Abstract
A significant consequence of climate change is the rising incidence of wildfires. When wildfires occur close to wine grape (Vitis vinifera) production areas, smoke-derived volatile phenolic compounds can be taken up by the grape berries, negatively affecting the flavor and aroma profile of the resulting wine and compromising the production value of entire vineyards. Evidence for the permeation of smoke-associated compounds into grape berries has been provided through metabolomics; however, the basis for grapevines’ response to smoke at the gene expression level has not been investigated in detail. To address this knowledge gap, we employed time-course RNA sequencing to observe gene expression-level changes in grape berries in response to smoke exposure. Significant increases in gene expression (and enrichment of gene ontologies) associated with detoxification of reactive compounds, maintenance of redox homeostasis, and cell wall fortification were observed in response to smoke. These findings suggest that the accumulation of volatile phenols from smoke exposure activates mechanisms that render smoke-derived compounds less reactive while simultaneously fortifying intracellular defense mechanisms. The results of this work lend a better understanding of the molecular basis for grapevines’ response to smoke and provide insight into the origins of smoke-taint-associated flavor and aroma attributes in wine produced from smoke-exposed grapes.
Similar content being viewed by others
Introduction
Climate change creates numerous challenges for the worldwide production of crops, including grapes. In addition to the emergence of new pests, diseases, and abiotic-stress-induced physiological disorders brought about by extreme weather1, the combined effects of heat and drought create conditions conducive to wildfires2. One of the world’s major wine grape-producing areas, the Western United States, has been heavily impacted by fires3. The hot dry conditions in this region often overlap with the grape maturation and harvest season, during which compounds contained in wildfire smoke infiltrate the plants and the fruit. These compounds are primarily volatile phenols (VPs), which arise from the combustion of lignin-rich wood and plant material, and thiophenols, which appear to arise during the grapes’ metabolism of phenolic compounds4,5. Together, these compounds are associated with undesirable “smoky” or “ashy” profiles that diminish the quality and value of the wine produced from smoke-exposed berries4,6, leading to substantial annual losses for vineyard and winery owners7.
Efforts to mitigate smoke taint include a variety approaches, from preventive measures in the vineyard to various amelioration techniques applied at the winery and during post-production8,9,10,11. Several metabolic studies have provided a better understanding of the chemical basis for smoke taint in wine4,12,13,14. These studies have primarily focused on the detection of smoke-taint-associated compounds at various stages of wine grape development and processing (e.g., during grape berry development, postharvest, and following fermentation in the winemaking process), as well as the establishment of strategies for detecting VPs and other smoke-taint associated metabolites. They have also provided insight into the variability of smoke taint susceptibility across grape developmental stages and varieties15,16,17,18. Based on the findings of these studies, VPs are primarily present in a conjugated state post-smoke exposure (e.g., via enzyme-mediated addition of sugar, glutathione, or other groups)12,19,20,21. Gene expression analysis-guided biochemical studies have facilitated the identification of several UDP-glycosyltransferases involved in the conjugation of specific smoke-derived volatiles21, and there are likely numerous other contributors to the detoxification of VPs and other smoke-associated compounds, as conjugation of phytotoxic compounds into less reactive forms is a major plant defense strategy used to mediate oxidative stress and damage. Once in a conjugated form, VPs may be transported to specialized compartments within the cell, such as cell walls or vacuoles, for storage22. In grapes, it has been demonstrated that, while conjugation renders VPs temporarily inert, enzymatic cleavage during fermentation releases the bound VPs, making them detectable in the final wine product and contributing to smoke taint12,14,22,23. In addition to VPs, additional metabolic work indicates that thiophenols and other sulfur-containing compounds, potentially derived from the VPs absorbed during smoke exposure, significantly contribute to the ashy flavors associated with smoke taint4. A major outcome of this body of research, in addition to identifying the basis for smoke taint manifestation in smoke-exposed wine grapes, is the identification of specific markers or signatures that can be used to enable the early detection of smoke taint in grapes, allowing for timely intervention before the wine production process begins.
Despite advances in understanding the chemical basis of smoke taint, a comprehensive understanding of grapevine responses to smoke at the gene expression level is still lacking. Identifying genes involved in redox stress responses, detoxification processes, and other defense mechanisms is expected to provide a novel perspective on grapevine’s concerted response to smoke at the berry level. To address this gap, we examined the transcriptomic response of Merlot wine grapes subjected to smoke treatments to identify genes and pathways involved in defense against smoke-induced stress. This study represents a novel analysis of the gene-expression-level responses of grapevine to smoke exposure, complementing existing metabolic knowledge and laying the groundwork for developing new smoke taint mitigation strategies.
Results
Berry metrics
The onset of veraison in Merlot grapes at the Roza vineyard was August 4th, 2021 with 50% veraison occurring 13 days later, on August 17th, 2021. During the pre-veraison stage, the average berry weight was 0.52 ± 0.06 g, Brix was 6.35 ± 0.32, and pH was 2.76 ± 0.02. In the post-veraison stage, the average berry weight increased to 0.99 ± 0.05 g, Brix to 20.20 ± 0.37, and pH to 3.22 ± 0.02. The growing degree days (GDD) base 50°F from April 1st for the Roza vineyard in 2021 was 1527 GDD, which is higher than the six-year average (2018–2023) of 1236 GDD (data from AgWeatherNet | Washington State University, accessed 07/16/2024). Photographs of pre- and post-veraison berries from these smoke exposures are shown in Supplementary File 1.
Grape transcriptome assembly and annotation
The de novo transcriptome assembly generated 212,936 contigs that passed the filtering criteria (Supplementary File 2). Functional annotation in OmicsBox resulted in the assignation of blast IDs and gene ontologies to 82,694 contigs, 61,860 of which were functionally annotated. The decision to perform read mappings to a de novo assembly, rather than use one of the available grape reference genomes, is based on our observations in previous transcriptomic studies that large numbers of reads often fail to map to a reference genome when the reference is a genotype different from that of the study samples24,25. However, to facilitate the translatability of our results to other grape studies employing a reference-based mapping approach, we have matched our list of functionally annotated contigs with the corresponding genes from the PN40024 reference genome, where matches were available (Supplementary File 3).
