Abstract
Growing research has suggested an association between chronic inflammation and Intervertebral disc degeneration (IVDD), but whether there is a causal effect remains unknown. This study adopted two-sample Mendelian randomization (MR) approach to explore the etiological role of chronic inflammation in IVDD risk. Here, summary statistics for C-reactive protein (CRP), interleukin (IL)-1\(\mathcal {\alpha }\), IL-1\(\mathcal {\beta }\), IL-6 expression and IVDD were obtained from genome-wide association studies (GWAS) of European ancestry. MR analyses were conducted by using inverse variance weighted (IVW), Wald Ratio, weighted median, and MR-Egger method. Sensitivity analyses were conducted to assess the robustness of the results. The MR analyses suggested a lack of causal association of CRP, IL-6 , and IL-1\(\mathcal {\alpha }\) levels on IVDD (CRP-IVDD: odds ratio [OR] = 0.97, 95% confidence interval [CI] 0.86–1.09, P = 0.583; IL-6-IVDD: OR = 1.04, 95% CI 0.86–1.27, P = 0.679; IL-1\(\mathcal {\alpha }\)-IVDD: OR = 1.09, 95%CI 1.00–1.18, P = 0.058). However, there was a sign of a connection between genetically elevated IL-1\(\mathcal {\beta }\) levels and a decreased IVDD incidence (OR = 0.87, 95%CI 0.77–0.99, P = 0.03). Our findings suggest a connection between IL-1\(\mathcal {\beta }\) levels and the risk of IVDD. However, due to the support of only one SNP, heterogeneity and pleiotropy tests cannot be performed, the specific underlying mechanisms warrant further investigation.
Similar content being viewed by others
Introduction
Intervertebral Disc Degeneration (IVDD) is a common degenerative disease that is considered a major cause of low back pain (LBP), imposing a significant burden on patients and society1,2,3. Most patients can alleviate symptoms independently without significant functional impairment. However, 10–20% of patients may require more than 12 weeks for recovery, with a slow and uncertain progress that often results in residual functional loss4. Although surgical treatment is effective, it is only applicable to the advanced stage of IVDD. Neither non-surgical nor surgical strategies can effectively reverse or stop the IVDD process, and the pathogenic mechanism of IVDD is still unclear. Relevant research has proved that excessive inflammatory responses, exacerbated aging, apoptosis of the intervertebral disc (IVD) cells, and degradation of the extracellular matrix are considered key pathological features of IVDD5. In recent years, an increasing amount of evidence has shown that chronic inflammation is closely related to the progression of IVDD.
IVD consists of the annulus fibrosus (AF), the nucleus pulposus (NP) and the cartilaginous endplate (CEP), which provides mobility and compressive resistance in the spine. At in vitro experiments, degenerative changes such as AF rupture, vascular formation, and degradation extracellular matrix (ECM) within NP were observed after IVD was injected with inflammatory factors6. Inflammatory factors have been proved to promote the destruction of IVD structure by reducing ECM gene expression and promoting the expression of ECM degradation enzymes7. research have confirmed that chronic inflammation can induce oxidative damage to IVD cells, promoting apoptosis and aging of IVD cells8,9.
Various inflammatory factors (e.g. interleukin (IL)-6, IL-1\(\mathbf {\alpha }\), IL-1\(\mathbf {\beta }\), C-reactive protein (CRP)) are significantly increased in degenerated intervertebral discs. The interplay among inflammatory indicators such as interleukin (IL)-6, IL-1\(\mathbf {\alpha }\), IL-1\(\mathbf {\beta }\), and C-reactive protein (CRP) involves a mutually reinforcing process, resulting in a progressively expanding inflammatory cascade and a sustained inflammatory state. For instance, IL-1\(\mathbf {\beta }\) significantly upregulates the expression of IL-6 in intervertebral disc (IVD) cells10,11. Members of the IL-1 family, including IL-1\(\mathbf {\alpha }\) and IL-1\(\mathbf {\beta }\), are important factors mediating chronic inflammation. IL-1\(\mathbf {\beta }\) promotes apoptosis of nucleus pulposus cells (NPC) and annulus fibrosus cells (AFC) by upregulating the expression of caspase-3 and caspase-912. In addition, in clinical observational experiments, Jacobse et al.8 observed that the IL-1 and IL-6 levels of the IVDD group were significantly higher than the normal group. CRP is a serum marker of chronic inflammation, produced in the liver under the stimulation of IL-1 and IL-6. IL-1 and IL-6, as upstream stimulating factors of CRP, play a key role in the occurrence of IVDD. Studies on the pathological features of various bacterial diseases have found that the expression of CRP, IL-1, and IL-6 in plasma is mutually related13. Clinical studies have suggested that elevated serum CRP levels are associated with the occurrence of IVDD14.
