Abstract
Catalytic hairpin assembly (CHA)-DNA walker allows nanostructures to spontaneously hybridize to the nucleic acids. The localized surface plasmon resonance provides the ability of color-shift for Au nanoparticles (AuNPs) to design a colorimetric biosensor by implementing CHA-DNA walker reaction on AuNPs. A target gene in Klebsiella pneumoniae as the reaction cascade trigger, was selected. H1 and H2 oligonucleotides as the components of the system were designed and verified by NUPACK. The AuNPs were conjugated to H1. The conjugation of the probes to the AuNPs was evaluated using FT-IR. The signal amplification process was conducted at 25℃. TEM imaging, zeta potential, spectroscopy, and gel-electrophoresis were used to examine the conduction of the reaction cascade and specificity. The sensitivity of the method was analyzed using serial dilution of the target. The formation of over-52 bp intermediate secondary structures (which only exist when the reaction happens) was confirmed by gel-electrophoresis. The color distinction between positive (0.08 to 0.058) and negative samples (0.098 to 0.05) was evidenced instantly and in a period of 90 min of the reaction as a drop change of 520 nm intensity absorbance. TEM imaging confirmed the further distance of AuNPs in the positive sample in comparison to that of the negative sample which reveals effective detection of the pathogen. The LOD of the technique was measured as 2.5 nM of the target sequence. The diagnostic approach is a label-free, enzyme-independent approach and can be executed in a single step. It has been designed by employing the CHA-DNA walker system along with the colorimetric properties of AuNPs for the first time, thereby paving the way for more rapid and accurate diagnostic kits.
Similar content being viewed by others
Introduction
The vital significance of accessing an affordable, easy, and swift diagnostic technique has been a demanding hotspot all around the world, especially in the two-year pandemic of SARS-CoV2 infection1. Nano-biosensing of biomarkers seems to serve effective approaches to diagnose biomolecules. DNA nanostructures e.g., DNA walkers have attracted high attention due to their programmability and stability2,3. DNA walkers are able to automatically and continuously move along specific tracks on nucleic acid strands which contributes to the signal amplification. The designable nature of DNA walkers turns them into a valuable and precise tool with a wide range of applications in nanoscale biosensing approaches due to their ability of nonenzymatic signal amplification. The fundamental specification in DNA walker structures is the strand displacement mechanism which makes the nucleic acid (NA) strands able to bind and separate themselves, to and from other NA structures, autonomously.
The DNA walker mechanism has been applied in several biosensing strategies3,4,5,6,7. A DNA walker mechanism was designed successfully to detect miRNA-21 by Wu et al. and the hairpin-structured DNA tracks on magnetic beads were triggered by the target miRNA-21 as the walker strand8. Beyond their use in biosensing, DNA walkers have been exploited in molecular computing and Drug delivery (in DNA origami constructs) according to their potential to contribute to encoding algorithms and processing molecular information, and programmability, respectively9,10.
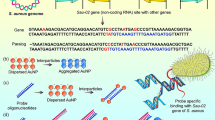
DNA walkers are capable of being applied to isothermal conditions. Even though several molecular detection techniques such as NASBA11, LAMP12, RCA13, SDA14, etc. have been developed during the last two decades to increase the availability and affordability of diagnostic methods by omitting their dependence on sophisticated equipment (i.e. thermal cycler)15,16, they still rely on a biocatalyst e.g., an enzyme to run. In that regard, hybridization chain reaction (HCR) and catalytic hairpin assembly (CHA) could be mentioned as two techniques potential of being used in DNA walkers with respect to hybridization property to conduct a recycling cleavage reaction and also the characteristics of utilizing enzyme-free circuits for target amplification17. In a CHA circuit, two (or more) engineered hairpins are used to detect a specific target18. The first hairpin is unfolded only in the presence of the target according to its complementary region. The linearized hairpin-1 is ready to open the second hairpin via the hairpin-2’s sticky end because of its complementary region. This reaction goes on by the strand displacement activity which allows the target to stay dissociated from the complementary sequences and stick to the other hairpins in the reaction mixture. The whole process is conducted in a constant reaction condition at room temperature which provides a promising system for biomolecule diagnosis18,19,20,21.
