Abstract
Pseudomonas aeruginosa, a common pathogen in nosocomial infections, presents significant global health challenges due to its high prevalence and mortality rates. However, the origins and distribution of this bacterium remain unclear, partly due to the lack of effective gene typing methods. This situation necessitates the establishment of trustworthy and high-resolution protocol for differentiating closely related P. aeruginosa strains. In this context, the present study attempted to undertake a comparative genomic analysis of multiple P. aeruginosa strains available in the public database NCBI, with the goal of identifying potential genetic markers for measuring the genetic diversity. The preliminary comparative analysis of 816 P. aeruginosa strains revealed notable variations in two genes—specifically, the CDF family iron/cobalt efflux transporter AitP and the protease modulator HflC—across 44 strains. These variations were associated with single amino acid repeats (SHRs) that responsible for encoding histidine residue. Additionally, comparative gene map analysis revealed differential clustering patterns in the Rsx and TAXI genes among 16 strains. Interestingly, the gene structure pattern observed in TAXI groups displayed a strong correlation with the SHRs pattern in the CDF and HflC groups. In addition, the SHRs pattern of CDF and HflC were strongly correlated with MLST sequence type number. Overall, the study present a novel genetic markers based on SHRs and gene cluster patterns, offering a reliable method for genotyping of P. aeruginosa.
Similar content being viewed by others
Introduction
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen and a common cause of hospital-acquired infections, accounting for nearly 10% of all nosocomial infections1. This pathogen is particularly alarming due to its high mortality and morbidity rates, reaching up to 40% in immunocompromised patients2. P. aeruginosa’s adaptability to adverse environments and production of various virulence factors enable it to cause diverse infections, including in patients with cystic fibrosis, pulmonary disease, sepsis, traumas, and burn wounds1,3. Among its virulence factors, biofilm production is notably problematic as it acts as a barrier, enhancing the bacterium’s survival and persistence in harsh environments4. Additionally, P. aeruginosa’s rapid mutation and adaptation confer resistance to a broad range of antibiotics, posing a significant challenge for therapeutic treatments5. In 2017, the World Health Organization (WHO) listed P. aeruginosa as a high-threat antibiotic-resistant pathogen, necessitating global concern6.
This situation demands innovative and effective methods for early epidemiological identification and genotyping, which can facilitate more precise antibiotic therapy and prevent subsequent colonization and chronic infection by P. aeruginosa in medical settings. Despite advances in molecular techniques, current methods for identifying and classifying P. aeruginosa are limited. The gold standard techniques for genotyping P. aeruginosa include pulsed-field gel electrophoresis of SpeI-restricted genomic DNA (PFGE-SpeI)7, single nucleotide polymorphism (SNP) analysis8, and core genome multilocus sequence typing (cg-MLST)9. Although these methods provide valuable information, their wide spread application are hampered by high costs, labor intensity, technical complexity, and insufficient discriminatory power at the strain level10,11. In addition to that, high genomic similarity among P. aeruginosa strains, even from different niches, further complicates strain-level genotyping12,13.
Recently, whole-genome sequencing combined with bioinformatics analysis has become increasingly prevalent for analyzing microbial diversity and tracing pathogen origins14,15,16. As of April 5, 2024, there are 30,192 whole-genome sequences of P. aeruginosa registered in GenBank. Nevertheless, to the best of our knowledge, none of study has yet utilized the publicly accessible genome sequences of P. aeruginosa for its genomic, evolutionary, and diversity analysis. As a result, many deposited P. aeruginosa genome sequences are remains in text files without a comprehensive genomic analysis.
In this context, our study aims to conduct a comparative genomic analysis of multiple P. aeruginosa strains available in the GenBank database, with the goal of improving conventional methods for measuring genetic diversity without relying on whole-genome sequencing (WGS) techniques. Initially, we downloaded and compared the genome data of P. aeruginosa strains registered in genebank to identify intraspecific genes. We discovered some protein-encoding genes differ in single-amino-acid repeats (SARs) of histidine (H). SARs, or homopolymeric amino acid tracts, are more abundant in eukaryotes than in prokaryotes and account for nearly one-fifth of all human gene products17. Despite their high distribution, the main function and evolution of SARs remain unclear. Generally, SARs are dynamic elements present in various patterns and locations throughout the genome, varying from strain to strain, making them unique to each organism12,18. Henceforth, several studies have demonstrated the use of these unique SARs for bacterial strain identification19,20.