Identification of differentially expressed genes in smoke-exposed grapes
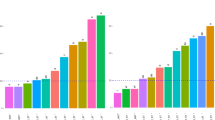
Sampling for gene expression analysis was conducted at 0, 2, 4, 6, 12, 24, and 36 h following smoke exposure. Pairwise differential expression (DE) analysis identified 1616 contigs that were differentially expressed in pre-veraison or post-veraison grapes for at least one-time point; of these, 854 were functionally annotated with Gene Ontology (GO) IDs (Supplementary File 3). At the pre-veraison stage, 1361 genes were differentially expressed; at the post-veraison stage, 547 were differentially expressed; 292 genes were DE at both developmental stages (Fig. 1A). The overall DE gene expression response for the pre-veraison grapes was characterized by a large number of downregulated DE genes 2 h following smoke exposure and a subsequent increase in the number of upregulated DE genes, peaking at 6 h post-smoke exposure (Fig. 1B). In contrast to the pre-veraison grapes, the DE gene expression of the post-veraison grapes was characterized by a peak in upregulated DE genes at 2 h of smoke exposure, with relatively few genes displaying reduced expression in the smoked vs control grapes (Fig. 1C).
Summary of global gene expression of differentially expressed (DE) genes in response to smoke. (A) Venn diagram showing the number of DE genes shared between, and unique to, pre-veraison and post-veraison stages. (B) Total number of up- and down-regulated DE genes in pre-veraison smoke-exposed grapes. (C) Total number of up- and down-regulated DE genes in pre-veraison smoke-exposed grapes.
Whole genome co-expression network analysis
Whole Genome Co-expression Network Analysis (WGCNA) revealed two distinct modules of co-expressed DE genes that were significantly associated with the response to smoke exposure in grape berries (Fig. 2A,B) (p < 0.05). The first significant module (yellow, p < 0.001) was associated with smoke exposure at the pre-veraison stage, while the second module (green, p < 0.01) was associated with smoke exposure at the post-veraison stage. Several additional co-expression modules were identified that, while not strongly associated with smoke exposure, displayed high association with either pre-veraison or the post-veraison developmental stage (i.e., gray, blue, brown) (Fig. 2A,B). The lists of DE genes pertaining to the smoke-associated modules, as well as the additional significant modules that were not strongly associated with smoke, are provided in Supplementary File 4.
WGCNA results and gene ontology (GO) enrichment of smoke-associated co-expressed DE genes. (A) Cluster dendrogram from WGCNA analysis showing color-coded DE gene co-expression modules. (B) Heatmap showing correlations between the co-expression modules and smoke exposure or developmental stage. Color intensity indicates the strength of the correlation, with red indicating a strong positive correlation and blue indicating a strong negative correlation; asterisks mark statistically significant correlations (p < 0.005). The two significant smoke-associated modules (yellow and green), were selected for GO enrichment analysis. (C) Enriched GOs pertaining to the smoke-associated co-expression modules. Color IDs correspond to color modules identified in A.
Gene Ontology (GO) enrichment analysis of the DE genes within the significant smoke-associated modules revealed biological processes, molecular functions, and cellular components that were overrepresented as a result of smoke exposure (Fig. 2C). While the yellow and green modules, corresponding to pre-veraison smoke exposure and post-veraison smoke exposure, respectively, contained different DE genes, many enriched gene ontologies were shared between pre-veraison and post-veraison smoke-exposed grapes. The included GO terms associated with the metabolism of glutathione and other sulfur-containing compounds (‘glutathione metabolic process, ‘glutathione transferase activity’, ‘sulfur compound metabolic process’); terms associated with metabolism of nitrogen-containing organic compounds (‘organonitrogen compound metabolic process’, ‘cellular modified amino acid metabolic process’, ‘amide metabolic process’, ‘FMN binding’); and terms associated with secondary metabolism (‘secondary metabolic process’, ‘phenylpropanoid biosynthetic process’). Additional shared GO terms included ‘response to chemical’, ‘catalytic activity’, ‘transferase activity’, and ‘oxidoreductase activity’. Enriched GOs that were uniquely associated with the pre-veraison stage included terms associated with glycosyltransferase activity (‘UDP-glucosyl/glycosyltransferase activity’), ‘glucosyl/glycosyltransferase activity’, ‘hexosyltransferase activity’, and ‘quercetin-3/7-O-glucosyltransferase activity’; terms associated with metabolism of toxins (‘detoxification’, ‘toxin catabolic process’, and ‘toxin metabolic process’); and terms associated with biosynthesis of lignin and cellular structural components (‘lignin biosynthetic process’, ‘lignin metabolic process’, ‘cinnamyl-alcohol dehydrogenase activity’, and ‘intracellular anatomical structure’). No GOs were unique to the post-veraison stage.
Following identification and GO enrichment of smoke-associated co-expression modules, we conducted intramodular analysis within the WGCNA pipeline to identify key drivers of the response to smoke exposure or hub genes. Hub genes are those that display high connectivity with other genes with regard to expression patterns and are, therefore, most likely to play central roles in the function of a given co-expression network. A total of 117 hub genes were identified with both high module membership and a gene significance p-value of 0.05 or less for their association with smoke exposure (Supplementary File 5). Observation of the expression patterns of the hub genes revealed notable upregulation of genes involved in four key processes: 1.) detoxification, including 27 glutathione S-transferases, 12 UDP-glycosyltransferases, six anthocyanidin glucosyltransferases, two crocetin glucosyltransferases, and 2 hydroquinone glucosyltransferases; 2.) maintenance of redox homeostasis, including 3 nucleoredoxins and 4 mannitol dehydrogenases; 3.) cell wall structural integrity, including 4 β-glucosidases, glycine-rich protein DC7.1-like, dirigent protein 19, repetitive proline-rich cell wall protein, and 2 cinnamoyl-CoA reductases (2); and 4.) vacuolar function, including 2 zinc-induced facilitators and serine carboxypeptidase-like 46. A subset of the smoke-associated hub genes pertaining to detoxification, redox homeostasis, cell wall fortification, and vacuolar function is shown in Fig. 3. All hub genes were expressed in both pre- and post-veraison grapes, particularly after the 6 h time point; however, many of the hub genes were only significantly differentially expressed at the pre-veraison stage.
Expression heatmap containing four key groups of hub genes involved in Merlot grape berries’ response to smoke over time at pre-veraison and post-veraison developmental stages. Red indicates increased fold change expression in the smoke-exposed versus the control grape berries, while blue indicates reduced fold change expression. Asterisks indicate differential expression (p < 0.05).
