Abstract
Plasma epidermal growth factor receptor mutation (EGFRm) circulating tumor DNA (ctDNA) dynamics exhibit promise in predicting outcomes in patients with EGFRm-advanced non-small cell lung cancer (NSCLC). However, there remains limited trial-level data on integrating ctDNA monitoring into clinical practice. We performed a prospective, multicenter trial to investigate the relationship between EGFRm ctDNA dynamic changes and clinical outcomes in NSCLC patients with EGFRm. Ninety-eight treatment-naive EGFRm-advanced NSCLC patients were recruited and administered icotinib until disease progression. Plasma samples were collected at baseline and four weeks after icotinib administration. ctDNA was analyzed using a droplet-digital polymerase chain reaction. At baseline, 71.4% of patients had detectable EGFRm ctDNA. Among them, 45.9% of patients’ ctDNA became undetectable within four weeks of treatment. These patients demonstrated significantly longer progression-free survival (PFS) and overall survival (OS) than those with detectable ctDNA after treatment (P = 0.004 and < 0.001, respectively) and were comparable to those with undetectable ctDNA at both baseline and four weeks. ctDNA detectable at four weeks emerged as a poor independent risk factor for PFS and OS. Patients whose ctDNA became undetectable after treatment had outcomes similar to those with initially undetectable ctDNA, underscoring the predictive value of ctDNA dynamics in treatment efficacy.
Registry and the Registration No. of the study/trial: ChiCTR-DDD-17013131. Date of registration: 2017-10-27.
Similar content being viewed by others
Introduction
Epidermal growth factor receptor mutations (EGFRm) are the most common targetable oncogenic driver mutation in advanced non-small cell lung cancer (NSCLC)1,2. Over the last two decades, there has been a significant change in the treatment landscape of advanced sensitive EGFRm NSCLC. Multiple EGFR-tyrosine kinase inhibitors (TKIs) have been approved for clinical use, including first-generation inhibitors erlotinib3, gefitinib4, and icotinib5; second-generation inhibitors afatinib6 and dacomitinib7; and third-generation inhibitors, osimertinib8. Additionally, the combination strategies of EGFR-TKI with other anticancer agents, such as chemotherapy or angiogenesis inhibitors, are also investigated to prolong patient survival. The Flaura 2 trial demonstrated that osimertinib plus chemotherapy provided a statistically significant and clinically meaningful progression-free survival (PFS) benefit over osimertinib monotherapy9.
Despite initially promising response rates to EGFR-TKIs (50–80%)3,4,5,6,7,8, significant heterogeneity in the duration of clinical benefit and pattern of progression has been observed. Therefore, searching for predictive biomarkers is one of the main issues to enhancing patient outcomes10. Moreover, the optimal sequence of targeted therapies remains unclear11. Currently, no clinical or molecular predictive markers are available to identify patients who could benefit from different therapeutic approaches. Finally, the timing and instigation of drug combinations require further investigation9,11.
Circulating tumor DNA (ctDNA) consists of DNA fragments shed by cancer cells into the plasma of patients with malignancies and represents a small fraction of total circulating cell-free DNA12,13. Significant advantages of ctDNA include its non-invasive nature and a presumed better representation of tumor heterogeneity compared to tissue biopsies. The use of ctDNA with particular and sensitive techniques, such as droplet digital polymerase chain reaction (ddPCR) and next-generation sequencing (NGS), has shown the potential for longitudinal monitoring of treatment outcomes, detection of minimal residual disease (MRD), prognosis, and selection of appropriate treatments10,12,14. Serial plasma analysis of advanced NSCLC patients with undetectable EGFRm ctDNA after three to six weeks of treatment showed a longer median PFS than those with detectable ctDNA15. However, although these applications show promise, their routine integration into clinical practice needs further evidence. There remains limited trial-level data in advanced NSCLC regarding the association between dynamic changes in ctDNA detection and EGFR-TKI treatment outcomes.
ddPCR is a well-validated ctDNA testing method for specific target genes and offers more time and cost-effectiveness than NGS. Here, we reported our real-world, prospective, multi-center phase II trial with a cohort of advanced NSCLC patients with EGFRm undergoing icotinib as first-line treatment. Serial plasma samples were taken from participants for EGFRm ctDNA analysis and were analyzed using ddPCR. The study aims to explore correlations between baseline and on-treatment EGFRm ctDNA dynamics and clinical outcome data at a trial level.
