Abstract
Paqr5b is a gene encoding membrane progesterone receptor γ (mPRγ), which is one of five mPR subtypes. Paqr5b belongs to the progestin and adipoQ receptor (PAQR) family, which consists of 11 genes. To elucidate the physiological functions of the mPR subtypes, we established gene knockout (KO) zebrafish strains by genetically editing seven paqr genes and analyzed their phenotypes. The null-mutant strain of paqr5b (paqr5b−/−) that we established in this study showed low fecundity, reduced chorion elevation and a high percentage of abnormal embryos. Embryos showed curvature of the spine and an abnormal head morphology. Individuals with abnormal head morphology continued to develop a phenotype of markedly abnormal palatine bone. The length of the brain of paqr5b−/− zebrafish was short, and the position of the cerebellum moved to the front and overlapped with that of the midbrain. Micro-CT scans revealed that the olfactory rosettes (ORs) were so shrunken that they were difficult to identify and connected with the olfactory bulbs (OBs) by thread-like structures. Immunohistochemical staining of OR with an anti-Paqr5b antibody revealed that Paqr5b was extensively expressed in neurons in the OR in wild-type zebrafish, whereas signals were not detected in paqr5b−/− zebrafish. In histological sections, the neurons disappeared, and the lamellar layer of the OR became thinner. These results indicate that Paqr5b is required for the formation of neurons in the OR. This is the first report demonstrating a distinct role for the mPR gene.
Similar content being viewed by others
Introduction
Membrane progesterone receptors (mPRs) were first identified in the ovary as receptors for oocyte maturation-inducing hormone, and subsequent genomic analyses revealed that the genes encoding these receptors belonged to a novel receptor family comprising 11 genes1. These genes are named Paqr7 (mPRα), Paqr8 (mPRβ), Paqr5 (mPRγ), Paqr6 (mPRδ) and Paqr9 (mPRϵ)2. Paqr7 and 5 are paralogs in zebrafish. Thus, there are seven genes encoding paqrs in zebrafish. Although mPRs were first found in fish ovaries3, they are distributed in various tissues and organs throughout the body. Thus, it is predicted that mPRs are responsible for a variety of rapid cell surface-induced progesterone actions in various intracellular signaling pathways. Previously, the induction of oocyte maturation in KO zebrafish was found to be affected by the seven paqr genes4. All seven genes in the KO zebrafish delayed but not blocked oocyte maturation. However, there have been no reports of further phenotypic analysis of the paqr genes in KO zebrafish. Thus, we also performed further phenotypic analysis of each paqr gene in KO zebrafish. Of particular interest is the role of mPRs in the brain. High expression of Paqr6 and Paqr9 has been reported in the brain5. The expression of paqr5b in the olfactory rosette (OR) has been reported in zebrafish6. In the goldfish brain, mPRα, mPRβ, mPRγ1, and mPRγ2 are also expressed7,8. European eel (Anguilla anguilla), channel catfish (Ictalurus punctatus) and rainbow trout express various mPRs and/or PGMRCs in the brain and pituitary9,10,11.
The olfactory system is an important organ for the survival of animals that plays a role in smell detection, social behavior, sexual behavior, hormone and pheromone detection, and danger recognition 12 13. Furthermore, the ability of olfactory organs to detect heavy metals, pollutants, chemical neurotoxicants, contagious agents, and trauma is limited, which increases the importance of olfactory plasticity14. In the fields of neurobiology, human physiology, disease, and toxicity, the zebrafish (Danio rerio), a small freshwater teleost fish, has become one of the most useful, beneficial and applicable model organisms15,16,17.
The zebrafish had two nasal holes in front of its two eyes in the anterior part of the snout. Each hole consisted of a frontal nostril, where water enters the nasal cavity, and a posterior nostril, where water is expelled from the nasal cavity. These two nostrils consist of olfactory epithelium (OE) arranged systematically in several lamellae that fuse into a middle/main raphe and form a cup-shaped formation called an olfactory rosette (OR)18,19. The central and medial regions of the OR consist of lamellae, which is composed of the sensory epithelium and the lateral nonsensory epithelium. The sensory epithelium consists of pseudostratified columnar epithelium composed of olfactory sensory neurons (OSNs) and basal and supporting cells 20,21,22.