Discussion
The global expression profiles of the DE genes revealed distinct responses to smoke exposure in pre- and post-veraison grape berries, indicating variability in the magnitude and timing of their response to smoke. In pre-veraison grapes, the substantial downregulation of genes 2 h after smoke exposure, followed by a peak in the number of upregulated genes at 6 h (Fig. 1B), suggests an initial shock phase, followed by a subsequent phase characterized by an elevation in expression of genes associated with stress response and detoxification. In contrast, the early peak in upregulated genes at 2 h observed in post-veraison grapes, with relatively few genes displaying reduced expression (Fig. 1C), indicates a potentially more immediate and sustained transcriptional adjustment to smoke in the mature berries. This rapid initial response to smoke is not surprising, as similar responses at the transcriptome level have been observed in grapes exposed to various types of abiotic stress, including temperature and heavy metal stress26,27,28. Notably, the total number of DE genes was reduced in post-veraison grapes compared to pre-veraison grapes, indicating a less pronounced initial transcriptional response overall at this stage of development. While it is expected that distinct berry developmental stages will respond differently to smoke, it is worth noting that a large degree of variation was present between the sequencing replicates at the post-veraison developmental stage for both smoke-exposed and control berries; this could partly explain the reduced number of genes that were statistically significant in terms of their differential expression. A possible explanation for the high degree of inter-replicate variation in the post-veraison grapes is that, at veraison, the transcriptional program in different berries in a cluster proceeds at different rates29. This is expected to result in a different baseline transcriptomic response as well as potential differential responses to smoke across replicates after veraison. Furthermore, this variability in the developmental stage of the post-veraison berries could also potentially impact the ability of to uptake smoke, contributing to the reduced concordance in transcriptome profiles between the replicates. Supplementary File 1 provides visual evidence of the variability in berries harvested at post-veraison, compared with the consistency of those harvested at the pre-veraison stage.
Application of WGCNA revealed networks of differentially expressed genes associated with pre-veraison and post-veraison smoke exposure. The GO enrichment analysis of the co-expression modules associated with significant correlation to smoke provides a high-level understanding of the biological processes, molecular functions, and cellular components underlying the response of grape berries to smoke. Shared enriched gene ontologies between pre-veraison and post-veraison are of particular interest as they underscore the common mechanisms of response to smoke that are independent of the developmental stage. These include GOs related to glutathione and other sulfur-containing compounds, indicating a response to oxidative stress, with glutathione playing a critical role in detoxification. Additionally, changes in the metabolism of nitrogen-containing organic compounds suggest essential adjustments in amino acid metabolism for the maintenance of cellular homeostasis under stress. The presence of GOs linked to secondary metabolism, particularly phenylpropanoid biosynthesis, implies an upregulation of processes associated with the production of secondary metabolites involved in plant defense, stress response, and signaling. In addition to the enriched GOs common to both pre-and post-veraison fruit, grape berries at the pre-veraison stage displayed unique enrichment of GOs associated with glycosyltransferase activity, toxin metabolism, and lignin biosynthesis. Increased glycosyltransferase activity suggests heightened modification of small molecules and metabolites, likely as a detoxification strategy for harmful compounds introduced by smoke, while activation of toxin metabolic processes supports the engagement of robust detoxification mechanisms by pre-veraison grape berries to mitigate smoke-derived toxins. Additionally, the enrichment of lignin biosynthetic processes indicates a strengthening of structural components, with lignin reinforcing cell walls against physical and chemical damage from smoke. Interestingly, there were no unique GOs associated with the post-veraison stage. This finding, along with the high number of GOs unique to the pre-veraison stage, highlights the more robust response to smoke that appears to occur earlier in development. Moreover, the strategic prioritization of detoxification and structural integrity in younger berries could be crucial for their survival and development under stress conditions, particularly at a stage of development in which fewer antioxidant pigments are present.
While GO enrichment analysis provides a broad overview of smoke response mechanisms, the analysis of hub genes facilitates a more targeted focus on the specific drivers of these mechanisms. The general expression trend shared among the hub genes was increasing expression over time, beginning particularly at the 6 h time point. Figure 3 displays the expression profiles of the major groups of hub genes identified, which we can classify into four major categories: 1) genes associated with cellular detoxification, 2) genes associated with redox homeostasis and osmoprotection, 3) genes associated with cell wall structure and fortification, and 4) genes associated with vacuolar function. The first major category of DE hub genes that were upregulated in response to smoke exposure, those associated with cellular detoxification, includes GSTs, UGTs, and various other glycosyltransferases, as well as nucleoredoxins. GSTs play a pivotal role in cellular detoxification processes by facilitating the conjugation of toxic molecules into less reactive forms via the addition of glutathione (GSH) to stress-inducing, xenobiotic, and/or phytotoxic compounds30. Similarly, UGTs attach sugar molecules to other reactive molecules via a glycosylation step. These conjugation processes enhance the water solubility and reduce the volatility and toxicity of VPs, aiding in their detoxification and storage31. The high number of GSTs and UGTs expressed in the grape berries during smoke exposure is consistent with previous work indicating the role of volatile phenol conjugation in the detoxification of smoke-derived compounds4,12,21,22,32. Moreover, the elevated expression, particularly from 6–36 h of smoke exposure, is likely directly related to the levels of VPs and other smoke-derived xenobiotics that are taken up and conjugated into non-toxic forms by the plant following smoke exposure. This, in turn, is expected to correlate with the amount of smoke-derived phenolic compounds that are released when the bonds of their conjugated forms are cleaved during the fermentation process, altering the flavor and aroma profile of the resulting wine and leading to smoke taint19,33,34.