Using this paradigm, in this study, we aim to differentiate strains of P. aeruginosa by analyzing variations in their SARs repeat patterns. We also created gene maps for genomic segments of strains exhibiting SARs variations. Notably, strains with similar gene mapping structures demonstrated consistent SARs patterns, leading to uniform genetic profiles. The findings from this research will offer valuable insights for the development of novel, highly discriminative, and easy-to-manage genetic markers based on SARs and gene cluster patterns. This approach has the potential to reduce both the cost and time required for conventional strain typing methods.
Materials and methods
Sampling collection
The in-house P. aeruginosa strains used in the study were isolated from agricultural produce distributed in South Korea. The collection of contaminated produce samples and use were carried out in accordance with the “Detection methods of foodborne pathogens in agricultural produces” of Rural Development Administration (RDA, Jeonju, South Korea, 2021) which is responsible for the management of foodborne pathogen-contaminated crops. The source of produce samples is listed in the Table 1.
Bacterial cultures and DNA extraction
The used P. aeruginosa strains were isolated from peppers, carrots, radishes, and Chinese cabbage obtained from markets in South Korea. The surface area of agricultural produce was cut into 25 g pieces, which were then placed into sterilized bags containing 250 mL of buffered peptone water. Following this, the samples were incubated at 37 ℃ for 24 h. After incubation, 100 µl of the macerated samples were streaked onto Pseudomonas Isolation Agar (PIA) and further incubated at 37 ℃ for 24 h21. Next, a single colony of P. aeruginosa was selected by comparing the obtained colonies from the positive control, and then streaked repeatedly to obtain a pure culture. To further confirm whether the colonies are P. aeruginosa, specific DNA regions were amplified by C1000 Touch Thermal Cycler (Bio-Rad, Inc., Germany) with PA431CF/R primers22.
The bacterial cultures were stored with 15% glycerol in a 1:1 ratio at -80 ℃ for subsequent DNA extraction. P. aeruginosa isolates were cultured in Luria-Bertani Broth at 37 ℃ and 180 rpm for 24 h, and the cell pellets of the culture were used to extract genomic DNA using a DNA extraction Kit (IncloneTM Genomic Plus DNA Prep Kit, Inclone Biotech, Inc., Korea), according to the manufacturer’s instructions.
Primer design
Two sets of primers were designed to directly analyze amino acid tandem repeats in the isolates of P. aeruginosa using PrimerSelect software (Version 15.1.0 (155); DNASTAR Inc., Madison, WI, USA). From the nucleotide sequences of the gene encoding CDF family iron/cobalt efflux transporter AitP and protease modulator HflC, forward and reverse primers were designed more than 40 bp outside of target region. The primers cdfa_F/R (5’–GGCCGGGCTGATGCTCTACCAAT–3′, 5′–CCACCGGCGCGTCCATCTGC–3′) for the CDF family iron/cobalt efflux transporter AitP protein and hflc_F/R (5′–CACTCCCACTCGCACAGCCACCAC–3′, 5′–TCCAGCGCCGAGCCGACGAA–3′) for the protease modulator HflC were selected. The PCR mixture for the amplification was as follows: 25 ng of genomic DNA, 10 pM of each primer, 1.25 unit of Taq Polymerase (Takara Bio, Inc., Japan) 1X Ex Taq Buffer, and 0.25 mM of dNTPs in a total volume of 50 µl. PCR was performed according to the previous study with slight modifications as follows: predenaturation at 95 ℃ for 3 min; 40 cycles of 95 ℃ for 60s, 68 ℃ for 30s, and 72 ℃ for 30s; and final extension at 72 ℃ for 10 min (Choi et al., 2013). The final products were 443 bp and 488 bp for AitP and HflC genes, respectively. The amplicons of strains BS0003PA, BS0009PA, BS0018PA, BS0022PA, and BS0028PA were cloned and sequenced (Macrogen, Daejeon, South Korea) to determine amino acid repeats.
Targeted region sequencing
The PCR amplification products obtained using cdfa_F/R and hflc_F/R primers were cloned and then sequenced. Following the sequencing on the Illumina platform, the reads were assembled into contiguous sequences using SPAdes version 3.1.3.0. Gene annotation and prediction were processed using Prokka version 1.13. The annotation process included the prediction of coding sequences (CDS), ribosomal RNA (rRNA) genes, transfer RNA (tRNA) genes, and other regulatory elements in the genome23.