Recent findings from in vitro experiments and clinical observational trials have elucidated the role of chronic inflammation in IVDD6,8,14,15, but there is still a lack of related randomized controlled studies. It is not currently clear whether chronic inflammation is a result or cause of IVDD, and observational studies cannot rule out the effects of reverse causality and confounding factors. Therefore, it is necessary to introduce some stronger alternative methods, such as Mendelian randomization (MR). Mendelian randomization solves these problems by using genetic variations as instrumental variables for testing exposures. The allele of genetic variation related to exposure is randomly assigned and is not affected by reverse causality. Therefore, this study uses the two-sample MR (TSMR) analysis method to explore the causal relationship between IL-6, IL-1\(\mathcal {\alpha }\), IL-1\(\mathcal {\beta }\), CRP, and IVDD.
Materials and methods
Study design
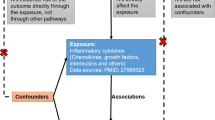
In this study, we conducted TSMR analysis to investigate potential causal associations between various inflammatory factors (i.e. CRP, IL-1\(\mathbf {\alpha }\), IL-1\(\mathbf {\beta }\) and IL-6) and IVDD. The research of this study was built upon three assumptions: (1) The instrumental variables (IVs) should be robustly associated with exposure; (2) The IVs should not be associated with potential confounders; and (3) the IVs should not have direct association with the outcomes of interest but affect the outcome exclusively through the exposure not via other biological pathways16. The schematic outline of this study is shown in Fig. 1.
Data sources
Summary data for CRP, IL-1\(\mathbf {\alpha }\), and IL-1\(\mathbf {\beta }\)-related single nucleotide polymorphisms (SNPs) were download from the genome-wide association study (GWAS) summary data (https://gwas.mrcieu.ac.uk/). The variants associated with CRP were obtained from a GWAS of up to 204,402 individuals of European ancestry17. Genetic variants for IL-1\(\mathbf {\alpha }\) were collected from a GWAS of European individuals18. The IL-1\(\mathbf {\beta }\)-related SNPs were studied from the INTERVAL study, including 3301 normal subjects19. Variants associated with IL-6 were obtained from a GWAS of up to 67,428 individuals of European ancestry20. The details of these GWAS data sources are listed in Supplementary Table 1.
The summary results for Intervertebral Disc Degeneration (IVDD) were acquired from the FinnGen consortium, including 29,508 cases and 227,388 controls21. The diagnosis of IVDD was based on ICD-10 code M51, ICD-9 code 722 and ICD-8 code 275. Other detailed information of the outcome is presented in Supplementary Table 2.
Selection of genetic instruments
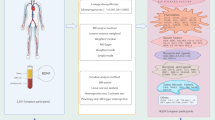
In the present TSMR analysis, we selected IVs based on consistent criteria. To identify IVs that fit the three MR assumptions, this study used the following methods to select SNPs: First, genome-wide significance analysis was performed with threshold of \(\mathbf {P < 5e-8}\) to select SNPs associated with exposures (IL-1\(\mathbf {\alpha }\), IL-1\(\mathbf {\beta }\), IL-6, and CRP)22. Second, to address linkage disequilibrium (LD), a clumping procedure with \(R^2 < 0.001\) and a window size of 10000 kb was conducted using data from the European ancestry-based 1000 Genomes Project23. Third, in order to assess weak instrumental variable bias, the F-statistics for each SNP were calculated using the equation \(\mathbf {Beta^2/SE^2}\)24, where Beta and SE represent genetic association with the exposure and standard deviation, respectively. IVs with the F-statistics values \(< 10\) were considered unreliable and were excluded from the subsequent MR analysis. Subsequently, after removing SNPs associated with IVDD at a threshold of \(P < 5e-8\)25, the palindromic SNPs were removed during harmonization to ensure that their association effects were linked with the same alleles in both the exposure and outcome. Then, the MR Pleiotropy RESidual Sum andOutlier (MR-PRESSO) method were used to eliminate potential outliers before each MR analysis26. Finally, in order to satisfy the second key assumption of MR (independence from confounders), we scanned each of the SNPs used as IVs for their potential secondary phenotypes using the GWAS Catalog (https://www.ebi.ac.uk/gwas), then the significant associations of the selected SNPs with IVDD (\(P < 5e-8\)) were excluded, including BMI, obesity and serum leptin levels27,28. After a thorough screening, the remaining SNPs were used for subsequent analyses. The study frame diagram is presented in Fig. 2.