In addition to GSTs and UGTs, the upregulation of other glycosyltransferases, including crocetin glucosyltransferases, anthocyanidin glucosyltransferases, and hydroquinone glucosyltransferases, indicates a multifaceted detoxification and redox mitigation strategy in response to smoke exposure. Like UGTs, these additional glycosyltransferases conjugate various compounds to alter their function or activity. Hydroquinone is a specific phenolic constituent of wildfire smoke and a known phytotoxin at high concentrations13,35. Thus, hydroquinone glycosyltransferases likely act to stabilize and detoxify hydroquinone in smoke-exposed berries, representing a targeted strategy for amelioration of a specific smoke-derived phenolic compound. On the other hand, while the mode of action of crocetin and anthocyanidin glucosyltransferases is similar to that of the UGTs and hydroquinone glycosyltransferases, the targeted molecules are antioxidant pigments that serve important functions in ameliorating oxidative stress. The glycosylation of crocetin and anthocyanins stabilize these secondary metabolic compounds and makes them more easily mobilized within plant cells36, which is crucial for effectively mitigating oxidative stress caused by smoke exposure. This differential function of the latter two glycosyltransferases highlights a dual strategy in the glycosyltransferase-driven response to smoke: detoxifying harmful compounds while simultaneously reinforcing the plant’s antioxidant capacity to respond to smoke-induced oxidative stress.
A second group of DE hub genes, those involved in redox homeostasis and osmoprotection, includes nucleoredoxins (NRXs) and mannitol dehydrogenases (MDHs). NRXs are a subclass of thioredoxin proteins that play a crucial role in maintaining redox homeostasis within plant cells37. They function by facilitating thiol-disulfide exchange reactions, which are essential for the proper folding and functioning of many cellular proteins. The upregulation of nucleoredoxins in smoke-exposed grape berries suggests their involvement in mitigating oxidative stress. By regulating the redox state of various cellular components, NRXs help protect the grape berries from the damaging effects of ROS generated during smoke exposure. Notably, mannitol dehydrogenase (MDH) also displayed increasing expression over time in the smoke-exposed vs control grape berries, which was strongly correlated with the expression pattern displayed by the nucleoredoxin genes. MDH, which converts mannitol to fructose, serves as a ROS scavenger, as well as a strong osmotic regulator during stress responses, directly engaging in the mitigation of oxidative damage through the scavenging of hydroxyl radicals38. While it may seem counterintuitive to increase the activity of an enzyme that converts mannitol—given the role of mannitol as an antioxidant—the shift might represent a strategy to utilize mannitol reserves more effectively under stress, enabling the mobilization of mannitol reserves to fuel energy-demanding processes essential for repair, growth, and the synthesis of additional antioxidants and detoxification agents.
A third category of DE hub genes, those associated with cell wall fortification, includes genes involved in both general cell wall structure and lignin biosynthesis. The increased expression of several β-glucosidases suggests a shift towards the modification of cell wall polysaccharides in response to smoke. Additionally, the upregulation of glycine-rich protein (GRP) DC7.1-like, which encodes key components of the cell wall39, indicates a strategy to enhance the cell wall and membrane barrier properties, potentially reducing the permeation of volatile phenols and other smoke-derived compounds into the cells. Among the genes associated with lignin biosynthesis were cinnamoyl-CoA reductases (CCRs), which catalyze the first committed step in the formation of monolignols, the building blocks of lignin40, and dirigent protein 19, which impacts lignin biosynthesis to modulate cell wall structure in response to stress41. One possible scenario to explain the elevation in lignin biosynthesis-associated genes observed in smoke-exposed grape berries is that volatile phenols from smoke may be converted into lignin precursors. Given the structural similarities between many VPs and lignin monomers, it is plausible that modified VPs could enter the phenylpropanoid pathway and be further processed by lignin biosynthesis enzymes. This potential integration into the lignin polymer could contribute to the reinforcement of cell walls, providing additional protection against smoke-induced stress. Further experimental research is needed to validate this hypothesis and understand the exact metabolic fate of VPs in smoke-exposed grapes. Overall, the differential expression of lignin biosynthesis-associated genes, as well as general cell wall-associated genes, likely serves as a defensive response to reinforce cell walls against the potential damage from smoke exposure, thereby limiting the entry of harmful volatile phenols and other xenobiotics.
A fourth category of hub genes includes those involved in vacuolar storage and processing. The increased expression of Zinc-Induced Facilitator (ZIF)-like genes, which are involved in the transport of zinc into vacuoles, suggests their importance in managing osmotic conditions within the cell during smoke-induced stress. ZIF-like proteins may also create ion gradients that facilitate the movement of conjugated VPs into the vacuole for storage. In this way, these proteins may enhance the vacuole's capacity to sequester toxic compounds42. In addition to ZIF-like proteins, the increased expression of serine carboxypeptidase (SCP), a vacuolar marker associated with the breakdown of proteins in the vacuoles, suggests enhanced vacuolar activity associated with protein turnover43, potentially removing damaged proteins and maintaining cellular function under smoke-induced stress conditions.
The targeted insights provided by DE gene co-expression analysis, complemented by the broad findings of the GO enrichment analysis, provide a more in-depth understanding of the genes and pathways at play in response to smoke exposure in Merlot grapes at the pre-veraison and post-veraison stages. Overall, the observed upregulation of detoxification and structural-associated genes signifies a targeted response to smoke, characterized by the activation of pathways involved in redox homeostasis, detoxification, and structural composition. These pathways balance the immediate benefits of direct ROS scavenging and toxic compound inactivation and storage with the necessity to support the plant’s overall health and recovery from smoke exposure.
Proposed mode of action for detoxification of smoke-derived compounds in grapes
Based on the findings of the DE and GO enrichment analyses, we propose a mode of action for the processing of smoke-derived compounds in grapes (Fig. 4). Due to the relatively small size and lipophilic nature of VPs from smoke, these compounds are able to diffuse across the berry cuticle and membrane. Once smoke-derived compounds reach the cytosol, they can generate ROS, leading to oxidative stress. In response, the grape berry initiates a series of coordinated responses to mitigate the potential damage caused by reactive VPs. This includes the activation of key enzymes, such as GSTs and UGTs, which deactivate volatile phenols by conjugating them with glutathione or sugar, respectively, rendering them more water-soluble and less reactive34,44. NRX further supports the detoxification process, and mitigation of oxidative damage, by scavenging ROS. Other glycosyltransferases, including anthocyanidin and crocetin glucosyltransferases, stabilize their corresponding antioxidant substrates, enhancing their solubility and facilitating their mobilization within the cell where they may be used for further ROS scavenging. Glycosylated VPs are transported into the vacuole for storage, preventing their interaction with critical cellular processes. ZIF-like proteins may assist in this transport by creating electrochemical gradients and supporting osmotic balance within the cell. In the vacuole, SCPs support protein turnover, aiding in the maintenance of cellular homeostasis under smoke-induced stress conditions. In parallel with the detoxification processes, the fortification of the cell wall is another critical response to smoke-induced stress. Dirigent proteins and CCRs contribute to lignin polymerization and deposition, reinforcing the cell wall. This lignification, along with the modification of cell wall components by β-glucosidases and GRPs, enhances the structural integrity of the cell wall, providing a strengthened barrier against infiltration of xenobiotic compounds.