Results
Patient characteristics
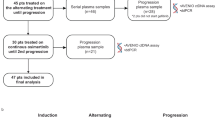
From January 2017 to November 2018, 100 eligible participants were enrolled, while two withdrew their informed consent. Finally, 98 patients were included in the analysis. Figure 1 displays the study flow diagram. All patients received icotinib as their initial treatment. Fifty-eight patients (59.2%) were female, and the median age of the patients was 61 years (interquartile range, IQR 53–69). 86% of patients were never smokers. Eighty-one patients (82.7%) had an adenocarcinoma histologic subtype. Most patients had stage IV NSCLC (93.9%). A total of 40 patients (40.8%) had brain metastasis. Bone metastasis was identified in 39 (39.8%) patients. The EGFR Ex 19del was present in 50 (51%) patients, and the EGFR L858R mutation was present in 48 (49%) patients. Seven (7.1%) patients had positive EGFR T790M mutations in plasma at baseline. At the time of progression on first-line therapy, 36 (36.7%) patients detected acquired-resistant T790M, while 45 (45.9%) patients had no acquired-resistant T790M. Moreover, 17 (17.3%) patients did not undergo further genetic testing. After first-line progression, 32 patients had received osimertinib, two patients had received furmonertinib, 16 had not received treatment with third-generation EGFR-TKIs, and it was unknown whether 48 patients had received third-generation EGFR-TKIs. Key demographic and baseline clinical characteristics are summarized in Table 1.
Clinical outcomes
The data cut-off was January 2, 2023. After a median follow-up time of 57.5 (95% CI 49.2–65.8) months, 78 patients (79.6%) had experienced disease progression and 60 patients (61.2%) had died. No patients achieved complete response (CR); sixty-seven patients had partial response (PR); 24 had stable disease (SD); and seven had progressive disease (PD). ORR was 68% (95% CI 58–77%), and DCR was 93% (95% CI 85–97%). The median PFS was 10.7 months (95% CI 8.9–12.6), with a 12 month PFS probability of 43.9% (95% CI 34.4–55.9%) and a 24-month PFS of 9.6% (95% CI 4.6–20.0%). Moreover, the median OS was 29.9 months (95% CI 23.6–36.2 months), with a one-year OS probability of 76.2% (95% CI 67.8–85.7%), a two-year OS of 58.4% (95% CI 48.5–70.5%), and a three-year OS of 40.1% (95% CI 30.1–53.3%).
Detection rate of baseline ctDNA and clearance of ctDNA after treatment
Of the 98 patients, seventy (71.4%) had baseline detectable EGFRm ctDNA with a median VAF of 4.1% (IQR 1.2–16.8). Nver-smokers had a baseline EGFRm ctDNA detectability of 77.1% (64/83), compared to 40.0% (6/15) in ever-smokers (P = 0.010) (Table 1). EGFRm ctDNA detectability was higher in patients with pleural or bone metastasis than those without and in non-postoperative recurrence patients than in postoperative recurrence patients (P = 0.011, 0.005 and 0.041, respectively). There was no significant difference in the baseline ctDNA detectability among the clinical stages or EGFR mutation (P = 0.205, 0.443, respectively). Spearman’s rank correlation coefficient analysis confirmed a positive correlation (r = 0.238, P = 0.0183) between patients’ baseline EGFRm ctDNA VAF and the sum of target lesions size (Fig. S1).
After four weeks of icotinib administration, forty-five (45.9%) patients with baseline detectable plasma EGFRm became undetectable; 25 (25.5%) patients with baseline detectable plasma EGFRm remained detectable; 26 (26.5%) patients with baseline undetectable plasma EGFRm remained undetectable; and two (2.0%) patients with baseline undetectable plasma EGFRm changed to detectable.