Statistical analyses
To evaluate the causal effects of exposure on outcome, MR Analyses were performed using several methods, including inverse variance weighting (IVW)29, MR-Egger regression30, weighted median31, and Wald Ratio (for IL-1\(\mathbf {\alpha }\) and IL-1\(\mathbf {\beta }\),because single SNP is available), with the IVW method or Wald Ratio method (for a single SNP) being the primary analysis, and the MR-Egger regression and the weighted median served as an auxiliary method to enhance the reliability of our finding. The IVW assumes that all SNPs are valid genetic variants and that there is no pleiotropy32. The MR-Egger method has the advantage of being less susceptible to directional pleiotropy and can provide causal estimates even if all IVs are invalid, but the statistical power of MR-Egger is low30. The weighted median method is less sensitive to outliers but generally less efficient31.
Experimental conditions, analysis platforms and different study subjects may contribute to heterogeneity, resulting in biased causal effect estimates. In this study, Cochran’s Q statistics were used to test heterogeneity in causal estimates. A P value \(< 0.05\) of Cochran’s Q statistics was considered significant heterogeneity, thereby random-effects model was employed; otherwise there was no heterogeneity and a fixed effects model was used33. Pleiotropy was evaluated using the MR-Egger intercept test based on the intercepts and P value30. If the MR-Egger regression intercept was close to \(0 (< 0.1)\) and P \(> 0.05\), there was no evidence of horizontal pleiotropy in the test. Additionally, leave-one-out analysis was applied to measure the dependability of the results. Namely, each SNP was removed sequentially and then reperformed MR analysis on the remaining SNPs to identify potential influential SNPs.
All statistical analyses were performed using the Two-Sample MR and MR-PRESSO packages in R version 4.2.1. To account for multiple testing, the Bonferroni method was used to adjust the test level as \(\mathbf {\alpha }\) = 0.05/number of statistical tests. P value below 0.0125 (where P = 0.05/4) represented strong evidence of causal association, and P value below 0.05 but above 0.0125 were considered suggestive evidence of association in MR Analysis.
Results
Causal effects of CRP on IVDD
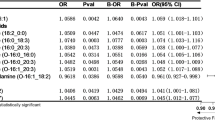
In our study, 46 SNPs of CRP-IVDD were incorporated as IVs for the analysis of CRP and the risk of IVDD. Detailed information on the selected IVs for CRP is listed in Supplementary Table 3. No SNP was excluded after comprehensive lookup of the GWAS Catalog (Supplementary Table 4). The F-statistics values for CRP-related SNPs ranged from 29 to 1245, indicating a strong association between IVs and exposure. The results for the causal analysis of CRP on IVDD are presented in Fig. 3. The main results of IVW found no evidence of a causal effect between IVDD and CRP (OR, 0.97; 95% CI 0.86–1.09; P = 0.583). No significant relationship result was yielded using the weight median method (OR, 1.04; 95% CI 0.96–1.12; P = 0.290). Noted that the MR Egger method showed a suggestive associations between CRP levels and risk of IVDD (OR, 1.11; 95% CI 1.01–1.22; P = 0.038). Cochrane’s Q results suggested some significant differences in the causal estimates between CRP levels and IVDD risk (Cochrane’s Q in MR-Egger = 74.62, P = 2.67E-03; Cochrane’s Q in IVW = 78.83, P = 1.34E−03, Table 1). In this scenario, the IVW random effects method was employed, because it addresses the heterogeneity issue of variant-specific causal estimates, compared to the fixed effects model34. P value \(> 0.05\) was obtained for MR-Egger intercept test, suggesting the absence of horizontal pleiotropy(intercept = 4.94E−03, P = 0.122, Table 1). As is shown in Fig. 4A, the leave-one-out analysis revealed that causal estimates for CRP and IVDD were not influenced by a single SNP, indicating that the MR results were robust. The funnel plot showed general symmetry, suggesting the absence of directional horizontal pleiotropy (Fig. 4B). The scatter plot and forest plot were shown in Supplementary Figs. S1 and S2.