Specific gene genomic analyses
We collected genomic FASTA files of the coding DNA sequences (CDSs) of the 816 P. aeruginosa strains, from the NCBI bacterial genome database (https://www.ncbi.nlm.nih.gov/genome/). We then confirmed the identity of the obtained sequences by verifying their taxonomy and Average Nucleotide Identity results against the NCBI Genome Assembly database. All collected sequences were compared to mine for species-specific genes, focusing on those with more than five amino acid differences per gene. The nucleotide and amino acid sequences of these genes were compared across P. aeruginosa strains using MUSCLE module of the Megalign Pro software (Version 15.1.0 (155); DNASTAR Inc., Madison, WI, USA). As a result, we identified variations in amino acid repeats within these genes among the P. aeruginosa strains. For the comparative analysis, the MLST sequence types of strains exhibiting variations were also retrieved using the software developed by Torsten Seemann that rely on PubMLST.
Structural analysis of the genome mapping
To identify and compare nucleotide sequences, a BLAST (Basic Local Alignment Search Tool) search was conducted using the NCBI (National Center for Biotechnology Information) online platform. We compared and analyzed the CDS regions located from the TAXI family TRAP transporter solute-binding subunit to the DNA polymerase III subunit chi and Rsx family gene regions in P. aeruginosa strains. The following specific settings were used to adapt the search according to our research requirements. The “Nucleotide collection (nr/nt)” was selected from the Standard databases available on the BLAST site. We excluded sequences from uncultured or environmental samples. The search was optimized for “somewhat similar sequences (blastn)” to improve the specificity and relevance of our query results.
Result and discussion
Understanding the genetic variability among different strains of P. aeruginosa is crucial for identifying distinct strains and gaining deeper insights into their local and global transmission patterns and distribution. This knowledge aids in the development and implementation of new, targeted (strain-specific) treatment strategies to combat this pathogen in clinical settings. Various methods, including PFGE-SpeI, SNP analysis, and cg-MLST, have been employed to study the phylogeny and genomic traits of different P. aeruginosa strains7,8,9. However, these methods face challenges in examining the complete genome in a single, reliable experiment. Thus, establishing a trustworthy and high-resolution protocol for differentiating closely related strains used in commercial or scientific applications is essential. In this study, we introduce a novel genotyping technique that enables the differentiation of P. aeruginosa at the strain level by analyzing variations in single-histidine repeats (SHRs) and gene structure (gene mapping).
Initially, we obtained a total of 816 distinct P. aeruginosa sequences from NCBI and subjected them to a thorough examination to identify any genotypic markers using comparative genomic analysis. Detailed information about these strains is available in the NCBI database (https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/Pseudomonas%20aeruginosa). Our results revealed significant variations in two protein-encoding genes, the CDF family iron/cobalt efflux transporter AitP and protease modulator HflC, across most of the strains (44 strains). These variations were primarily due to a trinucleotide tandem repeat (i.e., 5’-CAC or CAT-3’) that encoded histidine residues (H).
Classification based on single amino acid repeat (SAR) patterns
Single amino acid repeats (SARs) are short sequences in proteins that consist of one amino acid repeated several times in a row12,18. Due to high variability and genetic diversity among bacterial populations, these repeats are commonly used in bacterial identification, allowing unique genetic fingerprints to be generated24. Hence, a multitude of researchers has employed these SARs for the purpose of bacterial identification25,26,27. For instance, Subirana et al. (2021)26 found that tandem repeats in Bacillus exhibit distinct characteristics, including length, sequence composition, and distribution throughout the genome. These features can be used for taxonomic classification and molecular typing of Bacillus species. Another study demonstrated the utility of internet-based resources for developing and analyzing tandem repeats-based bacterial strain typing in E. coli25. Following this paradigm, our study presents unique genotyping characteristics based on SHRs to facilitate the identification of P. aeruginosa strains at the strain level.