Gold nanoparticles (AuNPs) are largely employed in sensing target biomolecules and suitable candidates in colorimetric molecular assays according to their phenomenal optical properties, possibility of diverse surface modifications, and biocompatibility which makes them a good candidate in colorimetric molecular assays22. Localized surface plasmon resonance (LSPR) is the key feature in AuNPs that provides the color distinction between the positive and negative samples23. The proximity between gold nanoparticles determines the solution’s color. The AuNPs’ aggregation leads to a blue-shift and a far-away status between them allows the solution to remain red.
In the present study, we designed an autonomous enzyme-free signal-amplification strategy, capable of genosensing in a constant reaction condition at room temperature. The method has been applied to diagnose a specific pathogen gene in Klebsiella pneumoniae by calorimetric differentiation between the target-containing sample and a negative sample lacking the target sequence. The negative sample contained a distinct same-length oligonucleotide by taking advantage of the programmability of DNA circuits, the spontaneous behavior of DNA walkers and, the optical traits of AuNPs.
Materials and methods
Materials
Gold-nanoparticles were prepared by Arminano, Co. (Iran). The required oligonucleotides were ordered from Metabion (Germany). Chemicals including Monosodium phosphate-monohydrate (NaH2PO4), Sodium chloride (NaCl), Disodium phosphate- heptahydrate (Na2HPO4), and Magnesium chloride were of molecular biology grade and obtained from Merck (Germany).
Target sequence and oligonucleotide design
The target sequence was selected from the chromosome in the genome of Klebsiella pneumonia KPHS_51180 (Gene ID: 11850189) using the NCBI database. A BLAST was run for the selected gene and the target sequence was chosen by GeneRunner software. The target sequence was checked for a unique specificity for Klebsiella pneumonia using BLAST online tool. Hairpin sequences were designed using the online bioinformatics tool “NUPACK” (nupack.org).
Characterization and conjugation of AuNPs
AuNP was characterized using TEM imaging, FT-IR, DLS size, and Zeta potential assays. In order to investigate the conjugation of nanoparticles and oligonucleotides, salt induction was used to evaluate the nanoprobe solution’s stability which leads to color change. The formation of appropriate AuNP-probe was examined by FT-IR. TEM imaging was also used to compare the distance between the nanoprobes and nanoparticles.
The gold nanoparticles conjugated with thiol-modified probes were prepared according to the following steps. 0.5 µL of 10X TCEP was exposed to 5 µL of 100 µM thiolated oligonucleotide and 5 µL of deionized water. The solution was incubated for an hour at room temperature (RT). Then, 95 µL AuNP was added, followed by a 16-hour incubation at RT. Furthermore, the solution was salt-aged in ten steps with 10-minute intervals between each step to reach the final concentration of 0.01 M NaCl and 0.01 M phosphate buffer (pH = 7.2). After adding the buffer, the solution was incubated for 40 h at room temperature, followed by a washing step where the supernatant was discarded after 20 min of centrifugation at 14,000 g. The washing step was repeated three times. The synthesized nanoprobe suspension was stored at 4 °C.
CHA–DNA walker assay
Firstly, to evaluate the thermodynamic feasibility of the reactions’ \(\:\varDelta\:G\) was calculated by bioinformatic tools (i.e. NUPACK). To optimize the method protocol, different modifications were implemented such as changing the concentration ratio of each reaction component (namely H1, H2, and target sequence) and time gradient. The two hairpins were heated to 95₅ ºC for 3 min and then gradually cooled to the room temperature preceding the CHA-DNA walker reaction. The reaction mixture was composed of hairpin 1 (conjugated with the AuNP) (500 nM), hairpin 2 (500 nM), and the target sequence (250 nM). The reaction was conducted at the room temperature in less than an hour. The CHA-DNA walker results were visualized using the color differentiation between the positive and negative samples with the naked eye. TEM imaging was used to investigate the distance between the oligonucleotides. Zeta potential was measured to analyze the electrical potential on the surface of nanostructures. An agarose gel electrophoresis (2%) was applied to assess the molecular weight of hybridized and non-hybridized oligonucleotides.
