in HPC (G,H) and DLS (I,J) sections expressing either Mito-GFP (G,I) or Mito-GFP + PstI (H,J). ImageJ quantification of Iba1 area (K) and intensity (L). Each data point is a single FOV taken from 1 to 4 brain slices from 3 to 5 mice, male and female, per condition per brain region (HPC and DLS). Data expressed as mean ± SEM, p-values are based on a two-sample t-test or Mann Whitney, based on normality. For GFAP, n = 29 FOV per condition in the HPC, 25 FOV per condition in the DLS. For Iba1, n = 18 FOV per condition in the HPC, 27 FOV per condition in the DLS.
Mito-PstI elicits differential microglial activation in the HPC and DLS
Microglia, the resident immune cells of the CNS, respond to perturbations of their surroundings, such as injury or neurodegenerative disease, by increasing their numbers and undergoing a series of morphological changes. This response, known as microgliosis, is very dynamic and heterogeneous, depending on multiple variables such as: brain region, age, sex, and specific pathological agent23. Chronic loss of astrocytic mtDNA triggers astrogliosis, and secondarily, microgliosis15. Therefore, we asked whether our astrocyte-targeted Mito-PstI tool would elicit microglia reactivity. We performed immunostaining for the microglial marker ionized calcium-binding adapter molecule 1 (Iba1) in both HPC and DLS slices, which received either a Mito-GFP or a Mito-GFP + Mito-PstI AAV injection (Fig. 5G–J). Quantification showed that Iba1 staining area significantly increased only in the PstI-treated HPC (Fig. 5K, Mito-GFP: 3370 ± 391 µm2, Mito-GFP + PstI: 7952 ± 918 µm2, p = 1.06E-4, Mann Whitney test), but not in the PstI-treated DLS, while Iba1 intensity (Fig. 5L) remained unchanged following PstI treatment in both brain regions. This may indicate increase recruitment of microglia to the HPC due to Mito-PstI expression in astrocytes, without alterations of Iba1 expression per cell. Thus, together, these data suggest that targeting PstI to astrocytic mitochondria triggers microgliosis in the HPC, but perhaps not in the DLS.
Discussion
The data show that PstI targeted to astrocytic mitochondria induces mtDNA loss and recombination in vivo. This has brain-region specific effects on astrocytic mitochondrial network dynamics and spontaneous Ca2+ influx events, with accompanying astrocytic activation and microglia reactivity, also brain-region dependent.
AAV-Mito-GFP, AAV-Mito-GCaMP6f and AAV-Mito-PstI all used the same genome/capsid pairing (AAV2/5), GfaABC1D promoter, and mitochondrial signal sequence to deliver their respective cargoes to astrocytic mitochondria (Fig. 1A). When used alone, AAV-Mito-GFP clearly labelled astrocytic mitochondria (Figs. 1B, 2A,C) without targeting mitochondria in any other cell, so was demonstrated previously for AAV-Mito-GCaMP6. Thus, there is little doubt that AAV-Mito-PstI selectively delivered this restriction enzyme to just astrocytic mitochondria either when used alone (Fig. 1), or in combination either with AAV-Mito-GFP (Fig. 2) or AAV-Mito-GCaMP (Figs. 3 and 4). The degree and nature of mtDNA depletion and deletion yielded by the AAV tool used here to deliver PstI to astrocytes, are remarkably similar to those reported by Moraes and colleagues, who delivered PstI to neurons12,13 and myofibers14, using tissue-specific transgenic approaches. This suggests that the mechanisms that lead to PstI-induced mtDNA loss and recombination, which likely involve DNA double-strand breaks, exonuclease activity and homologous and non-homologous DNA repair systems24, are conserved across these cell types. A different approach to generate mtDNA loss in astrocytes was the conditional deletion of Twinkle, a nuclear encoded gene necessary for mitochondrial genome replication15. In this case, mtDNA depletion was achieved by defective replication. Interestingly, this progressive astrocytic mtDNA attrition led to astrogliosis, secondary microgliosis and neuronal loss. AAV-Mito-PstI treatment targeted to astrocytes also led to astrocytic activation and microgliosis, particularly in the HPC. As hippocampal neuronal loss is a key component of Alzheimer’s disease25, but striatal neuronal loss is absent in classical Parkinson’s disease26,27, we tested for apoptosis in the granule cell layer of the HPC by staining for cleaved-caspase3 (CC3). Our data suggest no neuronal apoptosis in this region after 3wk of PstI treatment of astrocytic mtDNA. (Supplementary Material Fig. 2). We believe that future studies with longer postinfection periods (e.g., 3–6 months) may yield different results regarding neuronal loss or other neuronal-specific effects (e.g., alterations in synaptic markers).