Association of clinical responses with plasma EGFRm ctDNA
The clinical responses of patients with baseline detectable and undetectable EGFRm ctDNA are presented in (Table S1). ORR did not differ significantly between those with baseline detectable and undetectable EGFRm ctDNA (66 vs. 44%, P = 0.372). There was also no difference in DOR between patients with detectable and undetectable EGFRm ctDNA at baseline (91 vs. 96%, P = 0.385). In the 70 patients with baseline detectable plasma EGFRm, the baseline plasma EGFRm VAF distribution had no significant differences in patients with different subgroups of best overall responses (4.4, 3.8, and 13.1%, respectively; P = 0.495) (Fig. S2).
We classified patients into three groups: Group A, both baseline and four-week on-treatment ctDNA undetectable (n = 26), versus Group B, baseline ctDNA detectable but undetectable at four weeks (n = 45) (clearance), and Group C, detectable at four weeks (n = 27) (non-clearance). The clinical responses of patients in three groups are summarized in (Table S2). The best overall response, disease control, and objective response did not differ between patients in groups A and B (P = 0.787, > 0.999, and 0.501, respectively) (Table SS3 Patients in Group B had a higher ORR than those in Group C (80.0 vs. 44.4%, P = 0.002) but did not have a higher DOR (95.6 versus 85.2%, P = 0.188) (Table S4).
Correlation of progression-free survival with plasma EGFRm ctDNA
The median PFS for patients with baseline detectable plasma EGFRm was 10.1 months (95% CI 7.4–12.8 months), compared to 11.4 months (95% CI 7.4–12.8 months) for those with undetectable EGFRm (P = 0.384) (Fig. 2A). Among patients who achieved a PR, there were no significant differences in median PFS between the detectable and undetectable baseline EGFRm groups (12.3 vs. 11.4 months, P = 0.985). Similar outcomes were observed in patients with SD or PD (P = 0.240 and 0.432, respectively).
Survival analysis showed median PFS of 12.3 months (95% CI 10.2–18.2 months), 12.3 months (95% CI 9.4–16.4 months), and 7.6 months (95% CI 4.8–12.3 months) in Group A versus Group B and Group C (P = 0.003) (Fig. 2B). PFS did not differ significantly between Group A and Group B (HR 1.01 [95% CI 0.59–1.74], P = 0.958). Group A or Group B showed significantly better PFS than Group C (HR 0.42 [95% CI 0.22–0.78], P = 0.006; HR 0.43 [95% CI 0.25–0.76], P = 0.004, respectively). In univariable analysis, groups (Group A/B vs. Group C), in addition to pleural metastasis or bone metastasis and the best overall response (PR vs. PD), were found to be prognostic factors for PFS (Table 2). The multivariable analysis revealed that the best overall response (PR vs. PD), EGFR mutation (ex19del vs. L858R), age (< 61 vs.≥ 61), and groups (Group A/B vs. Group C) were independent prognostic factors for PFS. PFS plots for HR are shown in (Fig. 3).
Correlation of overall survival with plasma EGFRm ctDNA
The Kaplan-Meier analysis showed that the median OS for patients without baseline detectable plasma EGFRm was 34.0 months (95% CI 26.3–41.7 months) compared with 28.5 months (95% CI 19.5–37.6 months) for patients with baseline detectable plasma EGFRm (Fig. 4A). However, differences in survival outcomes between the baseline detectable and undetectable EGFRm subgroups were insignificant (hazard ratio (HR) 0.85; 95% CI 0.48–1.49; P = 0.573). Among patients achieving PR, there was no difference in median OS between the detectable and undetectable baseline EGFRm subgroups (37.0 vs. 34.0 months, P = 0.550). Patients with SD or PD gained similar results (P = 0.221 and 0.247, respectively).