Proposed model of action for detoxification of smoke-derived volatile compounds in grapes, based on the results of this study as well as previous metabolomic work4,21. Acronym definitions are as follows: CCR – cinnamoyl-CoA reductase, GRP – glycine-rich protein, GSH – glutathione, GST – glutathione-S-transferase, NRX – nucleoredoxin, ROS – reactive oxygen species, SCP – serine carboxypeptidase, UGT – UDP-glycosyltransferase, ZIF – zinc-inducible factor.
Conclusion
Plants, including grapevines, have evolved sophisticated mechanisms to manage volatile organic compounds, such as volatile phenols. These compounds can be crucial for plant defense and communication in certain contexts, while in others, they can be detrimental to overall plant health. Beyond their impact on plant health, the presence or absence of specific VPs can significantly influence the flavor and aroma profiles of plant-derived foods and beverages. With the growing incidence of wildfires, wine grapes are increasingly exposed to smoke and, consequently, smoke-derived VPs, posing a significant threat to both grapevine health and the quality of the resulting wine.
We have identified a set of key differentially expressed genes in pre-veraison and post-veraison Merlot grapes, as well as an associated set of key gene ontologies, which highlight the temporal activation of detoxification, redox homeostasis, structural modifications, and vacuolar sequestration of conjugated, smoke-derived volatile phenols. These findings are crucial for understanding both the grape response to smoke exposure and the basis for smoke taint in wine produced from smoke-exposed grapes. However, there remains a need to understand how smoke impacts different cultivars at all stages of fruit development and over multiple years.
This work is expected to complement metabolic research seeking to develop markers for detecting smoke taint prior to winemaking. The process for assessing smoke taint propensity in wine grapes could be further refined by incorporating assays developed through gene expression analysis. Such assays would target genes that are elevated in expression during smoke exposure, including those identified in this study, potentially offering an advanced warning system for wine grape smoke taint propensity before harvest. By combining metabolic and genetic insights, these integrated management solutions become increasingly effective in counteracting the impact of smoke on wine quality, addressing the challenge at both pre- and post-harvest levels. Overall, an improved understanding of the gene expression-level basis for smoke taint in wine grapes will equip wine scientists and winemakers with vital knowledge and tools to devise strategies for maintaining wine quality in regions prone to wildfires.
Methods
Experimental conditions
Field-planted Merlot grapevines were exposed to simulated wildfire smoke using a purpose-built system for creating and delivering smoke to the fruiting zone in exposed vines. The smoke exposures were conducted on 21 and 22 July 2021 (pre-veraison) and 24 and 25 August 2021 (post-veraison). Six modular hoop houses were set up in the Washington State University Roza experimental vineyard near the WSU Prosser Irrigated Agriculture Research and Extension Center. The Merlot blocks used for the pre-veraison and post-veraison experiments each consisted of 13 rows of vines, with 30 vines in each row. The outside rows were used as buffer rows to reduce the likelihood of smoke drifting from the experimental block into adjacent blocks. Four sets of double rows were used for experimental rows, with a buffer row between each set of double rows, again to reduce drift between adjacent experimental rows. Each set of double rows was divided in half, creating 8 potential locations for hoop houses. Actual locations for each hoop house were selected at random, leaving two unused locations after the houses were placed. The hoop houses and associated equipment were placed one day prior to each smoke exposure. Each house covered 30 grapevines—two rows of 15 vines each. Block maps for the smoke exposure trials are provided in Supplementary File 6. Just before beginning the smoke exposure, an 80/20 shade-cloth cover was pulled into place over the vine rows. In each smoke exposure trial, there were three smoke-exposed hoop houses and three control houses, which were covered with shade cloth but not exposed to smoke.
Smoke was generated using Green Mountain Grills BBQ smoke tubes (Green Mountain Grills, LLC, Reno, NV) placed in the firebox of Oklahoma Joe Side Firebox Smoker (Stillwater, OK). “Winemaker’s Reserve” hardwood blend pellets (Traeger, Salt Lake City, UT) were used as the fuel source for the smoke applied during the trial exposures. Each full tube of pellets provided smoke for 5–6 h. Smoke was delivered to the smokehouses through 4-inch ABS drainage lines using 4-inch inline fans. The drainage lines were zip-tied to the cordon wire to deliver smoke to the fruiting zone in the vine canopy. 2 mm holes were drilled at 25 cm intervals along the line to allow the smoke to ensure consistent smoke density along the length of each smokehouse. Smoke intensity was monitored throughout each smoke exposure using TSI SidePak AM 520 particle counters (TSI, Inc, Shoreview, MN), with the PM1.0 orifice installed. One particle counter was used in each hoop house, including control houses. Particle counters were calibrated at zero prior to each exposure. Particle density was recorded at 10 s intervals throughout each exposure. Data for pre-veraison particle density is available in Supplementary File 7. The post-veraison smoke exposure treatment was similar to the pre-veraison treatment. Particle count data for the post-veraison treatment is unavailable because it was overwritten by measurement of an actual wildfire soon after the treatment and sample collection.
Each smoke exposure was 36 h in duration. Samples were collected before the onset of smoke exposure, and then at 2, 4, 6, 8, 10, 12, 24, and 36 h from the start of the exposure trial. At each time point, ten primary clusters were selected at random, with 5 clusters collected from each side of the aisle between rows, and the harvested clusters at each time point were pooled into a single sample. Thus, there were 3 pooled samples per condition (smoke, control) per time point for each developmental stage (pre-veraison, post-veraison). The rows at the Roza vineyard run North to South, so there were equal numbers of clusters from the East and West sides of the rows. After collection, samples were immediately placed in a −20 ℃ freezer until frozen and then transported to the WSU Chateau Ste Michelle Wine Science Center, where the samples were held in a −80 ℃ freezer until further processing.