An advantage of the AAV-Mito-PstI tool over the conditional gene targeting approach used by Ignatenko and colleagues, is that it allows for brain-region-specific targeting of astrocytic mtDNA, which we have exploited here to show differential effects on mitochondrial network morphology and spontaneous Ca2+ influx in the DLS and HPC. Differences in the level and / or efficiency of AAV-mito-PstI infection or differences in the number of target astrocytic mitochondria between the two brain regions are two possible mechanisms that could account for heterogenous responses of astrocytes and microglia in the HPC and DLS following astrocytic mtDNA damage induced by Mito-PstI. Variations in infection efficiency of AAV-Mito-PstI between the HPC and DLS are unlikely because each of the two brain regions were injected with identical titers and volumes of AAVs (see “Materials and methods”). In addition, as shown in Fig. 1B, the extent of expression of Mito-GFP in the HPC and DLS is similar, suggesting that astrocytes in each of these brain regions are equally permissive to AAV infection and astrocyte-specific GfaABC1D promoter driven transgene expression from the AAV5 vector backbone. A second explanation is that the amount of target astrocytic mitochondria differs between the granule cell layer in the HPC and the DLS. In this regard, our quantitative data show that the HPC granule cell layer astrocytes possess significantly fewer and smaller astrocytic mitochondria than the DLS (Table 1). As further discussed in the paragraphs below, this observed quantitative difference in the baseline amount of target astrocytic mtDNA between the two brain regions could in part account for the observed heterogenous brain region-specific response to astrocytic mtDNA damage. However, it should be noted that despite a relatively smaller population of target astrocytic mitochondria in the granule cell layer of the HPC dentate gyrus, we observed PstI-specific, selective activation of microglia in HPC (Fig. 5), which point to underlying mechanisms different than just viral infection efficiency and/or baseline levels of mitochondrial targets fully accounting for the distinct responses of DLS and HPC to Mito-PstI treatment.Simple quantification of mitochondrial particle number (Fig. 2E) and size (Fig. 2F), which respectively showed an elevation and a diminution in Mito-GFP + PstI samples relative to Mito-GFP control, strongly suggested PstI-driven augmentation of mitochondrial fragmentation, consistent with elevated mitochondrial fission (or reduced fusion) in DLS, but not in HPC. This was confirmed by the more sophisticated analysis with Mitograph, which yielded a significant decrease in the DLS connectivity score, in particular (Fig. 2P), which has been experimentally linked to increased mitochondria fission18. In all cells28, in general, and astrocytes10, in particular, mitochondria fission, fusion and mitophagy form a tripartite quality control system that ensures eventual removal of damaged, non-functional mitochondria from the mitochondrial network of a cell. These processes are triggered by many cellular stresses including those that damage mtDNA like oxidative stress, and in our case, double-strand DNA breaks24,28. It is intriguing that astrocytes in DLS and HPC differ so significantly regarding this homeostatic response. In this context, consistent with our previous results in the HPC CA1 region6, control mitochondria in the astrocytes of the HPC dentate gyrus appeared smaller and less abundant than in the DLS (Fig. 2E,F; Table 1). It is tempting to suggest that these baseline morphological properties may account, in part, for the relative resilience of the HPC mitochondrial network to fission by PstI treatment. Interestingly, the connectivity index of the DLS astrocytic mitochondrial network was brought down to about the same value as that for the HPC after PstI treatment (Fig. 2P).