CDF family iron/cobalt efflux transporter AitP protein
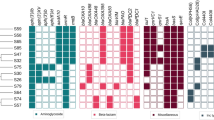
The gene encoding the CDF family iron/cobalt efflux transporter AitP (WP_023088961.1 of strain DHS01) was identified in 44 strains out of 816, displaying notable differences in gene size. The information about the 44 strain is illustrated in Table 2. The obtained variations were primarily attributed to a trinucleotide tandem repeat (i.e., 5′–CAC or CAT–3′) that encoded histidine residues (H), resulting in a single-histidine repeat (SHR) spanning from 6 to 19 units. Based on the histidine repeat number, the strains were categorized and named as CDF 6, 8, 9, 10, 12, 14, 15, and 19 (Table 2). The analysis revealed that the majority of strains, totaling 11, contained 6 units of SHRs, suggesting a high prevalence. On the other hand, only one strain each had 9 and 19 units of SHRs, showing a low prevalence. Interestingly, histidine (H) was randomly substituted with aspartic acid (D) in most strains, regardless of the SHRs length. These changes were found to be occurred by the substitution of G instead of C in the codon of histidine- CAU, CAC.
Besides, to directly assess variation, the SHR gene from five different in house P. aeruginosa strains (viz., BS0003PA, BS0009PA, BS0018PA, BS0022PA, and BS0028PA) was isolated using an PCR primers (cdfa_F/R), resulting in 443 bp amplicons. Following the purification and sequencing of the obtained amplicons, the strains were found to have SHRs unit of 6, 12, and 13 (Table 1). Interestingly, we found that the histidine residues was replaced with aspartic acid in in- house strains as well. However, we did not find the exact reason for this substitution in SHRs of CDF gene.
Protease modulator HflC protein
To gain further insights, the strains showed the SHRs variation in AitP protein were subjected to additional analysis. Interestingly, it was observed that all strains also exhibited another distinguishable SHRs repeats in gene responsible for encoding HflC protein. The units of SHRs ranged from 5 to 21 and strains were categorized based on the number of repeats, which included 5, 9, 11, 13, 15, 19, and 21 (Table 2). Notably, the SHRs patterns observed in the CDF gene displayed a strong correlation with SHRs pattern of HflC protein. This led to the hypothesis that both genes, CDF and HflC, might have a similar or single mode of function in the respective P. aeruginosa strains.
Origin of SHRs groups
The origin of each SHRs groups were analyzed. The result revealed that the isolates of SHRs groups originated from 12 countries, as listed in Table 2. Notably, the highest number of SHRs groups, totaling 12, were isolated from China. Conversely, the isolates of India, Netherlands, Japan, and Sweden had the lowest number of SHRs groups, with only 1 group each.
The isolates of South Korea were in narrow range of SHRs groups such as CDF6, CDF11, HflC 19 and HflC 21 (Table 2). Whereas Brazil strains were only in the group of CDF 10 and HflC 5. This suggesting the low level of genetic diversity in P. aeruginosa strains in both South Korea and Brazil. In contrast, there was significant diversity in SHRs group among strains isolated from the United Kingdom, USA, China, and Mexico City, indicating a high level of genetic diversity. Nevertheless, there was no apparent connection between the origin of isolation and SHRs groups of CDF and HflC.
Correlation of SHRs pattern with MLST
For the comparison, we analyzed the MLST type numbers of strains that exhibited variations in SHR repeats. Notably, most of the SHR patterns for both CDF and HflC showed a strong correlation with the MLST sequence types, underscoring the reliability of these patterns for sub-typing P. aeruginosa strains (Table 2). Interestingly, this method also has the potential to differentiate strains that are indistinguishable by MLST, including those lacking an MLST number, highlighting the novelty and significance of the current study (Table 2).
Genomic mapping and comparison of DNA segments in P. aeruginosa strains
Even though, the gene typing of P. aeruginosa strains according to SHRs yield significant genetic diversity, the resolution of this method is relatively low. Therefore, we conducted genomic mapping analysis to further explore genetic diversity among different P. aeruginosa strains. We compared the genetic composition of 16 selected P. aeruginosa strains for a specific genomic segment to identify the genetic diversity. The information of the selected strains for gene map analysis is illustrated in Table 3. The Peudomonas otitidis MrB4 and Peudomonas nitroreducens L4 were used as reference sequence for the comparison. According to the gene map, it was found that most of strains exhibited variations in two segments viz., the Rsx family gene and TAXI-TRAP gene.
Grouping of P. aeruginosa strains based on the variation in TAXI-TRAP
The genetic maps were anchored to a stable backbone structure, extending from the TAXI family TRAP transporter solute-binding subunit to the DNA polymerase III subunit chi. This structural alignment allowed for the classification of strains into six distinct types from TAXI 1 to TAXI 6 (Fig. 1).