RNA extraction
Fifty randomly selected grape berries per sample were pooled into a subsample and pulverized using a SPEX Freezer/Mill 6870 (Metuchen, NJ, USA). Total RNA was extracted using a DEPC-CTAB method modified from Reid et al.45 Briefly, for each extraction, 100 mg of frozen, pulverized grape tissue was added to 700 uL of pre-warmed (65 ℃) extraction buffer in (300 mM Tris- HCL pH 8.0, 25 mM EDTA, 2 M NaCl, 2% CTAB, 2% PVPP, 0.05% spermidine trihydrochloride, 2% B-mercaptoethanol to be added prior to use) 1.5 mL Eppendorf tubes and briefly vortexed, and incubated at 65 ℃ for 10 min, with intermittent vortexing every 10 min. Two chloroform extractions were performed with equal volumes of 24:1 chloroform: IAA, wherein Chl: IAA was added to the buffer/tissue mixture, vortexed for one minute, placed in a tube turner for 5 min, and centrifuged at 10,000 rpm for 15 min at 4 ℃ before transfer of the supernatant to a new tube. To the supernatant, 0.1 vol of 3 M NaOAc and 0.6 vol isopropanol were added, mixed by inverting the tubes gently 2–3 times, and stored at −80 ℃ for 30 min. Precipitated nucleic acids were pelleted by centrifuging tubes at 10,000 rpm for 30 min at 4 ℃, the supernatant was carefully removed and discarded, and the pellet was dissolved in 1 mL TE buffer. To selectively precipitate RNA, 0.3 vol 8 M LiCl was added to each sample, samples were stored for a minimum of 1.5 h at 4 ℃, and then RNA was pelleted by centrifuging tubes at 14,000 rpm for 30 min at 4 ℃. To wash the RNA pellets, 70% DEPC ethanol was added to each tube, and the tubes were centrifuged for 10 min at 14,000 rpm. Then, ethanol was removed and discarded by pipetting. Finally, the RNA pellets were dissolved in 40 μL DEPC-dH2O. Three RNA extractions were performed for each sample and pooled at the end of the extraction process to increase the total RNA obtained per sample.
RNA sequencing
RNA was quality-checked on an agarose gel and was quantified using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Following quality validation and quantification using a Qubit Fluorometer (Thermo Fisher Scientific) and Agilent Bioanalyzer (Santa Clara, CA, USA), cDNA libraries were prepared from the RNA using the TruSeq Stranded Total with Ribo-zero kit and sequenced on an Illumina HiSeq 2500 platform as 100 bp paired-end reads at the WSU Genomics Core (Spokane, WA).
Transcriptome assembly and read mapping
The fastq files containing the 100 bp paired-end sequencing reads were input into CLC Genomics Workbench for pre-processing and assembly according to published methods46,47. Reads were trimmed with a quality score limit of 0.001, corresponding to a Phred score of 30. The 13, 5’ terminal nucleotides were removed, as were any ambiguous nucleotides. Reads below length 33 (1/3 pre-trimming read length) were discarded. Overlapping pairs were merged. A de novo assembly was generated using the trimmed reads, and contiguous sequences (contigs) with less than 2 × coverage and of less than 200 bp in length were removed from the dataset, resulting in a final list of 212,937 contigs. For each treatment/replicate, the original non-trimmed reads were mapped back to the master assembly contig set to generate read counts per contig for each sample. The master transcriptome and the read count data were exported separately, the former as a fasta file for functional annotation and the latter as tab-delimited text files for differential expression analysis.
Functional annotation
The master transcriptome fasta file produced from the de novo assembly was imported into OmicsBox 3.1.0 (BioBam Bioinformatics S.L., Valencia, Spain) for functional annotation of expressed contigs. Using the Diamond Blast tool, contig sequences were identified by a blastx alignment against the NCBI ‘Viridiplantae’ database with an e-value specification of 10.0E-3. GO annotation was assigned using the ‘Mapping’ and ‘Annotation’ features using default parameters to generate a functionally annotated master assembly.
Differential expression analysis
To identify differentially expressed (DE) genes at each timepoint between smoke-exposed and control samples, the pairwise differential expression analysis feature in OmicsBox, which employs the EdgeR package, was used. Per the recommendations of the EdgeR manual, the TMM method was employed for read normalization, and the FDR cutoff was set to p ≤ 0.05.
In addition to pairwise DE analysis, the time-course, multi-series differential expression feature in the OmicsBox suite, which employs the maSigPro R package, was used to identify temporally significant genes (i.e., genes that display significantly different expression trends over time) for smoke-exposed vs control grapes. For this approach, the RPKM method was used for read normalization, the FDR-corrected cutoff was p ≤ 0.05, and the R2 cutoff was set to 0.5, as per recommendations of Nueda et al.48 The statistical analysis ensured that genes that did not meet the assumption of equal variances were eliminated from the analysis.
The resulting DE genes and their expression values were matched with their corresponding functional annotations for pairwise and time course DE analyses.
Analysis of co-expressed genes
To identify smoke-responsive gene networks, Weighted Gene Co-Expression Network Analysis (WGCNA) was performed on the raw RNA-seq count data corresponding to the 854 functionally annotated differentially expressed (DE) genes49. The read count dataset was pre-processed to remove outliers based on hierarchical clustering and principal component analysis (PCA). Normalization was performed using DESeq2, and variance stabilization transformation (VST) was applied to the filtered dataset.
To facilitate the construction of gene co-expression networks, a range of soft-thresholding powers (1–10, 12–70) was tested, and an optimal power of 20 was selected for network construction. The blockwiseModules function was used to identify modules of co-expressed genes. Module eigengenes (or representative expression patterns) were calculated to summarize the expression profiles of the identified modules. The correlations between module eigengenes and external factors, like smoke treatment and developmental stage, were calculated using Pearson correlation. The significance of these correlations was assessed using Student's t-test, and p-values were calculated for each correlation. Correlations with p-values less than 0.05 were considered significant and were annotated with asterisks in the heatmap. A cluster dendrogram with the identified modules, along with a module-trait association heatmap, is shown in Fig. 2. The gene modules with significant association with smoke exposure were further analyzed for functional enrichment based on associated GO terms, as described in the next section. This enrichment analysis identified the enriched biological processes, molecular functions, and cellular components among the hub genes (Fig. 2).