Our prior study comparing DLS and CA1 HPC astrocytes found interregional baseline differences in spontaneous Ca2+ influx events6. Even though, AQuA was not used for the analysis of the data in that report, it is likely that given the 10:1 predominance of Non-Propagating events reported here, most of the events analyzed in that previous study were Non-Propagating as well. Event frequency in CA1 HPC astrocytic mitochondria was about half of that in the DLS, which matches really well with what we found in the dentate gyrus here (Fig. 4A). The analysis here also revealed important baseline differences particularly in event duration, ~ 4 times as large in the DLS relative to the HPC, and in propagation distance, ~ 2 times shorter in the DLS (Fig. 4A,B; Table 2).
Lower mitochondrial abundance in the HPC relative to DLS (Fig. 2E; Table 1) might account for decreased baseline frequency of Non-Propagating Ca2+ events in the former relative to the latter (Fig. 4A). Likewise, longer event duration in the DLS might be related to increased baseline connectivity in its mitochondrial network. Increased mitochondria fission in PstI-treated DLS may also account for the decrease in Non-Propagating event duration and the increase in Non-Propagating event frequency (Fig. 4A), and perhaps, for the reduction of propagation distance in Propagating events (Fig. 4B). It is difficult to relate the increase in event amplitude in HPC, and the decrease in Non-Propagating event amplitude in the DLS, to network connectivity status. Ca2+ influx in astrocytic mitochondria strongly depends on ER Ca2+ stores and not extracellular Ca2+6. It is likely that Ca2+ movement from ER to mitochondria in astrocytes involves, like in other cell types, mitochondria-associated ER membranes (MAMs)29. MAMs are present in astrocytes, where they have been shown structurally and functionally influenced by manipulation of mitochondrial dynamics30. Hence, it is reasonable to suggest that the disruption of the mitochondrial network induced by PstI, particularly in the DLS (Fig. 2), is associated with alteration of MAMs, which in turn may underlie, in part, some of the more profound changes observed in spontaneous Ca2+ events in that brain region (Fig. 4).
The induction of astrocyte reactivity by mtDNA loss using PstI (Fig. 5A–F) is not surprising given that conditional deletion of Twinkle in astrocytes, which also generate mtDNA depletion, did so as well15. Somewhat unexpected is that our data only support secondary microgliosis in the HPC and not the DLS (Fig. 5G–L). In addition to highlighting another brain region-specific difference for astrocytic mtDNA loss, this result raises the possibility of uncharacterized distinctions in reactivity to mtDNA damage between HPC and DLS astrocytes, which in turn may lead to different downstream responses by surrounding microglia. Diversity of astrocyte and microglia reactivity as a response to multiple kinds of insults, brain region, age, sex and other factors, is a well-established concept in the field21,23.
The immediate fate of astrocytic mtDNA following PstI digestion is unclear. If some leaks into the cytosol, it could trigger an inflammatory immune response via interacting with the GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) pathway31. Upon binding cytosolic DNA, cGAS catalyzes the production of 2′3′ cyclic GMP–AMP (cGAMP), which acts as a second messenger by binding STING and mediating the transcription of inflammatory genes such as interferon stimulatory genes (ISGs) and type I interferon32. Additionally, if oxidized, cytosolic mtDNA could interact with the NLRP3 inflammasome leading to the activation of proinflammatory cytokines Interleukin-1β and Interleukin-18. Moreover, Interleukin-1β and other proinflammatory cytokines such as Tumor Necrosis Factor-α and Interleukin-6 could be induced downstream of the binding of mtDNA by Toll-like Receptor 9 in endosomes32. It will be interesting to investigate in the future whether these DNA sensing mechanisms could be involved in the activation of astrocytes following PstI treatment and whether their involvement is brain-region specific as well.
In summary, the data demonstrate the utility of the novel AAV-Mito-PstI in inducing astrocytic mtDNA damage in a brain-region-specific manner, which produces stark interregional differences in mitochondrial network dynamics and spontaneous Ca2+ fluxes in astrocytes from DLS and HPC, two brain areas relevant to major neurodegenerative diseases, Parkinson’s and Alzheimer’s diseases. It remains to be investigated what other mitochondrial functions, i.e., ATP, ROS, lactate production, glutamate uptake, blood–brain barrier regulation, are affected by PstI-treatment and how their alteration could be mechanistically connected to the observed changes in mitochondrial dynamics, spontaneous Ca2+ fluxes, and astrocytic and microglial reactivity reported here, and to potential neuronal effects germane to Parkinson’s and Alzheimer’s diseases.