There was no difference in overall survival between Group A and B (HR 0.87 [95% CI 0.46–1.65], P = 0.664). Patients with Group A or B had a more prolonged survival than Group C (34.0 months [95%CI 29.1-NA months], 38.2 months [95% CI 29.9–47.9 months] vs. 12.1 months [95% CI 9.9–28.5 months] P < 0.001) (Fig. 4B). Kaplan-Meier curves for OS according to the groups are shown in (Fig. 4B). Univariate analysis identified that groups (Group A/B vs. C), liver metastasis, bone metastasis, EGFR mutation (ex19del vs. L858R), and best overall response (PR vs. PD) were considered significant predictors for OS (Table 3). Multivariate analysis confirmed that groups (Group A/B vs. Group C), EGFR mutation (ex19del vs. L858R), and best overall response (PR vs. PD) remained independent prognostic factors for predicting favorable OS. Liver metastasis, bone metastasis, and unknown acquired T790M status were identified as independent factors for predicting worse OS (Table 3). OS plots for HR are shown in Fig. 5.
Discussion
Further studies are required to comprehend the potential implications of EGFRm ctDNA monitoring. In this prospective, multi-center trial, we explored the relationship between baseline and four-week on-treatment plasma EGFRm ctDNA dynamics and clinical outcomes in a real-world NSCLC patient population treated with icotinib. There was no significant correlation between baseline plasma EGFRm detectability, objective response, disease control, or the best overall response. Survival analysis revealed that plasma EGFRm ctDNA detectable at baseline did not lead to a shorter PFS or OS. Notably, patients with baseline detectable plasma EGFRm achieving clearance after four weeks of treatment (Group B) exhibited similar PFS and OS to those with baseline and four weeks of undetectable plasma EGFRm (Group A). Both groups showed longer PFS and OS than those with detectable EGFRm at four weeks (Group C). Detectable plasma EGFRm at week four was the independent inferior predictor of PFS and OS. Serial plasma EGFRm ctDNA testing could aid in predicting clinical outcomes in advanced NSCLC patients with EGFRm who received icotinib.
Previous studies mainly compared the differences in PFS and OS between patients with detectable and non-detectable EGFR ctDNA at the baseline or between the ctDNA clearance and non-clearance groups after treatment16,17,18,]. The WJOG8114LTR study19 is a prospective study that included 57 patients with EGFRm NSCLC receiving afatinib treatment. The results showed that ctDNA clearance within four weeks was associated with a longer PFS than patients whose ctDNA was unclear. Gray et al.15 analyzed data from the AURA3 and FLAURA trials to explore the potential of early clearance (3/6 weeks) of EGFRm in plasma as a predictive marker for treatment outcomes with osimertinib in advanced NSCLC and obtained similar results. Rare studies have compared the clinical outcome between patients with baseline detectable ctDNA but achieved early ctDNA clearance after treatment and those with baseline undetectable ctDNA. In our study, we investigated the differences in clinical outcomes among patients with both baseline and four weeks of treatment undetectable plasma EGFRm ctDNA, patients with EGFRm ctDNA detectable at baseline and undetectable after treatment, and those who failed to achieve EGFRm ctDNA clearance after treatment. Intriguingly, patients with EGFRm detectable at baseline and clearance after treatment showed fairly consistent clinical outcomes to those who had both baseline and four weeks of on-treatment EGFRm ctDNA undetectable and had experienced longer PFS and OS compared to those who failed to achieve ctDNA clearance. These results suggest that patients who achieve early ctDNA clearance may gain more clinical benefits from EGFR-TKI treatment.
After adjustment for clinical factors, patients with detectable ctDNA at four weeks of treatment remained a poor prognostic factor in PFS and OS. This subset of patients might be the optimal candidates for early switching to third-generation inhibitors or combination therapy. Confirmation will be needed from future clinical studies.