To identify hub genes within each module, the module membership (kME) and gene significance were calculated. Module membership, which quantifies the similarity of each gene’s expression profile to the module eigengene, was calculated using Pearson correlation. The significance of these correlations was assessed using Student’s t-test, and p-values were calculated for each correlation. Hub genes were identified as those genes with both high module membership and a gene significance p-value of 0.05 or less.
Gene ontology enrichment analysis
GO enrichment analysis using Fisher’s exact test was conducted in OmicsBox to identify the cellular components, molecular functions, and biological processes enriched in the smoked grape treatments compared to the control (FDR-corrected p-value < 0.05). This analysis was performed using the pairwise DE analysis results, and lists of the differentially expressed, functionally annotated genes at each time point were generated for the smoke and control datasets. These lists served as the treatment datasets for enrichment analyses, and the master annotated transcriptome was used as the reference dataset. The resulting enriched GO lists for each timepoint were reduced to the most specific parent terms using a p-value cutoff of 0.05.
Permissions
Grape berries were treated and collected at the Washington State University Roza experimental vineyard near the WSU Prosser Irrigated Agriculture Research and Extension Center. Permissions necessary to conduct the experiments were obtained by TC.
Data availability
The RNAseq datasets generated during the study are available in the NCBI Short Read Archive with the accession number PRJNA1096040: https://www.ncbi.nlm.nih.gov/sra/PRJNA1096040.
References
van Leeuwen, C. et al. Climate change impacts and adaptations of wine production. Nat. Rev. Earth Environ. 2024, 1–18. https://doi.org/10.1038/s43017-024-00521-5 (2024).
Madhusoodanan, J. Wildfires pose a burning problem for wines and winemakers. Proc. Natl. Acad. Sci. U. S. A. 118, e2113327118 (2021).
Jones, G. V. et al. 17 - Climate change and its consequences for viticulture. In Woodhead Publishing Series in Food Science, Technology and Nutrition (ed. Reynolds, A.G.B.T.-M.W.Q.) (Woodhead Publishing, 2022).
Tomasino, E. et al. A combination of thiophenols and volatile phenols cause the ashy flavor of smoke taint in wine. Food Chem. Adv. 2, 100256 (2023).
Kennison, K. R., Gibberd, M. R., Pollnitz, A. P. & Wilkinson, K. L. Smoke-derived taint in wine: The release of smoke-derived volatile phenols during fermentation of Merlot juice following grapevine exposure to smoke. J. Agric. Food Chem. 56, 7379–7383 (2008).
Kennison, K. R., Wilkinson, K. L., Williams, H. G., Smith, J. H. & Gibberd, M. R. Smoke-derived taint in wine: Effect of postharvest smoke exposure of grapes on the chemical composition and sensory characteristics of wine. J. Agric. Food Chem. 55, 10897–10901 (2007).
Summerson, V., Gonzalez Viejo, C., Pang, A., Torrico, D. D. & Fuentes, S. Review of the effects of grapevine smoke exposure and technologies to assess smoke contamination and taint in grapes and wine. Beverages 7, 7 (2021).
Szeto, C., Ristic, R. & Wilkinson, K. Thinking inside the box: A novel approach to smoke taint mitigation trials. Molecules 27, 1667 (2022).
Simos, C. The implications of smoke taint and management practices. Aust. Vitic. 1, 77–80 (2008).
Favell, J. W., Noestheden, M., Lyons, S. M. & Zandberg, W. F. Development and evaluation of a vineyard-based strategy to mitigate smoke-taint in wine grapes. J. Agric. Food Chem. 67, 14137–14142 (2019).
Mirabelli-Montan, Y. A., Marangon, M., Graça, A., Mayr Marangon, C. M. & Wilkinson, K. L. Techniques for mitigating the effects of smoke taint while maintaining quality in wine production: A review. Molecules 26, 1672 (2021).
Szeto, C. et al. Uptake and glycosylation of smoke-derived volatile phenols by Cabernet sauvignon grapes and their subsequent fate during winemaking. Molecules 25, 3720 (2020).
Szeto, C., Lloyd, N., Nicolotti, L., Herderich, M. J. & Wilkinson, K. L. Beyond volatile phenols: An untargeted metabolomic approach to revealing additional markers of smoke taint in grapevines (Vitis vinifera L.) cv. Merlot. J. Agric. Food Chem. 72, 2018–2033 (2023).
Liu, Z. et al. A simple GC-MS/MS method for determination of smoke taint-related volatile phenols in grapes. Metabolites 10, 294 (2020).
Ristic, R., Boss, P. K. & Wilkinson, K. L. Influence of fruit maturity at harvest on the intensity of smoke taint in wine. Molecules 20, 8913–8927 (2015).
Jiang, W. W. et al. The effect of pre-veraison smoke exposure of grapes on phenolic compounds and smoky flavour in wine. Aust. J. Grape Wine Res. https://doi.org/10.1155/2022/9820204 (2022).
Kennison, K. R., Wilkinson, K. L., Pollnitz, A. P., Williams, H. G. & Gibberd, M. R. Effect of timing and duration of grapevine exposure to smoke on the composition and sensory properties of wine. Aust. J. Grape Wine Res. 15, 228–237 (2009).
Kennison, K. R., Wilkinson, K. L., Pollnitz, A. P., Williams, H. G. & Gibberd, M. R. Effect of smoke application to field-grown Merlot grapevines at key phenological growth stages on wine sensory and chemical properties. Aust. J. Grape Wine Res. 17, S5–S12 (2011).
Hayasaka, Y., Dungey, K. A., Baldock, G. A., Kennison, K. R. & Wilkinson, K. L. Identification of a β-d-glucopyranoside precursor to guaiacol in grape juice following grapevine exposure to smoke. Anal. Chim. Acta 660, 143–148 (2010).
Krstic, M. P., Johnson, D. L. & Herderich, M. J. Review of smoke taint in wine: Smoke-derived volatile phenols and their glycosidic metabolites in grapes and vines as biomarkers for smoke exposure and their role in the sensory perception of smoke taint. Aust. J. Grape Wine Res. 21, 537–553 (2015).
Härtl, K. et al. Glucosylation of smoke-derived volatiles in grapevine (Vitis vinifera) is catalyzed by a promiscuous resveratrol/guaiacol glucosyltransferase. J. Agric. Food Chem. 65, 5681–5689 (2017).