Materials and methods
Mice
Male and female C57BL/6 WT mice were obtained at 5–7 weeks old from Jackson Laboratories. Mice were housed on a 12 h light/dark cycle with ad libitum access to food and water. All animal experiments were approved by the Texas A&M University Institutional Animal Care and Use Committee (IACUC Animal Use Protocol #2022-0252). All experiments in this study are reported in accordance with the ARRIVE guidelines.
Adeno-associated viral vectors (AAVs)
AAV 2/5 GfaABC1D-mito7-eGFP and AAV 2/5 GfaABC1D-mito7-GCaMP6f constructs were generated by Vector Builder (Chicago, IL) as described previously6. A third AAV, AAV 2/5 GfaABC1D-mito7- PstI, was created in a similar manner, fusing the PstI coding sequence, optimized for mammalian codon use, to an N-terminal 87 bp Mito7 signal sequence from the mitochondrial COX8A subunit, driven by the astrocyte specific GfaABC1D promoter (Fig. 1A). We have verified genomes for all AAVs by sequencing. These vectors were designed to express each target protein only within astrocytic mitochondria.
Stereotaxic surgery
Following prior protocols6, stereotaxic surgeries were performed under continuous isoflurane (induction at 5%, maintenance at 1–2% vol/vol), administered via a gas syringe injection system (Kent Scientific). Glass injection pipettes (Fisher Scientific) were pulled using a Sutter P-2000 laser puller and tips were beveled using a Narishige EG-45 grinder.
For immunohistochemistry, 2 × 1010 genome copies (gc) of AAV were stereotaxically injected into the HPC or DLS. Control mice were injected with 2 × 1010 gc of AAV2/5 GfaABC1D-mito7-eGFP. Test animals were injected with 1 × 1010 gc of AAV2/5 GfaABC1D-mito7-eGFP and 1 × 1010 gc of AAV2/5 GfaABC1D-mito7-PstI.
For Ca2+ imaging, control animals were injected with 2 × 109 gc of AAV2/5 GfaABC1D-mito7-GCaMP6f. Test animals were injected with 2 × 109 gc of AAV2/5 GfaABC1D-mito7-GCaMP6f and 1 × 1010 gc of AAV2/5 GfaABC1D-mito7-PstI.
All injections were done using the Stoelting Quintessential Stereotaxic Injector (QSI). Mice were sacrificed 2–4 weeks post-injection for either perfusions or live-slice imaging. HPC coordinates were -2.0 mm posterior to bregma, + 1.5 mm lateral to bregma, and − 1.6 mm ventral to pial surface. DLS coordinates were + 0.9 mm anterior to bregma, + 1.8 mm lateral to bregma, and + 2.5 mm ventral to the pial surface.
mtDNA content analysis
DLS from two groups of three mice, which either had a stereotaxic injection of mito-PstI or were uninjected controls, were microdissected and pooled. DNA was extracted using the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN; Germantown, MD; CAT# 69504). For quantitative PCR, Perfecta SYBR Green FastMix with ROX (QuantaBio; Beverly, MA; CAT# 95073) was used with Forward and Reverse Primers (Supplementary Material Table 1) for amplifying both Cytochrome b, a mtDNA gene, and Erk2, a nuclear DNA gene. The cycling parameters for amplification were as follows: 95 °C for 7 min, [95 °C for 10 s, 60 °C for 30 s] 50 cycles, 95 °C 15 s, 60 °C 1 min, 95 °C 15 s. Mitochondrial DNA content was calculated as described in17. Nested PCR was performed using SYBR Green FastMix (10 μL), Primers (Supplementary Material Table 1) were designed to sit outside the PstI cut sites (Fig. 1C). Cycles were 95 °C for 2 min, [94 °C for 45 s, 50 °C for 45 s, 60 °C for 1 min] 35 cycles, 60 °C for 7 min, 10 °C hold. PCR product was run on a 1.5% agarose gel at 110 V for 40 min. Bands were cut from the gel and sent to EtonBio (San Diego, CA) for sequencing.