Genome mapping from ‘TAXI family TRAP transporter solute-binding subunit’ gene to ‘DNA polymerase III subunit chi’ gene among the different Pseudomonas aeruginosa strains. DUF2165 family protein are colored in purple; T3SS effector bifunctional cytotoxin exoenzyme S, grey; ABC transporter family gene, yellow; and DUF2007 domain-containing protein, green.
The gene DUF2165 (highlighted in purple in Fig. 1) was found across all P. aeruginosa strain types. The TAXI types 2, 3, 4, 5 and 6 exhibited distinct clustering patterns in the downstream of the ABC transporter gene, while TAXI type 1 showed variation in the upstream of the ABC transporter.
For TAXI Type 1 strains, genetic differences were associated with proteins such as the zinc-dependent alcohol dehydrogenase family protein (WP_033939636.1) and the metalloregulator ArsR/SmtB family transcription factor (WP_03393963.1) (Fig. 2). In TAXI type 3 strains, variations were noted in the intergenic regions between the ABC transporter protein (WP_003092876.1), an FG-GAP-like repeat-containing protein (WP_23553742.1), and a hypothetical protein (WP_123823007.1). For TAXI type 6 strains, the variation was observed in the intergenic region between WP_153519859.1 and WP_023081550.1. However, in TAXI types 2, 4, and 5, the genetic variation was associated with a hypothetical protein, which did not provide sufficient distinction between these types.
Grouping of P. aeruginosa strains based on the variation in Rsx family operon
The genetic maps were aligned to a stable structural framework, ranging from the electron transport complex subunit E to the methionine-tRNA ligase. Following the gene mapping, all the strains were categorized into two groups such as Rsx A and group Rsx B (Fig. 3). The key difference between these groups was linked to the presence of three specific genes: the T6SS immunity protein Tli4 family protein, phospholipase D, and type VI secretion system tip protein Tssl/VgrG. Strains containing these three genes were categorized as Rsx B, while those without them were categorized as Rsx A.
Comparing the pattern of SHRs with gene mapping patterns
We also attempted to find out whether SHRs patterns of CDF and HflC groups could exhibited any correlation with genetic pattern identified in TAXI and Rsx groups. Interestingly, the gene cluster of TAXI groups were somewhat correlated with SHRs pattern of CDF and HflC groups, resulting in consistent genetic patterns (Table 2). However, not complete matching of patterns were observed between SHRs and CDF groups.
Conclusion
Overall, the present study propose a novel combination of genotyping markers based on SHRs (CDF and HflC genes) and gene structure (TAXI and Rsx genes) to enhance the robustness of strain typing by identifying specific variations within the same species. The comparison of the current approach with the MLST technique demonstrated that it produces reproducible results and can also distinguish strains that lack an MLST type number. This novel approach could serve as a useful genetic marker and reliable metric for genotyping of P. aeruginosa and could serve as one of the potential element for the understanding of their evolution and genetic diversity. However, elucidating the function of amino acid repeats and the biological significance of strains remains imperative. Further research and validation studies are needed to effectively implement this approach in practical applications.
Data availability
The datasets generated and analyzed during the course of this study are publicly available in the National Center for Biotechnology Information (NCBI) repository, which ensures they are accessible to the scientific community for further research and verification. These datasets include all the raw sequencing data, assembly files, and annotated features relevant to our study.
References
Qin, S. et al. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal. Transduct. Target. Therapy. 7(1), 199 (2022).
Sadikot, R. T., Blackwell, T. S., Christman, J. W. & Prince, A. S. Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit Care Med.171(11), 1209–1223 (2005).
Karthika, C. et al. Two novel phages PSPa and APPa inhibit planktonic, sessile and persister populations of Pseudomonas aeruginosa, and mitigate its virulence in zebrafish model. Sci. Rep.13(1), 19033 (2023).
Thi, M. T. T., Wibowo, D. & Rehm, B. H. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci.21(22), 8671 (2020).
Yin, R., Cheng, J. & Lin, J. Treatment of Pseudomonas aeruginosa infectious biofilms: Challenges and strategies. Front. Microbiol.13, 955286 (2022).
Denissen, J. et al. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ Health. 244, 114006 (2022).
Selim, S., Kholy, E., Hagagy, I., Alfay, N. E., Aziz, M. A. & S., & Rapid identification of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. Biotechnol. Biotechnol. Equip.29(1), 152–156 (2015).