The serial dilution was performed by diluting normal amounts of DNA content to 10− 4 to determine the lowest amounts of the target sequence that the designed method is capable of. The test was conducted three times to verify the consistency of the results. The color discrimination between the samples was detectable by the naked eye and it was confirmed by UV-Vis spectroscopy measurements at 520 nm wavelength by NanoDrop (EPOCh2, US).
To evaluate the specificity of the method, bacterial strains including Klebsiella pneumoniae, Yersinia enterocolitica, Shigella sonnei, Morganella morganii, Helicobacter pylori, and Citrobacter freundii were cultured overnight at 30–37 °C in Tryptic Soy Broth and then prepared for DNA extraction. Genomic DNA was retrieved by the boiling method, which involved centrifugation at 3000 g for 8 min, followed by two washing steps. The DNA was then heated to 100 °C for 20 min and centrifuged again at 3000 g for 3 min, resulting in a supernatant used as the template.
It is notable to mention that in the TEM imaging that was conducted both for the analysis of CHA-DNA walker and characterization of AuNPs, the NP and AuNP stocks were meticulously pipetted before being added. The AuNP addition was made from the same stock with a time interval of one second, thereby minimizing any potential differences that may exist. Moreover, the reaction was repeated multiple times, and each repetition yielded the same results. The TEM imaging was conducted twice, with each imaging involving three repetitions of the reaction. These steps were taken to ensure the accuracy and reproducibility of the results.
Results
The CHA-DNA walker colorimetric assay was designed using the CHA reaction cascades and a walking system on the nanoparticle core which led to the colorimetric differentiation of positive and negative samples. This strategy, allows the method to generate the signal, amplify and detect it all together. As illustrated in Fig. 1, the complementary regions between two auxiliary hairpin-shaped oligonucleotides are engaged when they are folded and closed. In the presence of the target DNA (DNA walker), the sticky end of hairpin1 is hybridized with the target sequence and a strand displacement phenomenon which is mediated by toeholed structures exposes the trapped sequences within the hairpin structures. The untrapped sequence could hybridize to the target sequence, which unfolds the hairpin 1 and enables the complementary regions between the two hairpins to hybridize one another. Hairpin1-hairpin 2 hybridization, displaces and releases the target sequence. The released target sequence is now able to walk to the neighbor hairpin 1 and repeat the abovementioned cascade autonomously. At the end, there are hairpin 1– hairpin 2 complexes on the surface of AuNPs (AuNPs-H1-H2 complexes). Simultaneously, a colorimetric assay is in progress on a larger scale. The hairpin 1-functionalized AuNPs are at a regular distance from each other. When the target sequence is present in the reaction and the cascade begins, at first a slight blue-shift appears in the positive sample which could be the result of the charge density alteration when the designed machinery is active. Yet, the reaction mixture becomes more reddish when the reaction continues. This indicates that the ion density on AuNPs is increased in the end-point color differentiation. Besides, AuNPs are more negatively charged which affects the regular distance of AuNPs from each other, leading to an increase in redshift.
Target sequence
Klebsiella pneumoniae is one of the most significant secondary infectious agents and its detection is highly needed. Several genes involved in the pathogenicity of the bacteria were analyzed using NCBI’s BLAST. The results indicated that the gene KPHS_51180 which encodes the “divergent polysaccharide deacetylase family” protein presented the required specificity. The target gene plays an important role in the capsule formation of the bacterium that helps the pathogenicity of K. pneumoniae24,25. This is the first report on the usage of this gene as a target for the detection of K. pneumoniae. A target sequence was selected from the gene and used as a trigger by which the machinery of the system is started.
Characterization and conjugation of AuNP
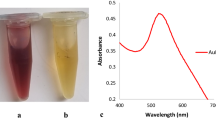
To ensure the functionality of the designed system, firstly, AuNP was characterized and its conjugation was evaluated. As it is shown in Fig. 2, UV-vis spectroscopy was used to characterize AuNPs before and after functionalization. Scanning the spectrum of 500–700 nm shows the absorbance peak at 528 nm confirming the proper distribution of the particles (Fig. 2A).