Immunostaining
Three-weeks after AAV injections, mice were anesthetized with isoflurane, transcardially perfused with 1X PBS, followed by 10% Formalin, and brains were extracted. Brains were post-fixed in 10% formalin at 4 °C overnight before being moved to 30% sucrose solution for cryoprotection. Brains were set using Tissue Tek O.C.T. Compound (Sakura Finetek) and 40 μm sections were obtained using a cryostat (Epredia, CryoStar NX50).
For NeuN staining (Fig. 1B), following a 1X PBS wash, HPC and DLS sections were blocked and permeabilized in 10% normal goat serum (NGS, Abcam, ab7481) and 0.5% Triton X-100 (Sigma, cas# 9002-93-1) rocking gently for 45 min at room temperature. Sections were washed twice with PBS before incubation overnight at 4 °C in primary mouse IgG2b anti-NeuN (1:500, Abcam cat# ab104224). Sections were washed twice the following day with 1X PBS followed by incubation in goat anti-IgG2b AlexaFlour 647 (1:1000, life technologies, ref# A21235). Slices were mounted using mounting media with DAPI (Fluoroshield, Abcam, ab104139).
The same protocol was used for GFAP and Iba1 staining (Fig. 5). Chicken anti-GFAP (1:2000, Abcam cat# ab4674) primary in 1% NGS+ 0.05% Triton X-100 was used. For HPC slices, the secondary incubation was done in donkey anti-chicken AlexaFluor 647 (1:2000, JacksonImmuno cat#706-605-155) for one hour at room temperature. For DLS slices, goat anti-chicken AlexaFluor 594 (1:2000, Abcam cat# ab150176) was used.
For Iba1 staining in the HPC, rabbit anti-Iba1 (1:750, Wako cat#019-19741) was used as a primary antibody, and goat anti-rabbit AlexaFluor 594 (1:1500, Abcam cat#ab150080) was used for secondary. For DLS, rabbit anti-Iba1 (1:1000, Wako cat#019-19741) was used as a primary antibody, and goat anti-rabbit AlexaFluor 594 (1:2000, Abcam cat#ab150080) for secondary.
For CC3 staining in the HPC (Supplementary Material Fig. 2), rabbit anti-CC3 (1:1000, Abcam cat#ab208003) was used as a primary antibody and goat anti-rabbit AlexaFluor 594 (1:1500, Abcam cat#ab150080) was used for secondary. Fluoroshield Mounting Medium with DAPI was used to stain cell nuclei.
Imaging
Imaging of fixed tissue
Fixed tissue sections of NeuN-stained HPC or DLS were imaged using an Olympus VS120 Virtual Slide Scanning System (Fig. 1B) equipped with a UPlanSApo 20X air objective.
For Figs. 2 and 5, fixed tissue sections of HPC and DLS were imaged using an Olympus FV3000 laser-scanning confocal microscope. 488, 561 and 647 nm LED-based excitation wavelengths were used for obtaining images. Images for MitoGraph analysis (Fig. 2) were obtained using a 60× oil immersion objective (N.A. 0.8), and 3× digital zoom. Z-stacks for these images consisted of 51 optical sections with a step size of 0.2 µm (Fig. 2). For HPC and DLS, GFAP and IBA1 images (Fig. 5) were obtained using a 60× oil immersion objective (N.A. 0.8) with 1× digital zoom. These images consisted of ~ 25 optical sections with a step size of 0.5 µm. To enable quantification, confocal parameters such as excitation intensity, HV, gain and offset were maintained constant for each brain region.