Icotinib is a widely used EGFR-TKI in certain regions (especially China). However, there are few trial-level studies on the related dynamic change of ctDNA in advanced NSCLC patients. Most research focused on using plasma EGFR ctDNA genotyping to guide icotinib treatment20. Different EGFR-TKIs may have subtle pharmacokinetic differences that could affect the dynamics of ctDNA clearance. Our study is a prospective, multicenter, biomarker-directed observation trial evaluating the dedicated predictive value of the dynamic change of plasma EGFRm ctDNA to icotinib in advanced EGFRm NSCLC patients. The time point of 4 weeks for ctDNA aligns with routine clinical follow-up and imaging assessments, facilitating implementation in actual clinical practice. We think the results of this study provide new perspectives and potential methods for individualized detection in patients administered icotinib.
In most patients (71.4%), plasma EGFRm ctDNA was detectable prior to treatment initiation with icotinib. cfDNA detectability may be influenced by many factors, such as age or smoking status21. In general, cfDNA detectability in older patients is higher than in younger ones. In oncological patients, the concentration of ctDNA may vary depending on the cancer type, stage of disease (especially tumor size), genotypes, and presence of local or distant metastases22,23. In our study, the results showed that smoking history, non-postoperative recurrence, the presence of baseline pleural metastases, and bone metastases were associated with EGFRm ctDNA detectability and a moderate but statistically significant correlation between EGFRm ctDNA VAF and tumor size. These data are consistent with those of previous studies.
Plasma ctDNA concentrations have been associated with tumor load and metastatic burden, suggesting it as a prognostic biomarker10,19,24,25. Early dynamic changes in plasma EGFRm may predict clinical outcomes in NSCLC patients under EGFR-TKI treatment. In the context of the AURA study15, patients with undetectable EGFRm in plasma after six weeks of osimertinib had better ORR and median PFS. Plasma EGFRm analysis can also predict outcomes as early as three weeks after starting treatment with osimertinib. Our study found that patients whose plasma EGFRm ctDNA was detectable after four weeks of treatment had a worse prognosis than those with undetectable EGFRm ctDNA. Monitoring plasma ctDNA changes over time may offer insight into treatment effects in patients with EGFRm NSCLC.
The strength of our study is the prospective, multi-center biomarker-directed trial that defined the collection of plasma samples at baseline and after that at clearly defined time points while on therapy. There are some limitations to the current study. First, due to budget limitations, our study did not incorporate plasma EGFRm ctDNA testing when patients experienced progress during TKI treatment. Second, the ctDNA tests only assessed the EGFRm. Co-existing mutations such as TP53 or others were not investigated, even though these subcloned mutations are well-known prognostic factors in NSCLC. Given the observational nature of this study, clinical treatment strategy data according to ctDNA status was not available and needs further investigation.
In conclusion, our results indicate that patients with baseline detectable plasma EGFRm ctDNA and undetectable at four weeks of icotinib treatment could achieve a similar prognosis to those with baseline and four weeks of undetectable plasma EGFRm. Detectable plasma EGFRm at four weeks of treatment is the independent poor prognostic risk factor for PFS and OS. These biomarkers may guide treatment decisions and follow-up approaches in daily clinical practice.
Materials and methods
Study design
This study is a prospective, multi-center, biomarker-directed observation trial evaluating the association between the prognosis and dynamic change of plasma EGFRm ctDNA to EGFR-TKI therapy in treatment-naive advanced NSCLC patients with EGFRm identified in tumor tissues. Patients were recruited from four tertiary hospitals in Fujian Province, China. Eligible patients, each with EGFR-sensitizing mutations such as exon 19 deletion (Ex 19del) or exon 21 L858R, received icotinib 125 mg three times a day until progressive disease (PD) or unacceptable toxicity. Blood samples were obtained pretreatment administration (the baseline) and four-week on-treatment for each participant, and liquid biopsy analyses were performed for quantitative assessment of EGFRm ctDNA dynamics.
The trial was conducted following the Declaration of Helsinki and the International Conference on Harmonization Guidelines for Good Clinical Practice and approved by the Institutional Review Board of Fujian Provincial Cancer Hospital (Fuzhou, China). All patients provided written informed consent. This study was registered at ChiCTR-DDD-17,013,131.