Causal effects of IL-6 on the risk of IVDD
A total of 2 SNPs were included as IVs for the analysis of IL-6 and the risk of IVDD. Detailed information on the selected IVs for IL-6 is listed in Supplementary Table 3. No SNP was excluded after comprehensive lookup of the GWAS Catalog (Supplementary Table 4). The range of F-statistics for SNPs related to IL-6 is between 36 and 398, suggesting a strong correlation between the IVs and the exposure. No evidence of a causal association between IL-6 and IVDD was identified (OR, 1.04; 95% CI 0.86–1.27; P = 0.679). The heterogeneity test revealed no evidence of heterogeneity (Cochrane’s Q in IVW = 1.77, P = 1.82E−01).
Causal effects of IL-1\(\mathbf {\alpha }\) on the risk of IVDD
One SNP was selected as the instrument variant for the analysis of IL-1\(\alpha\) and the risk of IVDD. Detailed information of the selected instrument variant was listed in Supplementary Table 3. No SNP was excluded after comprehensive lookup of the GWAS Catalog (Supplementary Table 4). F-statistics value for the IL-1\(\mathbf {\alpha }\)-associated SNP was 31.36, suggesting that the SNP was unlikely to be affected by weak instrument bias. MR Estimates showed no significant relationship between genetically predicted IL-1\(\mathbf {\alpha }\) levels and IVDD (OR, 1.09; 95% CI 1.00–1.18; P = 0.058). The heterogeneity test could not be performed at IL-1\(\mathbf {\alpha }\) because there was only a single SNP.
Causal effects of IL-1\(\mathbf {{\beta }}\) on the risk of IVDD
In the analysis of IL-1\(\beta\) and the risk of IVDD, one SNP was included, with Supplementary Table 3 providing detailed information on the instrument variant for IL-1\(\mathbf {\beta }\). No SNP was excluded after comprehensive lookup of the GWAS Catalog (Supplementary Table 4). With the F-statistic value of 34.20 for the SNP associated with IL-1\(\mathbf {{\beta }}\), it suggested that there was a strong correlation between the instrument variant and the exposure. The Wald Ratio result suggested suggestive association between genetically elevated IL-1\(\mathbf {{\beta }}\) levels and a decreased risk of IVDD(OR, 0.87; 95% CI 0.77–0.99; P = 0.03).
Discussion
In this study, TSMR analysis was employed to study a potential causal relationship between CRP, IL-6, IL-1\(\mathbf {{\alpha }}\) and IL-1\(\mathbf {{\beta }}\) on Intervertebral disc degeneration(IVDD). No evidence was discovered to support a causal relationship between IL-1\(\alpha\), IL-6 and CRP in the occurrence of IVDD. The present study discovered a correlation between genetically increased IL-1\(\beta\) expression and a lower risk of IVDD, but this finding is based on just one SNP and lacks heterogeneity and horizontal pleiotropy analysis. Sensitivity analyses also suggested that IVs affected outcomes only through exposure, not through confounding and other pathways. Therefore, this means that there is no pleiotropy.
Previous studies have confirmed a strong association between CRP, IL-6, IL-1\(\mathbf {\alpha }\), and IL-1\(\mathbf {\beta }\) with IVDD. CRP is a biomarker for monitoring infections and various diseases (including IVD protrusion, rheumatism, etc.). Clinical observational studies found that plasma CRP levels in patients with lumbar IVDD were significantly higher than those patients with lumbar pain who had normal MRI findings14. Similar findings were also noted in cervical IVDD35. In vitro experiments found that CRP had a significant pathogenic effect in the intervertebral disc. CRP can also cause a sharp increase in the expression of inflammatory mediators in normal IVD cells36. It had been revealed by in vitro experiments that there was a balance between IL-1 (IL-1\(\mathbf {\alpha }\) and IL-1\(\mathbf {\beta }\)) and its inhibitor IL-1Ra in normal IVD cells, which can maintain local homeostasis37. IL-1\(\mathbf {\beta }\) and IL-1\(\mathbf {\alpha }\) are significantly increased in degenerated intervertebral disc (IVD) tissues and cells, leading to an increase in degrading enzymes and a decrease in matrix protein gene expression38,39. Clinical observational studies found that IL-6 levels were significantly increased in IVDD patients15. A meta-analysis involving eight studies found that IL-6 levels were positively correlated with the degree of disc protrusion40.