Characterization and conjugation of AuNP. (A) The UV-Vis scanning absorbance. The figure depicts the absorbance in 300-800 nm range which shows the proper distribution of the gold nanoparticles and AuNP-H1 structures. (B) Salt induction colorimetric test. The left microtube contains AuNPs and microtube in right contains AuNProbes after salt induction. (C) FT-IR diagram comparison between AuNPs and AuNProbes. It represents two absorbance peaks at 1072 cm-1 and 995 cm-1 which shows that the successful S=O and C=C bond formation respectively. (D) Dynamic light scattering (DLS) analysis. 1: The DLS graph determines the average particle size of AuNPs is 95.74 nm. 2: The size distribution of AuNProbes. The mean size of the AuNProbes is 104.59 nm. (E) TEM Imaging of AuNPs and AuNProbes in the scale of 200 nm. 1: Before functionalization, 2: After functionalization. Separate figures are provided individually in the supplementary file.
A colorimetric differentiation was also observed after performing a salt induction assay for both AuNP and AuNProbe solutions. The assay evaluates the stability of the samples where a higher charge density around AuNProbes makes it more resistant to the ion shock that is caused by the salt induction and remains red when the salt is added. However, a blue-shift occurred in AuNP, immediately after the salt induction (Fig. 2B).
The conjugation of the probes to the AuNPs was evaluated using FT-IR. The corresponding diagram demonstrates two absorbance peaks at 1072 cm− 1 and 995 cm− 1, the former confirmed the S = O bond formation and the latter revealed the formation of C = C26 which indicated the fabrication of AuNP-H1 nanostructures. (Fig. 2C)
Dynamic light scattering (DLS) analysis was also performed to determine the size distribution of the particles. According to DLS results, the average diameter of the AuNPs was 127.96 nm and that of AuNProbes was 104.59 nm (Fig. 2D).
In Fig. 2E, TEM images confirmed the desired size and spherical morphology of nanoparticles. After adding the H1 oligonucleotides to the nanoparticles, the particles seem more dispersed in the TEM imaging. This could be as a result of an increased charge density on the surface of the particles which is as well evidenced by the zeta potential results.
Analysis of the CHA-DNA walker assay
The bioinformatic analysis of the practicability of the reaction cascade was primarily conducted by the software mentioned in Sect. 2. According to which, the thermodynamic probability of conformation of H1 (AuNP)-H2 which was measured by \(\:\varDelta\:G\), was − 167, indicating that the two main components show a significant affinity towards each other. Yet there could not be a bond except in the presence of the target. To confirm these results, agarose gel electrophoresis was performed to validate the existence of AuNP-H1-H2 structures in comparison to hairpin1 and hairpin2. AuNP-H1-H2 complexes are formed only when the target sequence exists, and the DNA circuit runs successfully. It is illustrated in Fig. 3A, that when the 51nt H1(= AuNProbe), the 46nt H2, and CHA-DNA walker system products (positive and negative samples) are each loaded individually, the positive CHA-DNA walker lane was heavier in the gel documentary. This indicated that the CHA-DNA walker system has started, and the complex nanostructures were formed. The size of the CHA-DNA walker system in the presence of the target DNA, was predicted to be somewhere between 97 and 120 bp which complies with the results showed in the gel electrophoresis lane 3. On the other hand, the negative CHA-DNA walker lane showed a smear-like pattern on the gel and was lighter than the positive sample. It is worth mentioning that the ladder was only used to check that the gel-electrophoresis was run properly and not to determine the precise molecular weight of the products. To help visualizing short H1 and H2 hairpins on lane 1 and lane 2 of the gel electrophoresis, a ten-fold final concentration was used.
Confirmation of CHA-DNA walker assay. (A) Agarose gel-electrophoresis results suggesting that the DNA walking-CHA procedure has occurred. Lane1: H2. Lane 2: H1. Lane3: positive. Lane4: ladder. Lane5: negative. Original gel and the gridded version are presented in Supplementary file. (B) The UV-Vis spectroscopy to compare the absorbance of the positive sample (red) with the negative sample (Blue) at 520 nm in 90 minutes. The color differentiation between positive (top) and negative (bottom) samples at the endpoint of CAH-DNA walker reaction. (C) TEM imaging of positive and negative samples at the scale of 200 nm. 1: Negative sample. 2: Positive sample. As depicted in the figure, at the 200 nm scale in the positive sample, the distance between AuNProbe clusters is approximately 346 nm while in the negative sample, the whole AuNProbes have become coagulated. (D) Zeta potential of positive and negative samples. 1: Positive sample. 2: Negative sample. Separate figures are provided individually in the supplementary file.