Noestheden, M., Dennis, E. G. & Zandberg, W. F. Quantitating volatile phenols in Cabernet Franc berries and wine after on-vine exposure to smoke from a simulated forest fire. J. Agric. Food Chem. 66, 695–703 (2018).
Caffrey, A. et al. Changes in smoke-taint volatile-phenol glycosides in wildfire smoke-exposed Cabernet Sauvignon grapes throughout winemaking. Am. J. Enol. Vitic. 70, 373–381 (2019).
Hewitt, S. L. S. L., Hendrickson, C. A. C. A. & Dhingra, A. Evidence for the involvement of vernalization-related genes in the regulation of cold-induced ripening in ‘D’Anjou’ and ‘Bartlett’ pear fruit. Sci. Rep. https://doi.org/10.1038/s41598-020-65275-8 (2020).
Williamson-Benavides, B. A. et al. Identification of Fusarium solani f. sp. pisi (Fsp) responsive genes in Pisum sativum. Front. Genet. 11, 950 (2020).
Rienth, M. et al. Temperature desynchronizes sugar and organic acid metabolism in ripening grapevine fruits and remodels their transcriptome. BMC Plant Biol. 16, 164 (2016).
Leng, X. et al. Comparative transcriptome analysis of grapevine in response to copper stress. Sci. Rep. 5, 17749 (2015).
Han, X. et al. Transcriptomics reveals the effect of short-term freezing on the signal transduction and metabolism of grapevine. Int. J. Mol. Sci. 24, 3884 (2023).
Gouthu, S. et al. A comparative study of ripening among berries of the grape cluster reveals an altered transcriptional programme and enhanced ripening rate in delayed berries. J. Exp. Bot. 65, 5889 (2014).
Dorion, S., Ouellet, J. C. & Rivoal, J. Glutathione metabolism in plants under stress beyond reactive oxygen species detoxification. Metab. 11, 641 (2021).
Harborne, J. B. Variation in and functional significance of phenolic conjugation in plants .in Biochemistry of plant phenolics (Springer, 1979).
Hayasaka, Y. et al. Glycosylation of smoke-derived volatile phenols in grapes as a consequence of grapevine exposure to bushfire smoke. J. Agric. Food Chem. 58, 10989–10998 (2010).
Ono, E. & Ohnishi, T. Volatile glycosylation—A story of glycosyltransferase for volatiles: Glycosylation determining the boundary of volatile and nonvolatile specialized metabolites .in Plant Specialized Metabolism (CRC Press, 2016).
Song, C., Härtl, K., McGraphery, K., Hoffmann, T. & Schwab, W. Attractive but toxic: Emerging roles of glycosidically bound volatiles and glycosyltransferases involved in their formation. Mol. Plant 11, 1225–1236 (2018).
Pandey, D. K., Mishra, N. & Singh, P. Relative phytotoxicity of hydroquinone on rice (Oryza sativa L.) and associated aquatic weed green musk chara (Chara zeylanica Willd.). Pestic. Biochem. Physiol. 83, 82–96 (2005).
Tiwari, P., Sangwan, R. S. & Sangwan, N. S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 34, 714–739 (2016).
Hazra, S., Chatterjee, A., Bhattacharyya, S. & Sen, P. Nucleoredoxin Vis-à-Vis a novel thioredoxin in regulating oxidative stress in plants: A review. Agric. Res. https://doi.org/10.1007/s40003-024-00737-3 (2024).
Jennings, D. B., Ehrenshaft, M., Mason Pharr, D. & Williamson, J. D. Roles for mannitol and mannitol dehydrogenase in active oxygen-mediated plant defense. Proc. Natl. Acad. Sci. 95, 15129–15133 (1998).
Ringli, C., Keller, B. & Ryser, U. Glycine-rich proteins as structural components of plant cell walls. Cell. Mol. Life Sci. C. 58, 1430–1441 (2001).
Lacombe, E. et al. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: Cloning, expression and phylogenetic relationships. Plant J. 11, 429–441 (1997).
Paniagua, C. et al. Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 68, 3287–3301 (2017).
Meena, V., Sharma, S., Kaur, G., Singh, B. & Pandey, A. K. Diverse functions of plant zinc-induced facilitator-like transporter for their emerging roles in crop trait enhancement. Plants 11, 102 (2021).
Yamada, K., Basak, A. K., Goto-Yamada, S., Tarnawska-Glatt, K. & Hara-Nishimura, I. Vacuolar processing enzymes in the plant life cycle. New Phytol. 226, 21–31 (2020).
Lushchak, V. I. Glutathione homeostasis and functions: potential targets for medical interventions. J. Amino Acids 2012, 1–26 (2012).
Reid, K. E., Olsson, N., Schlosser, J., Peng, F. & Lund, S. T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6, 1–11 (2006).
Hewitt, S. L., Ghogare, R. & Dhingra, A. Glyoxylic acid overcomes 1-MCP-induced blockage of fruit ripening in Pyrus communis L var ‘D’Anjou’. Sci. Rep. https://doi.org/10.1038/s41598-020-63642-z (2020).
Sharpe, R. M. et al. Concomitant phytonutrient and transcriptome analysis of mature fruit and leaf tissues of tomato (Solanum lycopersicum L. Cv. Oregon Spring) grown using organic and conventional fertilizer. PLoS ONE 15, e0227429 (2020).
Nueda, M. J., Tarazona, S. & Conesa, A. Next maSigPro: Updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics 30, 2598–2602 (2014).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Acknowledgements
This research was supported by a USDA ARS NACA #58-2072-0-033 awarded to TSC and AD. Work in the Dhingra lab was supported in part by the Washington State University Agriculture Center Research Hatch Grant WNP00011 and startup funds from Texas A&M AgriLife Research, Texas A&M University.
Author information
Authors and Affiliations
Contributions
TC and AD designed the study. TC, MA, and PLA performed the smoke treatments. SH performed the RNA work and, along with AD, performed the data analysis. SH and AD prepared the manuscript. All authors reviewed, edited, and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hewitt, S., Aragon, M., Ashmore, P.L. et al. Transcriptome analysis reveals activation of detoxification and defense mechanisms in smoke-exposed Merlot grape (Vitis vinifera) berries. Sci Rep 14, 21330 (2024). https://doi.org/10.1038/s41598-024-72079-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-72079-7