There has been a large amount of research on the mechanisms driving IVDD by inflammatory factors such as CRP, IL-1 (IL-1\(\mathbf {\alpha }\) and IL-1\(\mathbf {\beta }\)), and IL-6, but these mechanisms have not been fully revealed. The association between CRP, IL-1 (IL-1\(\mathbf {\alpha }\) and IL-1\(\mathbf {\beta }\)), and IL-6 is intricate, with these inflammatory factors reciprocally inducing one another and thereby leading to a stepwise amplification of the inflammatory response. For example, CRP can increase IL-6 and IL-8 expression in IVD AF cells36, and IL-1\(\mathbf {\beta }\) stimulation significantly enhances IL-6 and IL-8 expression in human IVD cells10,11. CRP is produced in the liver under the stimulation of IL-1 and IL-6. In vivo experiments, degenerative changes including ECM degradation and AF rupture can be observed after IVD injection of inflammatory factors6. In normal IVD cells, inflammatory factors and their inhibitors are in a dynamic equilibrium, but in IVDD cells, inflammatory factors such as IL-1\(\mathbf {\beta }\) are significantly increased, the expression of ECM degradation metabolites such as matrix metalloproteinases (MMP) is increased, and ECM synthesis metabolism does not significantly increase, resulting in ECM degradation and IVD structure destruction7,41,42. There was research proving that inflammatory factors can promote IVD cell aging and apoptosis. Inflammatory factors can promote an increase in blood vessels on the IVD surface43, as well as nerve growth into the IVD, increasing the risk of back pain44. All of these mechanisms contribute to the gradual instability of the IVD, leading to lumbar disc degeneration, protrusion, and back pain.
Although a large number of literature have proved the correlation between CRP, IL-1\(\mathbf {\alpha }\), and IL-6 with IVDD45, our research findings indicate that CRP, IL-1\(\mathbf {\alpha }\), and IL-6 have no significant causal relationship with IVDD. Our speculation is that these three inflammatory factors do not directly cause IVDD but may be reactive elements to other chronic inflammatory risk factors, which requires further in-depth research. A MR study45 found that increased IL-6 may be associated with a reduced risk of LBP. Our research findings differ from this MR study, possibly because IVDD is only a part of the spectrum of LBP diseases, with varying levels of IL-6 in different diseases within the LBP spectrum. Weber et al.15 found that compared to patients with LBP caused by disc herniation, patients with LBP resulting from spinal stenosis and IVDD had higher IL-6 levels.
Our research shows that elevated levels of IL-1\(\mathbf {\beta }\) may reduce the risk of IVDD, which is contrary to the conclusions of many observational studies. However, since only one SNP is supported, heterogeneity and gene pleiotropy tests cannot be performed, so we are more cautious in interpreting this result. Research found that the IL-1bT3954 allele is a common IL-1\(\mathbf {\beta }\) gene polymorphism, which has been proven to be associated with elevated levels of IL-1\(\mathbf {\beta }\)46 and has the potential to reduce the risk of disc degeneration47. However, the specific underlying mechanism needs to be further studied in future experiments.
This study conducted the first TSMR study to explore the etiological role of chronic inflammation in IVDD risk, which utilized the MR method to significantly reduce the common confounding factors and reverse causation bias in observational studies. However, our study also had some limitations. Firstly, we selected individuals of European ancestry as our study subjects to reduce population stratification bias. Consequently, the generalization of our findings to other racial groups may be subject to limitations. Second, we merely investigated the causal relationship between CRP, IL-1, IL-6 expression levels and IVDD, and did not analyze other biomarkers and transcription factors of chronic inflammation, which may also be critical for the development of IVDD. Thirdly, we only used genetic tools to assess the causal relationship between inflammatory biomarkers and IVDD risk, and further mechanistic studies are needed to elucidate our findings.
Conclusion
Our research used Mendelian randomization methods and found that there was no significant causal relationship between the levels of CRP, IL-1\(\mathbf {\alpha }\), and IL-6 and susceptibility to IVDD. A decrease in IL-1\(\mathbf {\beta }\) levels may increase the risk of IVDD.
Data availibility
All data generated or analysed during this study are included in this published article and its supplementary information files. Further inquiries can be directed to the corresponding authors.
References
Vos, T. et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the global burden of disease study 2019. Lancet 396, 1204–1222 (2020).