Patients
The critical eligibility criteria included: (1) age between 18 and 80 years; (2) histologically or cytologically confirmed stage IIIB/C or IV NSCLC; (3) Eastern cooperative oncology group performance status (ECOG PS) score between 0 and 2; and (4) harboring common activating EGFRm (Ex 19del or L858R mutation) identified in tumor tissues. Tissue biopsies were tested using the amplification refractory mutation system Polymerase Chain Reaction (ARMS PCR) method or gene sequencing. Participants with brain metastasis are allowed to undergo brain radiotherapy, as determined by the attending physicians. Patients who had received systemic therapy after being diagnosed with advanced NSCLC were excluded. Additionally, patients harboring rare EGFR mutations or those with co-mutations involving other targetable genes such as Anaplastic Lymphoma Kinase (ALK), c-ros oncogene 1, receptor tyrosine kinase (ROS1), or B-Raf proto-oncogene, serine/threonine kinase (BRAF V600E) were excluded from the study.
Demographic, clinical, and laboratory information, including age, sex, ECOG PS, smoking history, histology, prior surgical history, type of EGFR mutation detected using tumor tissue, and presence of liver, bone, or brain metastasis at baseline, were collected at baseline.
Blood sample collection and EGFR mutation analysis
Peripheral blood samples were collected in EDTA tubes according to a fixed schedule. The whole blood was centrifuged within 4 h of collection at 1200 g for 10 min, followed by further centrifugation at 3000 g for 10 min to clarify the plasma supernatant, which was then stored at − 80 °C until use. We employed custom ddPCR assays to detect the plasma activating EGFRm (Ex 19del or L858R) and the EGFR T790M resistance mutation. The custom ddPCR assays had a detection limit of ≤ 0.10% variant allele fraction (VAF), calculated as the ratio of observed variant alleles to the total reads. Samples from patients who withdrew consent were excluded from the analysis.
To evaluate plasma EGFRm ctDNA dynamic changes, patients needed a baseline plasma sample and a sample at four weeks of treatment that returned a valid ddPCR result for EGFRm. When assessing plasma EGFRm detectability and clearance, we only included plasma EGFR Ex 19del or L858R mutation ctDNA without plasma EGFR T790M resistance mutation.
Clinical endpoints
The best overall response (BOR) was evaluated by the local investigator using the response evaluation criteria in solid tumors (RECIST V.1.1). PFS was defined as the time from the start of icotinib treatment until disease progression, death from any cause or the last follow-up. Overall Survival (OS) was the time from the start of enrollment treatment to death or the last follow-up. The study’s primary endpoint was to explore the association of baseline and on-treatment plasma EGFRm ctDNA dynamics with PFS. Secondary endpoints included analyzing the correlations of ctDNA dynamics with radiographic response and OS.
Statistical analysis
The ddPCR method has a reported 70% sensitivity for detecting plasma EGFR10,26. Considering the 12 month PFS, around 60% of patients are expected to experience tumor progression within one year of follow-up. We need ninety-three patients to achieve a two-sided 95% confidence interval (CI) for sensitivity with a half-width of 0.12. Finally, we set the sample size at 98 in our study, assuming a dropout rate of 5% at 12 months.
We used descriptive statistics to summarize the patient’s characteristics. Categorical variable differences were assessed with the chi-square test or Fisher’s exact test, while the Mann-Whitney U nonparametric test was used for continuous, non-normally distributed variables. The 95% CI for the objective response rate (ORR) and disease control rate (DCR) were estimated via the Clopper-Pearson method. We calculated the median follow-up using the reverse Kaplan-Meier method. Survival analyses were conducted using the Kaplan-Meier method, and differences among groups were compared using the log-rank test. The Cox proportional hazards model calculated hazard ratios (HR) values with a 95% CI. We employed a backward stepwise regression method for the multivariate analysis to avoid overfitting the Cox regression model. The stepwise variable selection procedure was conducted using ‘stepAIC’ function in R package ‘MASS’. This method starts with a model that includes all potential variables identified in the univariate analysis. The selection process is based on minimizing the Akaike Information Criterion (AIC) value, which balances the goodness-of-fit and complexity of the model. Through this backward elimination approach, variables are iteratively removed to find the optimal combination that minimizes the AIC value.