Tissue preparation for live brain slicing
2 to 3-weeks following AAV injection, mice were decapitated and brains were extracted. 300 μm-thick coronal slices for HPC and DLS were obtained using a vibratome (Ted Pella Inc., D.S.K Microslicer ZERO 1 N). The slicing solution consisted of (in mM): 194 sucrose, 30 NaCl, 4.5 KCl, 1.2 NaH2PO4, 26 NaHCO3, 10 d-glucose, and 1 MgCl2 and saturated with 95% O2 and 5% CO2 (pH 7.2). All recordings of Ca2+ signals were performed in artificial cerebrospinal fluid (ACSF) made of (in mM): 126 NaCl, 2.5 KCl, 1.24 NaH2PO4, 26 NaHCO3, 10 d-glucose, 2.4 CaCl2, and 1.3 MgCl2 saturated with 95% O2 and 5% CO2 (pH 7.4).
Live imaging of acute mouse brain slices
An Olympus FV3000 upright laser-scanning confocal microscope equipped with a 40× water immersion objective (N.A. 0.45) was used for live slice imaging (Fig. 3). Confocal parameters (LED power, voltage, gain, offset, and aperture diameter) were maintained constant across all HPC or DLS GCaMP6f imaging sessions. All mitochondrial Ca2+ influx events were recorded at 1 frame per second for 5 min. During all live slice imaging sessions, temperature of the recording buffer and bath was maintained at 37 °C using an in-line and bead bath heaters.
Image analysis
Image analysis for Figs. 2E–G, 5 and Supplementary Material Fig. 2 were performed using ImageJ (ver. 1.54). For Fig. 2E–G, a z projection of images was created using the max intensity function. The polygon tool was used to manually create a region of interest (ROI) around a single astrocyte. The area outside the cell was cleared and manually thresholded to encompass all GFP expressing mitochondrial particles, then smoothed and ROIs for mitochondrial particles were obtained from these images using the analyze particles function in ImageJ. ROIs obtained in this way were utilized for deriving particle count, size and area. For Fig. 5, ROIs were similarly obtained as described above, except that the images used were entire fields of view consisting of multiple cells. ROIs in Fig. 5 were utilized to obtain areas as well as intensity measures for GFAP and Iba1 staining. For Supplementary Material Fig. 2C, the BIOP JACoP ImageJ plugin was used. DAPI and CC3 z-stacks were manually thresholded and the thresholding was applied to all planes of the z-stacks for each of the two channels. The BIOP JACoP plugin was then utilized to generate Mander’s Coefficient values of fraction of CC3 overlap with DAPI. For Supplementary Material Fig. 2F, a maximum intensity projection of the z-stack for the CC3 channel was generated, and manually thresholded to generate ROIs of CC3 fluorescence across entire fields of view. ROIs generated in this way were used to acquire integrated intensity values of CC3.
MitoGraph analysis
Single Mito-GFP expressing astrocytes within the HPC and DLS were analyzed using the MitoGraph package in ImageJ18. Maximum projections of single astrocytes were first obtained, and the astrocyte territory was manually delineated using the polygon tool. ROIs encompassing single astrocytes, obtained in this way were then applied to each of the 51 optical sections comprising the z-stack. Mito-GFP expressing mitochondria outside the ROI for a single astrocyte were then deleted in each of the 51 optical sections. For each astrocyte, z-stacks processed as described above were analyzed using the MitoGraph package run on the Texas A&M supercomputing cluster (Terra portal). MitoGraph outputs were analyzed using R statistical package. Mitochondrial network recreation files (skeleton, surface, and nodes) were visualized using the ParaView program, as shown in Fig. 2.
AQuA Ca2+ imaging in acute mouse brain slices
The Astrocyte Quantitative Analysis (AQuA)20 software plug-in for ImageJ was used to obtain and analyze Ca2+ events from Mito-GCaMP6f astrocytes. Ca2+ signals were analyzed using the AQuA data type presets (Supplementary Material Table 2). ROIs were drawn around the entire cell territory and around the cell soma to obtain Ca2+ events from single astrocytes. “Draw anterior” was selected to insert an arrow facing toward the top of the field of view, to indicate true north for analysis. For input in the Detection Pipeline box, we used the parameters shown in Supplementary Material Table 3. In the Export window, Events and features and Movies with overlay were selected before selecting Export/Save. For AquA analysis, we defined Non-Propagating events as those with a traveling distance of 0.99 μm or below, while calcium events with a propagation distance of greater than 1 μm were considered as Propagating events.