Moore, R. J., Vernon-Roberts, B., Fraser, R. D., Osti, O. L. & Schembri, M. The origin and fate of herniated lumbar intervertebral disc tissue. Spine 21, 2149–2155 (1996).
Weber, K. T. et al. Developments in intervertebral disc disease research: Pathophysiology, mechanobiology, and therapeutics. Curr. Rev. Musculoskelet. Med. 8, 18–31 (2015).
Andersson, G. B. Epidemiological features of chronic low-back pain. Lancet 354, 581–585 (1999).
Adams, M. A. & Roughley, P. J. What is intervertebral disc degeneration, and what causes it?. Spine 31, 2151–2161 (2006).
Kang, R. et al. Intervertebral disc degenerative changes after intradiscal injection of tnf-\(\alpha\) in a porcine model. Eur. Spine J. 24, 2010–2016 (2015).
Le Maitre, C., Pockert, A., Buttle, D., Freemont, A. & Hoyland, J. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem. Soc. Trans. 35, 652–655 (2007).
Jacobsen, H. E., Khan, A. N., Levine, M. E., Filippi, C. G. & Chahine, N. O. Severity of intervertebral disc herniation regulates cytokine and chemokine levels in patients with chronic radicular back pain. Osteoarthritis Cartilage 28, 1341–1350 (2020).
Song, J. et al. Exosome-transported circrna_0000253 competitively adsorbs microrna-141-5p and increases idd. Mol. Therapy-Nucleic Acids 21, 1087–1099 (2020).
Jia, J., Nie, L. & Liu, Y. Butyrate alleviates inflammatory response and nf-\(\kappa\)b activation in human degenerated intervertebral disc tissues. Int. Immunopharmacol. 78, 106004 (2020).
Jimbo, K., Park, J. S., Yokosuka, K., Sato, K. & Nagata, K. Positive feedback loop of interleukin-1\(\beta\) upregulating production of inflammatory mediators in human intervertebral disc cells in vitro. J. Neurosurg. Spine 2, 589–595 (2005).
Huang, J.-F. et al. Sinapic acid inhibits il-1\(\beta\)-induced apoptosis and catabolism in nucleus pulposus cells and ameliorates intervertebral disk degeneration. J. Inflamm. Res 13,883–895 (2020).
Pepys, M. B. et al. C-reactive protein: A critical update. J. Clin. Investig. 111, 1805–1812 (2003).
Durdag, E. et al. The importance of c-reactive protein in discogenic low back pain: The analysis of 444 patients. J. Turk. Spinal Surg. 29, 115–118 (2018).
Weber, K. T. et al. Serum levels of the proinflammatory cytokine interleukin-6 vary based on diagnoses in individuals with lumbar intervertebral disc diseases. Arthritis Res. Therapy 18, 1–14 (2016).
Emdin, C. A., Khera, A. V. & Kathiresan, S. Mendelian randomization. Jama 318, 1925–1926 (2017).
Ligthart, S. et al. Genome analyses of> 200,000 individuals identify 58 loci for chronic inflammation and highlight pathways that link inflammation and complex disorders. Am. J. Hum. Genet. 103, 691–706 (2018).
Suhre, K. et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 8, 14357 (2017).
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Ahluwalia, T. S. et al. Genome-wide association study of circulating interleukin 6 levels identifies novel loci. Hum. Mol. Genet. 30, 393–409 (2021).
Kurki, M.I. et al. Finngen: Unique genetic insights from combining isolated population and national health register data. MedRxiv . 2022–03 (2022).
Davies, N.M., Holmes, M.V. & Smith, G.D. Reading mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ. 362:k601 (2018).
Consortium et al. A map of human genome variation from population scale sequencing. Nature. 467, 1061 (2010).
Perry, B. I. et al. Associations of immunological proteins/traits with schizophrenia, major depression and bipolar disorder: A bi-directional two-sample mendelian randomization study. Brain Behav. Immunity 97, 176–185 (2021).
Ren, L. & Wang, G. Causality between sarcopenia and diabetic nephropathy: A bidirectional mendelian randomization study. Front. Endocrinol. 14, 1188972 (2023).
Verbanck, M., Chen, C.-Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Hsu, H.-T. et al. Immuohistochemical score of matrix metalloproteinase-1 may indicate the severity of symptomatic cervical and lumbar disc degeneration. Spine J. 20, 124–137 (2020).