All analyses were performed using the IBM SPSS Statistics software (version 25.0; IBM, Armonk, NY, USA) and R version 4.1.0 (www.r-project.org). A two-tailed test was set with statistical significance accepted at P-value < 0.05.
Data availability
The data that supports the findings of this study is available from the corresponding author upon reasonable request.
Abbreviations
- EGFRm:
-
Epidermal growth factor receptor mutations, NSCLC
- TKIs:
-
Tyrosine kinase inhibitors
- PFS:
-
Progression-free survival
- ctDNA:
-
Circulating tumor DNA
- ddPCR:
-
Droplet digital polymerase chain reaction
- NGS:
-
Next-generation sequencing
- MRD:
-
Minimal residual disease
- Ex 19del:
-
Exon 19 deletion
- PD:
-
Progressive disease
- ECOG PS:
-
Eastern cooperative oncology group performance status
- ARMS PCR:
-
Amplification refractory mutation system polymerase chain reaction
- ALK:
-
Anaplastic lymphoma kinase
- ROS1:
-
c-ros oncogene 1,receptor tyrosine kinase (ROS1)
- BFAF V600E:
-
B-raf proto-oncogene, serine/threonine kinase
- BOR:
-
Best overall response
- RECIST:
-
Response evaluation criteria in solid tumors
- OS:
-
Overall survival
- CI:
-
Confidence interval
- ORR:
-
Objective response rate
- DCR:
-
Disease control rate
- HR:
-
Hazard ratios
- CR:
-
Complete response
- PR:
-
Partial response
- SD:
-
Stable disease
References
Ciardiello, F. & Tortora, G. EGFR antagonists in cancer treatment. N Engl. J. Med.358, 1160–1174 (2008).
Shi, Y. et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J. Thorac. Oncol. Off Publ. Int. Assoc. Study Lung Cancer9, 154–162 (2014).
Wu, Y. L. et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann. Oncol. Off J. Eur. Soc. Med. Oncol.26, 1883–1889 (2015).
Maemondo, M. et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl. J. Med.362, 2380–2388 (2010).
Shi, Y. K. et al. First-line icotinib versus cisplatin/pemetrexed plus pemetrexed maintenance therapy for patients with advanced EGFR mutation-positive lung adenocarcinoma (CONVINCE): a phase 3, open-label, randomized study. Ann. Oncol. Off J. Eur. Soc. Med. Oncol.28, 2443–2450 (2017).
Wu, Y. L. et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol.15, 213–222 (2014).
Wu, Y. L. et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol.18, 1454–1466 (2017).
Soria, J. C. et al. Osimertinib in untreated EGFR-Mutated Advanced Non-small-cell Lung Cancer. N Engl. J. Med.378, 113–125 (2018).
Osimertinib with or without Chemotherapy. in EGFR-Mutated Advanced NSCLC | New England Journal of Medicine. https://www.nejm.org/doi/full/10.1056/NEJMoa2306434
Desai, A. et al. ctDNA for the evaluation and management of EGFR-Mutant non-small cell lung cancer. Cancers16, 940 (2024).
Araki, T., Kanda, S., Horinouchi, H. & Ohe, Y. Current treatment strategies for EGFR-mutated non-small cell lung cancer: from first line to beyond osimertinib resistance. Jpn. J. Clin. Oncol.53, 547–561 (2023).
Semenkovich, N. P. et al. Genomic approaches to cancer and minimal residual disease detection using circulating tumor DNA. J. Immunother. Cancer11, e006284 (2023).
Ettinger, D. S. et al. NCCN guidelines® insights: Non-small cell lung cancer, version 2.2023. J. Natl. Compr. Cancer Netw. JNCCN21, 340–350 (2023).
Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer. A Prospective Multicenter Cohort Study (LUNGCA-1). https://pubmed.ncbi.nlm.nih.gov/34844976/
Gray, J. E. et al. Early clearance of plasma epidermal growth factor receptor mutations as a predictor of outcome on osimertinib in advanced non-small cell lung cancer; exploratory analysis from AURA3 and FLAURA. Clin. Cancer Res. Off J. Am. Assoc. Cancer Res.29, 3340–3351 (2023).
Johnson, M. et al. Longitudinal circulating tumor DNA modeling to predict disease progression in first-line mutant epidermal growth factor receptor non-small cell lung cancer. Clin. Pharmacol. Ther.115, 349–360 (2024).
Li, N. et al. Perioperative circulating tumor DNA as a potential prognostic marker for operable stage I to IIIA non-small cell lung cancer. Cancer128, 708–718 (2022).
Visser, E. et al. Liquid biopsy-based decision support algorithms for diagnosis and subtyping of lung cancer. Lung Cancer Amst. Neth.178, 28–36 (2023).
Akamatsu, H. et al. Clinical significance of monitoring EGFR mutation in plasma using multiplexed digital PCR in EGFR mutated patients treated with afatinib (West Japan Oncology Group 8114LTR study). Lung Cancer131, 128–133 (2019).
Xu, J. et al. Evaluation of clinical outcomes of Icotinib in patients with clinically diagnosed advanced lung cancer with EGFR-sensitizing variants assessed by circulating tumor DNA testing. JAMA Oncol.8, 1328–1332 (2022).
Shen, H. et al. Potential clinical utility of liquid biopsy in early-stage non-small cell lung cancer. BMC Med.20, 480 (2022).
Dao, J. et al. Using cfDNA and ctDNA as oncologic markers: a path to clinical validation. Int. J. Mol. Sci.24, 13219 (2023).
Aucamp, J., Bronkhorst, A. J., Badenhorst, C. P. S. & Pretorius, P. J. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol. Rev. Camb. Philos. Soc.93, 1649–1683 (2018).
Ernst, S. M. et al. Clinical utility of circulating tumor DNA in patients with advanced KRASG12C-mutated non-small cell lung cancer treated with sotorasib. J. Thorac. Oncol. Off Publ Int. Assoc. Study Lung CancerS1556-0864 (24), 00165–00165. https://doi.org/10.1016/j.jtho.2024.04.007 (2024).
Thress, K. S. et al. Complete clearance of plasma EGFR mutations as a predictor of outcome on osimertinib in the AURA trial. J. Clin. Oncol.35, 9018–9018 (2017).
Kitazono, S. et al. Barcode sequencing identifies resistant mechanisms to epidermal growth factor receptor inhibitors in circulating tumor DNA of lung cancer patients. Cancer Sci.110, 3350–3357 (2019).
Funding
This study was supported by the Natural Science Foundation of Fujian Province (Grant No. 2023J01180), the Fujian Provincial Health Technology Project (Grant No. 2022QNA046), the Startup Fund for Scientific Research of Fujian Medical University (Grant No. 2020QH1218), and the Leading Project Foundation of Science and Technology, Fujian Province(No.2016Y0101).
Author information
Authors and Affiliations
Contributions
Yunjian Huang and Gen Lin: study conception and manuscript preparation, writing—review & editing. Yaping Hong and Wu Zhuang: Data curation, formal analysis, resources, investigation, software, writing—original draft, writing—review & editing. Jinhuo Lai, Haipeng Xu, Yueming He, Jinlan Lin, Qin Shi, Shengjia Chen, Zhangzhou Huang, Shijie Chen and Dongzhu Lu: data curation, writing—review & editing. All authors have reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
Approval of the research protocol by an Institutional Reviewer Board: Institutional Review Board of Fujian Provincial Cancer Hospital.
Informed consent
All informed consent was obtained from the subjects and/or guardians.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hong, Y., Zhuang, W., Lai, J. et al. Plasma EGFR mutation ctDNA dynamics in patients with advanced EGFR-mutated NSCLC treated with Icotinib: phase 2 multicenter trial result. Sci Rep 14, 23115 (2024). https://doi.org/10.1038/s41598-024-73749-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-73749-2