Ajayi, T., Allmond, L. R., Sawa, T. & Wiener-Kronish, J. P. Single-nucleotide-polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. J. Clin. Microbiol.41(8), 3526–3531 (2003).
de Sales, R. O., Migliorini, L. B., Puga, R., Kocsis, B. & Severino, P. A core genome multilocus sequence typing scheme for Pseudomonas aeruginosa. Front. Microbiol.11, 518183 (2020).
Tang, Y. et al. Detection methods for Pseudomonas aeruginosa: History and future perspective. RSC Adv.7(82), 51789–51800 (2017).
Chen, J. W., Lau, Y. Y., Krishnan, T., Chan, K. G. & Chang, C. Y. Recent advances in molecular diagnosis of pseudomonas aeruginosa infection by state-of-the-art genotyping techniques. Front. Microbiol.9, 378358 (2018).
Lee, D. G. et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol.7, 1–14 (2006).
Gómez-Martínez, J. et al. Comparative genomics of Pseudomonas aeruginosa strains isolated from different ecological niches. Antibiotics. 12(5), 866 (2023).
Oakeson, K. F., Wagner, J. M., Mendenhall, M., Rohrwasser, A. & Atkinson-Dunn, R. Bioinformatic analyses of whole-genome sequence data in a public health laboratory. Emerg. Infect. Dis.23(9), 1441 (2017).
Olkkonen, E. & Löytynoja, A. Analysis of population structure and genetic diversity in low-variance Saimaa ringed seals using low-coverage whole-genome sequence data. STAR. Protocols. 4(4), 102567 (2023).
Gan, Y. et al. Analysis of whole-genome as a novel strategy for animal species identification. Int. J. Mol. Sci.25(5), 2955 (2024).
Orsi, R. H., Bowen, B. M. & Wiedmann, M. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genom.11, 1–12 (2010).
Kumar, A. S., Sowpati, D. T. & Mishra, R. K. Single amino acid repeats in the proteome world: Structural, functional, and evolutionary insights. PLoS One, 11(11), e0166854. (2016).
Siwach, P., Pophaly, S. D. & Ganesh, S. Genomic and evolutionary insights into genes encoding proteins with single amino acid repeats. Mol. Biol. Evol.23(7), 1357–1369 (2006).
Widmer, G. Diverse single-amino-acid repeat profiles in the Genus Cryptosporidium. Parasitology. 145(9), 1151–1160 (2018).
Amankwah, F. K. D., Gbedema, S. Y., Boakye, Y. D., Bayor, M. T. & Boamah, V. E. Antimicrobial potential of extract from a Pseudomonas aeruginosa isolate. Scientifica, 2022. (2022).
Choi, H. J. et al. Improved PCR for identification of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol.97, 3643–3651 (2013).
Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 30(14), 2068–2069 (2014).
Subirana, J. A. & Messeguer, X. Unique features of tandem repeats in bacteria. J. Bacteriol.202(21), 10–1128 (2020).
Denœud, F. & Vergnaud, G. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: A web-based resource. BMC Bioinform.5, 1–12 (2004).
Subirana, J. A. & Messeguer, X. Tandem repeats in Bacillus: Unique features and taxonomic distribution. Int. J. Mol. Sci.22(10), 5373 (2021).
Lee, S. et al. Individual identification with short tandem repeat analysis and collection of secondary information using microbiome analysis. Genes. 13(1), 85 (2021).
Acknowledgements
This work was supported by the Research Program for Agriculture Science and Technology Development (Project No. PJ016298) of the National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
Chaerin Kim: Design the research plan, carried out experiments, writing original draft and analyzed the data; Kwang-Kyo Oh: Analyzed the data, review the original draft; Ravi Jothi: Writing and review the original draft; Dong Suk Park: Design the research plan, analyzed the data, investigation, formal analysis, software, resources, project administration and review the original draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, C., Oh, KK., Jothi, R. et al. An innovative approach to decoding genetic variability in Pseudomonas aeruginosa via amino acid repeats and gene structure profiles. Sci Rep 14, 22610 (2024). https://doi.org/10.1038/s41598-024-73031-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-73031-5
Keywords
This article is cited by
-
Multi-omics driven biomarker discovery and pathological insights into Pseudomonas aeruginosa pneumonia
BMC Infectious Diseases (2025)