Sampling and statistics
Statistical analyses were performed in Origin Lab, using the Shaprio-Wilk test to test data for normality. Normally distributed datasets were compared using two-sample t-tests. Non-normally distributed datasets were tested using Mann-Whittney for unpaired data. A p-value less than 0.5 was used to determine statistically significant. Unless otherwise indicated, data were expressed as mean ± SEM. Figure legends contain sample sizes and statistical tests used for each experiment.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Stogsdill, J. A., Harwell, C. C. & Goldman, S. A. Astrocytes as master modulators of neural networks: Synaptic functions and disease-associated dysfunction of astrocytes. Ann. N. Y. Acad. Sci. 1525, 41–60. https://doi.org/10.1111/nyas.15004 (2023).
John Lin, C. C. et al. Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396–405. https://doi.org/10.1038/nn.4493 (2017).
Batiuk, M. Y. et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11, 1220. https://doi.org/10.1038/s41467-019-14198-8 (2020).
Khakh, B. S. & Deneen, B. The emerging nature of astrocyte diversity. Annu. Rev. Neurosci. 42, 187–207. https://doi.org/10.1146/annurev-neuro-070918-050443 (2019).
Zimmer, T. S., Orr, A. L. & Orr, A. G. Astrocytes in selective vulnerability to neurodegenerative disease. Trends Neurosci. 47, 289–302. https://doi.org/10.1016/j.tins.2024.02.008 (2024).
Huntington, T. E. & Srinivasan, R. Astrocytic mitochondria in adult mouse brain slices show spontaneous calcium influx events with unique properties. Cell Calcium https://doi.org/10.1101/2020.06.21.163618 (2021).
Bazargani, N. & Attwell, D. Astrocyte calcium signaling: The third wave. Nat. Neurosci. 19, 182–189. https://doi.org/10.1038/nn.4201 (2016).
Jackson, J. G. & Robinson, M. B. Regulation of mitochondrial dynamics in astrocytes: Mechanisms, consequences, and unknowns. Glia 66, 1213–1234. https://doi.org/10.1002/glia.23252 (2018).
Fecher, C. et al. Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nat. Neurosci. 22, 1731–1742. https://doi.org/10.1038/s41593-019-0479-z (2019).
Stephen, T. L., Gupta-Agarwal, S. & Kittler, J. T. Mitochondrial dynamics in astrocytes. Biochem. Soc. Trans. 42, 1302–1310. https://doi.org/10.1042/BST20140195 (2014).
Bacman, S. R., Williams, S. L., Pinto, M. & Moraes, C. T. The use of mitochondria-targeted endonucleases to manipulate mtDNA. Methods Enzymol. 547, 373–397. https://doi.org/10.1016/B978-0-12-801415-8.00018-7 (2014).
Pickrell, A. M., Pinto, M., Hida, A. & Moraes, C. T. Striatal dysfunctions associated with mitochondrial DNA damage in dopaminergic neurons in a mouse model of Parkinson’s disease. J. Neurosci. 31, 17649–17658. https://doi.org/10.1523/JNEUROSCI.4871-11.2011 (2011).
Fukui, H. & Moraes, C. T. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 18, 1028–1036. https://doi.org/10.1093/hmg/ddn437 (2009).
Srivastava, S. & Moraes, C. T. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Genet. 14, 893–902. https://doi.org/10.1093/hmg/ddi082 (2005).
Ignatenko, O. et al. Loss of mtDNA activates astrocytes and leads to spongiotic encephalopathy. Nat. Commun. 9, 70. https://doi.org/10.1038/s41467-017-01859-9 (2018).
Lee, Y., Messing, A., Su, M. & Brenner, M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia 56, 481–493. https://doi.org/10.1002/glia.20622 (2008).
Venegas, V., Wang, J., Dimmock, D. & Wong, L. J. Real-time quantitative PCR analysis of mitochondrial DNA content. Curr. Protoc. Hum. Genet. https://doi.org/10.1002/0471142905.hg1907s68 (2011).