There are five OSNs in the zebrafish OR, including ciliated, microvillous, crypt, kappe, and pear-shaped neurons23. OSNs are dissimilar to each other in their structure, anatomy, and relative location to the olfactory epithelium, with ciliated and microvillous OSNs being more abundant than other cells. The deeper layers of the OR are where the ciliated OSNs are present, and these neurons are spread through a large number of cilia along elongated dendrites into the lumen of the rosette cavity. In the superficial layers, microvillous OSNs are present and consist of small dendrites and small microvilli. Crypt cells are oval-shaped cell bodies composed of microvilli and small cilia within the same cells that reside in the uppermost layer of the OE1922,24. Kappe neurons are the fourth most common olfactory sensory neuron population within the zebrafish peripheral olfactory system (OS); kappe neurons consist of microvilli and are known for their characteristic shape as well as their distinct spatial distribution in the OE, but they are distinct from crypt neurons25.
The zebrafish olfactory system is similar in structure and function to that of other vertebrates and comprises a pair of peripheral olfactory organs or rosettes, in the nasal cavity that are linked to the olfactory bulbs (OBs) in the rostral-most forebrain region21,26,27,28,29,30. The sensory epithelium is part of the olfactory organ and is composed of olfactory sensory neurons (OSNs) that respond to odorant molecules or odorants. The olfactory nerve is an amazing connecting fiber that extends the axonal projections of OSNs to the OB31,32. OSN axons, which extend into the olfactory bulb, form distinct structures called glomeruli, where they synapse with bulbar mitral cells33,34,35.
In our present study, we investigated the role of paqr5b by establishing paqr5b-KO zebrafish. Head region abnormalities were detected in adult paqr5b−/− fish. Micro-CT revealed an abnormal structure of the OR. Olfactory sensory neurons (OSNs), including ciliated, microvillous and crypt cells, were absent in paqr5b−/− zebrafish. These results indicated that paqr5b is responsible for the formation of neurons in the OR.
Results
Generation of the paqr5b −/− strain by genomic editing
Paqr5b gene-edited fish were generated using CRISPR/Cas9 as previously described36(Supplementary Fig. S1). F0 and F1 adults were checked before pairing through heteroduplex mobility analysis (HMA) (Supplementary Fig. S2). Edited F0 adults were crossed with wild-type (WT) fish to produce heterozygous paqr5b+/− F1 fish. The F1 zebrafish that showed the same pattern upon HMA analysis were crossed with each other to obtain homozygous F2 mutants. DNA sequencing was conducted to confirm the insertion and/or deletion induced at the target site. A strain with a 13-nucleotide deletion (-13) at the target site of the paqr5b gene was established (Fig. 1A).