The UV-Vis spectroscopy demonstrates that at the 520 nm wavelength and after 90 min, the absorption of the positive sample changed from 0.08 to 0.058 while the absorption of the negative sample changed from 0.098 to 0.05 with a sharp decrease in the first 10 min. It significantly showed that the red color of the positive samples (Fig. 3B) was more stable than the negative sample. The result was also distinguishable by the naked eye at the reaction endpoint.
The TEM images in Fig. 3C revealed that the particles are at a further distance in positive samples compared to the negative samples suggesting that the CHA-DNA walker system was performed productively. The distance between the particles is due to the increased charge density around the positive sample particles compared to the negative sample. The results were also affirmed by zeta potential analysis, shown in Fig. 3D.
To realize the lowest amount of nucleic acid content that our design is able to detect, a serial dilution was performed. The naked eye and UV-Vis spectroscopy were used to evaluate the power of the method to detect minute amounts of the target sequence. A red color is still visible in the reaction in which the target sequence is diluted 102 folds in comparison to the normal amounts of DNA content which was 0.25 µM, while in reactions with more diluted targets (i.e. 10− 3 and 10− 4), the blue-shift occurred. This suggests that the designed technique is capable of diagnosing the small quantities of target DNA, as little as 2.5 nM. As it is shown in Fig. 4A, this result was also confirmed with the UV-Vis spectroscopy where the absorbance of each dilution was measured in 520 nm wavelength.
Sensitivity and specificity of the designed technique. (A) Sensitivity. The UV-Vis absorption of each dilution along with their error bars (error bars represent mean value of three independent identical replicates). Respectively, from the left: 100, 10-1, 10-2, 10-3, and 10-4. This suggests that the smallest DNA target that the designed technique is capable of detecting is as low as 2.5 nM. (B) Specificity. The colorimetric differentiation between the positive and five other negative samples: 1- Klebsiella pneumonia (positive), 2- Morganella morganii, 3- Yersinia enterocolitica, 4- Shigella sonnei, 5- Citrobacter freundii, 6- Helicobacter pylori.
To evaluate the specificity of the method, the results were compared against five additional bacterial strains. As depicted in Fig. 4B, the designed technique successfully differentiated between the positive sample and the five other bacteria calorimetrically.
Figure 4.
Discussion and conclusion
The experimental findings have elucidated the functional capacity of the engineered CHA-DNA walker system in harnessing nanoparticles to establish a colorimetric methodology through the conjunction of AuNPs with H1. The validation of the formation of transient intermediate structures, exclusive to the completion of the reaction, was confirmed through agarose gel electrophoresis, zeta potential assay, and TEM imaging. The discernible color differentiation between positive and negative samples, perceptible to the naked eye, was substantiated via spectroscopic analysis. The study has demonstrated the viability of leveraging the colorimetric properties of AuNPs in conjunction with the signal amplification characteristics of a CHA-DNA walker reaction cascade for the detection of a clinically significant biomarker. The developed biosystem represents a diagnostic approach that integrates the machinery of CHA-DNA walkers with the colorimetric attributes of gold nanoparticles for identifying a crucial prokaryotic gene in K. pneumoniae. This investigation has introduced an enzyme-free, tag-free, one-pot colorimetric technique by engineering a single-stranded DNA walker based on the CHA system for sensing and diagnosing a prokaryotic gene. By simplifying equipment requirements, streamlining procedures, and reducing the need for specialized expertise, this method emerges as a promising candidate for biosensing applications, particularly for short nucleo-biomarkers. While the utilization of nanoparticles in colorimetric detection methodologies is well-documented in literature27,28, and they have been applied to sense numerous biomolecules including cancerous and infectious bacterial biomarkers29,30 in different forms such as DNA dendrimers31, DNA tweezers32, DNA walkers33, and DNA circuits34, and even the most recent studies employing three pedal DNA walker using the FAM tag to detect the early cancerous biomarkers35, the concurrent utilization of CHA-DNA walker signal amplification and colorimetric properties of AuNPs to diagnose a prokaryotic pathogen biomarker without employing any tag, dye, and enzyme remains unprecedented. It is noteworthy that the technique designed here is not meant to be able to discriminate the various strains of Klebsiella pneumoniae at this point. As it is repeatedly mentioned before, the primary focus of this research at this stage was the design of a novel detection method to recognize the nucleic acid content of K. pneumonia at the species level, without the need for complex equipment and using enzymes. Although the ultimate objective of this project is to develop an efficient method that is applicable to more complex samples, the optimization of the designed method, is being pursued in subsequent stages. The method is seemingly preferable over the aggregation-based AuNP biosensors because of its signal amplification process during the CHA reaction. With respect to a promising simple design pattern in comparison with the other DNA-walking mechanisms, the method has a strong potential to be applied in the next generation detection techniques.