Chen, H.-H., Hsu, H.-T., Liao, M.-H. & Teng, M.-S. Effects of sex and obesity on lep variant and leptin level associations in intervertebral disc degeneration. In. J. Mol. Sci. 23, 12275 (2022).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments effect estimation and bias detection through egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Hemani, G. et al. The mr-base platform supports systematic causal inference across the human phenome. eLife. 7, e34408 (2018).
Burgess, S. et al. Guidelines for performing mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 4: 186 (2019).
Zhou, T. et al. Causal association between whole-body water mass and sleep apnea: A mendelian randomization study. Ann. Am. Thoracic Society 19, 1913–1919 (2022).
Ethemoğlu, K.B. & Erkoç, Y.S. Is there any relationship between cervical disc herniation and blood inflammatory response? Cureus. 12(8): e10161 (2020).
Ruiz-Fernández, C. et al. Monomeric crp regulates inflammatory responses in human intervertebral disc cells. Bone Joint Res. 12, 189–198 (2023).
Le Maitre, C. L., Freemont, A. J. & Hoyland, J. A. The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res. Therapy 7, 1–14 (2005).
Burke, J. et al. Intervertebral discs which cause low back pain secrete high levels of proinflammatory mediators. J. Bone Joint Surg. Br. 84, 196–201 (2002).
Li, P.-B., Tang, W.-J., Wang, K., Zou, K. & Che, B. Expressions of il-1\(\alpha\) and mmp-9 in degenerated lumbar disc tissues and their clinical significance. Eur. Rev. Med. Pharmacol. Sci. 21: 4007-4013 (2017).
Fan, H.-T., Jiang, W.-H., Zhang, B., Wang, L. & Liu, S.-B. Elevated il-6 levels correlate with the development of intervertebral disc degeneration: A case-control study and meta-analysis. Int. J. Clin. Exp. Med. 9, 1015–1026 (2016).
Wang, W.-J. et al. Mmps and adamtss in intervertebral disc degeneration. Clinica Chimica Acta 448, 238–246 (2015).
Ma, H. et al. Mfg-e8 alleviates intervertebral disc degeneration by suppressing pyroptosis and extracellular matrix degradation in nucleus pulposus cells via nrf2/txnip/nlrp3 axis. Cell Death Discov. 8, 209 (2022).
Kwon, W.-K., Moon, H. J., Kwon, T.-H., Park, Y.-K. & Kim, J. H. The role of hypoxia in angiogenesis and extracellular matrix regulation of intervertebral disc cells during inflammatory reactions. Neurosurgery 81, 867–875 (2017).
Miyagi, M. et al. Increased innervation and sensory nervous system plasticity in a mouse model of low back pain due to intervertebral disc degeneration.: O56 (issls prize paper). in Spine Journal Meeting Abstracts, 39–40 (LWW, 2014).
Li, W. et al. Assessing the causal relationship between genetically determined inflammatory biomarkers and low back pain risk: A bidirectional two-sample mendelian randomization study. Front. Immunol. 14, 1174656 (2023).
Pociot, F., Mølvig, J., Wogensen, L., Worsaae, H. & Nerup, J. A taql polymorphism in the human interleukin-1\(\beta\) (il-1\(\beta\)) gene correlates with il-1\(\beta\) secretion in vitro. Eur. J. Clin. Investig. 22, 396–402 (1992).
Solovieva, S. et al. Intervertebral disc degeneration in relation to the col9a3 and the il-1 ß gene polymorphisms. Eur. Spine J. 15, 613–619 (2006).
Acknowledgements
We are grateful to the IEU Open GWAS project and the GWAS Catalog for supplying data on summary statistics for MR analyses. We also want to acknowledge the original studies for sharing the GWAS data used in this project.
Author information
Authors and Affiliations
Contributions
X.Y. conceived the presented idea and drafted the manuscrip. A.C. and E.S. performed the analysis. Y. Z. and F.Z. reviewed the manuscript. All authors discussed the results and contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yan, Xa., Shen, E., Cui, A. et al. Assessing the causal relationship between CRP, IL-1α, IL-1β, and IL-6 levels and intervertebral disc degeneration: a two-sample Mendelian randomization study. Sci Rep 14, 23716 (2024). https://doi.org/10.1038/s41598-024-73205-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-73205-1
Keywords
This article is cited by
-
The interleukin gene landscape: understanding its influence on inflammatory mechanisms in apical periodontitis
Molecular Biology Reports (2025)