Harwig, M. C. et al. Methods for imaging mammalian mitochondrial morphology: A prospective on MitoGraph. Anal. Biochem. 552, 81–99. https://doi.org/10.1016/j.ab.2018.02.022 (2018).
Helassa, N., Podor, B., Fine, A. & Török, K. Design and mechanistic insight into ultrafast calcium indicators for monitoring intracellular calcium dynamics. Sci. Rep. 6, 38276. https://doi.org/10.1038/srep38276 (2016).
Wang, Y. et al. Accurate quantification of astrocyte and neurotransmitter fluorescence dynamics for single-cell and population-level physiology. Nat. Neurosci. 22, 1936–1944. https://doi.org/10.1038/s41593-019-0492-2 (2019).
Escartin, C. et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 24, 312–325. https://doi.org/10.1038/s41593-020-00783-4 (2021).
Yang, Z. & Wang, K. K. Glial fibrillary acidic protein: From intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 38, 364–374. https://doi.org/10.1016/j.tins.2015.04.003 (2015).
Paolicelli, R. C. et al. Microglia states and nomenclature: A field at its crossroads. Neuron 110, 3458–3483. https://doi.org/10.1016/j.neuron.2022.10.020 (2022).
Rong, Z. et al. The mitochondrial response to DNA damage. Front Cell Dev. Biol. 9, 669379. https://doi.org/10.3389/fcell.2021.669379 (2021).
Kumari, S., Dhapola, R. & Reddy, D. H. Apoptosis in Alzheimer’s disease: Insight into the signaling pathways and therapeutic avenues. Apoptosis 28, 943–957. https://doi.org/10.1007/s10495-023-01848-y (2023).
Witzig, V. S., Komnig, D. & Falkenburger, B. H. Changes in striatal medium spiny neuron morphology resulting from dopamine depletion are reversible. Cells https://doi.org/10.3390/cells9112441 (2020).
Deutch, A. Y., Colbran, R. J. & Winder, D. J. Striatal plasticity and medium spiny neuron dendritic remodeling in parkinsonism. Parkinsonism Relat. Disord. 13(Suppl 3), S251-258. https://doi.org/10.1016/s1353-8020(08)70012-9 (2007).
Youle, R. J. & van der Bliek, A. M. Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. https://doi.org/10.1126/science.1219855 (2012).
Lee, S. & Min, K. T. The interface between ER and mitochondria: Molecular compositions and functions. Mol. Cells 41, 1000–1007. https://doi.org/10.14348/molcells.2018.0438 (2018).
Gӧbel, J. et al. Mitochondria-endoplasmic reticulum contacts in reactive astrocytes promote vascular remodeling. Cell Metab. 31, 791-808.e798. https://doi.org/10.1016/j.cmet.2020.03.005 (2020).
West, A. P. et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557. https://doi.org/10.1038/nature14156 (2015).
De Gaetano, A. et al. Molecular mechanisms of mtDNA-mediated inflammation. Cells https://doi.org/10.3390/cells10112898 (2021).
Acknowledgements
We thank Grace Hall for assistance with Fig. 1B. The authors acknowledge the assistance of the Integrated Microscopy and Imaging Laboratory at the Texas A&M College of Medicine. RRID:SCR_021637. This work was funded by a research grant from the National Institutes of Health (NIH)/NINDS, R21 NS121959 to MR and RS. AM and DA were partially funded by NIH T32 GM135115 and NIH T32 GM135748 grants respectively.
Author information
Authors and Affiliations
Contributions
DAA and AM performed all experiments, analyzed all the data and created the figures. BLS performed experiments and analyzed data. MP, ET, CM, GB, AB and AT analyzed data. MR and RS conceived the project, trained and supervised DAA, AM, BLS, MP, ET, CM, GB, AB and AT, coordinated and designed experiments, and provided resources and funding. MR and RS wrote the manuscript along with DAA and AM, and all authors contributed to editing the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ayala, D.A., Matarazzo, A., Seaberg, B.L. et al. Heterogeneous brain region-specific responses to astrocytic mitochondrial DNA damage in mice. Sci Rep 14, 18586 (2024). https://doi.org/10.1038/s41598-024-69499-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-69499-w