To encapsulate the findings, this investigation has revealed the prospective employment of AuNPs’ colorimetric qualities alongside the signaling intensification attributes of a CHA-DNA walker reaction sequence for the identification of a medically significant biomarker. This approach presents itself as a compelling alternative within the realm of biosensing endeavors, notably for the recognition of brief nucleo-biological markers, while also displaying considerable promise for inclusion in forthcoming generations of analytical tools.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Pecora, N. D. & Pettengill, M. A. The role of laboratory-based Viral Testing in the COVID-19 Pandemicp. 33–35 (Oxford University Press, 2022).
Krishnan, Y. & Seeman, N. C. Introduction: nucleic acid nanotechnology. ACS Publications pp. 6271–6272. (2019).
Jiang, Y. et al. Real-time detection of isothermal amplification reactions with thermostable catalytic hairpin assembly. J. Am. Chem. Soc.135 (20), 7430–7433 (2013).
Liu, J. et al. Applications of catalytic hairpin assembly reaction in biosensing. Small15 (42), 1902989 (2019).
Dong, G. et al. A rapid room-temperature DNA amplification and detection strategy based on nicking endonuclease and catalyzed hairpin assembly. Anal. Methods11 (19), 2537–2541 (2019).
Dai, J. et al. Self-replicating catalyzed hairpin assembly for rapid signal amplification. Anal. Chem.89 (22), 11971–11975 (2017).
Ebrahimi, A., Ravan, H. & Mehrabani, M. Multiplex monitoring of Alzheimer associated miRNAs based on the modular logic circuit operation and doping of catalytic hairpin assembly. Biosens. Bioelectron.170, 112710 (2020).
Zhang, P. et al. Highly ordered and field-free 3D DNA nanostructure: the next generation of DNA nanomachine for rapid single-step sensing. J. Am. Chem. Soc.140 (30), 9361–9364 (2018).
Chen, X. & Ellington, A. D. Shaping up nucleic acid computation. Curr. Opin. Biotechnol.21 (4), 392–400 (2010).
Schuller, V. J. et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano5 (12), 9696–9702 (2011).
Mollasalehi, H. & Yazdanparast, R. An improved non-crosslinking gold nanoprobe-NASBA based on 16S rRNA for rapid discriminative bio-sensing of major salmonellosis pathogens. Biosens. Bioelectron.47, 231–236 (2013).
Shahbazi, E., Mollasalehi, H. & Minai-Tehrani, D. Development and evaluation of an improved quantitative loop-mediated isothermal amplification method for rapid detection of morganella morganii. Talanta191, 54–58 (2019).
Ali, M. M. et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev.43 (10), 3324–3341 (2014).
Mollasalehi, H. & Yazdanparast, R. Non-crosslinking gold nanoprobes for detection of nucleic acid sequence-based amplification products. Anal. Biochem.425 (2), 91–95 (2012).
Zhao, Y. et al. Isothermal amplification of nucleic acids. Chem. Rev.115 (22), 12491–12545 (2015).
Yan, L. et al. Isothermal amplified detection of DNA and RNA. Mol. Biosyst.10 (5), 970–1003 (2014).
Wu, H. et al. Label-free and enzyme-free colorimetric detection of microRNA by catalyzed hairpin assembly coupled with hybridization chain reaction. Biosens. Bioelectron.81, 303–308 (2016).
Yin, P., Choi, H. M., Calvert, C. R. & Pierce, N. A. Programming biomolecular self-assembly pathways. Nature451 (7176), 318–322 (2008).
Miao, P. & Tang, Y. Gold nanoparticles-based multipedal DNA walker for ratiometric detection of circulating tumor cell. Anal. Chem.91 (23), 15187–15192 (2019).
Li, Y. et al. A visual method for determination of hepatitis C virus RNAs based on a 3D nanocomposite prepared from graphene quantum dots. Anal. Chim. Acta1203, 339693 (2022).
Oishi, M. & Saito, K. Simple single-legged DNA walkers at diffusion-limited nanointerfaces of gold nanoparticles driven by a DNA circuit mechanism. ACS Nano14 (3), 3477–3489 (2020).
Mollasalehi, H. & Shajari, E. A colorimetric nano-biosensor for simultaneous detection of prevalent cancers using unamplified cell-free ribonucleic acid biomarkers. Bioorg. Chem.107, 104605 (2021).
Elghanian, R. et al. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science277 (5329), 1078–1081 (1997).
Lau, H. Y., Clegg, S. & Moore, T. A. Identification of Klebsiella pneumoniae genes uniquely expressed in a strain virulent using a murine model of bacterial pneumonia. Microb. Pathog.42 (4), 148–155 (2007).
Dong, D. et al. Survey and rapid detection of Klebsiella pneumoniae in clinical samples targeting the rcsA gene in Beijing, China. Front. Microbiol.6, 519 (2015).
Nandiyanto, A. B. D., Oktiani, R. & Ragadhita, R. How to read and interpret FTIR spectroscope of organic material. Indonesian J. Sci. Technol.4 (1), 97–118 (2019).
Hua, Z., Yu, T., Liu, D. & Xianyu, Y. Recent advances in gold nanoparticles-based biosensors for food safety detection. Biosens. Bioelectron.179, 113076 (2021).
Du, Y. & Dong, S. Nucleic acid biosensors: recent advances and perspectives. Anal. Chem.89 (1), 189–215 (2017).
Mollasalehi, H. & Hamidi, A. Early-phase nano-genosensing of cell-free Nucleobiomarkers in the Plasma of Cancerous Patients32p. 102344 (Nanotechnology, Biology and Medicine, 2021).
Zhang, Y., McKelvie, I. D., Cattrall, R. W. & Kolev, S. D. Colorimetric detection based on localised surface plasmon resonance of gold nanoparticles: merits, inherent shortcomings and future prospects. Talanta152, 410–422 (2016).
Xuan, F., Fan, T. W. & Hsing, I. M. Electrochemical interrogation of kinetically-controlled dendritic DNA/PNA assembly for immobilization-free and enzyme-free nucleic acids sensing. ACS Nano9 (5), 5027–5033 (2015).
Xu, X. et al. A smart DNA tweezer for detection of human telomerase activity. Anal. Chem.90 (5), 3521–3530 (2018).
Jung, C., Allen, P. B. & Ellington, A. D. A simple, cleated DNA walker that hangs on to surfaces. ACS nano11 (8), 8047–8054 (2017).
Ang, Y. S. & Yung, L. Y. L. Engineering self-contained DNA circuit for proximity recognition and localized signal amplification of target biomolecules. Nucleic Acids Res.42 (14), 9523–9530 (2014).
Li, J. & Li, H. W. Ultrasensitive miRNA detection by AuNP-based 3D DNA walker and Catalytic Hairpin Assembly (CHA) Cascade amplification for Early Cancer diagnosis. Chemistry–An Asian J.18 (15), e202300367 (2023).
Acknowledgements
The authors kindly appreciate supports of Faculty of Life Sciences & Biotechnology, Shahid Beheshti University.
Author information
Authors and Affiliations
Contributions
ESH: Investigation, experimental analysis, Writing- Original draft preparation. HM: Conceptualization, Methodology, Supervision, Writing- Reviewing and Editing, Project administration. DMT: Validation of the spectrophotometric analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shahbazi, E., Mollasalehi, H. & Minai-Tehrani, D. A gold nanoparticle conjugated single-legged DNA walker driven by catalytic hairpin assembly biosensor to detect a prokaryotic pathogen. Sci Rep 14, 22980 (2024). https://doi.org/10.1038/s41598-024-74227-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74227-5