Fertility and embryonic development of the established paqr5b−/− line. (A) DNA sequence and predicted protein structure of an established paqr5b gene-edited strain. (Uppest panel) DNA sequences around the target site (in red) for CRISPR/Cas9 digestion in wild-type (paqr5b+/+) and paqr5b−/− mutants are indicated. A 13-nucleotide deletion was induced in the target site in the selected mutant. (Middle panel) The predicted protein sequences of the paqr5b+/+ and paqr5b−/− mutants are indicated. It was expected that a peptide of 74 amino acids in length with an N-terminal 18 amino acid sequence produced in the paqr5b−/− mutant was the same as that produced in wild-type Paqr5b zebrafish. Thus, only an N-terminal short fragment without a transmembrane region is present in the Paqr5b protein produced in the paqr5b−/− mutant. (Lowest panel) Diagrams of the predicted protein structures of the paqr5b+/+ and paqr5b−/− mutant strains are shown. The predicted transmembrane regions and the altered amino acid regions in the paqr5b−/− mutant are indicated by black boxes and diagonal boxes, respectively. (B) The relative expression levels of paqr5b in the olfactory rosette (OR) of the wild-type and the paqr5b mutants were compared. mRNA abundance was measured in triplicate for each sample from 10 fish, and all data from three preparations were normalized by the number of elongation factor 1α (EF1α) transcripts in each sample. The data indicated the expression level of the wild type set as 100. (C) Fecundity of paqr5b+/+ and paqr5b−/− zebrafish. The total number of eggs crossing each line was counted (n = 4, ** P ≤ 0.001). (D) Representative photograph of developing embryos with different sizes of chorions. The scale bar is 1 mm. Comparison of choroidal diameter between paqr5b+/+ and paqr5b−/− embryos (n = 4, **** P ≤ 0.00001). (E) Representative photograph of embryos at 5 dpf. Typical head abnormalities and spine curvatures in the paqr5b−/− embryos are indicated by arrows. The scale bars in the upper panels are 1 mm. The scale bars in the lower panels are 500 μm. (F) Comparison of the percentage of abnormal embryos between the paqr5b+/+ and paqr5b−/− embryos (n = 4, **** P ≤ 0.00001). (G) Comparison of brain transverse sections of paqr5b+/+ and paqr5b−/− embryos. The ventricle of the embryo brain is indicated by arrows. The scale bars are 20 μm.
The strain was assumed to be a null mutant with 18 amino acids without a predicted transmembrane region. Although the original mutant line was established in the transparent zebrafish strain roy (mpv17a9/a9), the mutation was introduced into the AB wild-type and albino (golden-2; brsb2/b2) strains by crossing for removal of the off-target gene. Thus, the phenotypic analysis described below was conducted in F5 or later generations. qPCR analysis revealed that the mRNA expression levels of paqr5b−/− were not different from those of the wild type (Fig. 1B).
Knockout of paqr5b resulted in head abnormalities, especially in the olfactory rosette (OR)
The null-mutant strain of paqr5b (paqr5b−/−) that we established in this study showed low fecundity (Fig. 1C), reduced chorion elevation and a high percentage of abnormal embryos (Fig. 1D). A high percentage of the embryos showed curvature of the spine and an abnormal head morphology (Fig. 1E) (Fig. 1F). The brains of the paqr5b−/− embryos were small in size, and abnormal morphology was observed; in particular, holes were formed as a result of disconnection between midbrain lobules (Fig. 1G). Paqr5b−/− embryos did not move during cultivation in Petri dishes (Supplementary Movie 1). Despite these head abnormalities, paqr5b−/− zebrafish survived to adulthood (Fig. 2A). Adult paqr5b−/− zebrafish (Fig. 2B) exhibited distinct palatine bone abnormalities (Fig. 2C, Supplementary Movies 2 and 3).
Abnormality of head structure in the paqr5b−/− mutant. (A) Survival rates of adult paqr5b+/+ and paqr5b−/− mutant fish (F6 generation). The number of surviving adult paqr5b+/+ and paqr5b-/- fish (left panel) and the percentage of head abnormalities (right panel) in each of the three batches were compared. Each value represents the mean of the data, and the vertical lines indicate the standard deviations. Asterisks represent significant differences between paqr5b+/+ and paqr5b−/− fish (* P ≤ 0.01). (B) Representative morphologies of the head region of adult paqr5b+/+ and paqr5b−/− fish are shown. The scale bars in the upper panels are 1 cm. The scale bars in the lower panels are 1 mm. (C) Micro-CT scan of the head skeleton of paqr5b+/+ and paqr5b−/−. The upper panels are volume-rendered images, and the lower panels are maximum-intensity projection images. Scale bars are 1 mm.
Paqr5b−/− zebrafish exhibited a large frontal concavity, which resulted in the upper lip being furrowed. This morphology was presumed to be caused by the loss of internal structures in the frontal region. The length of the brain of paqr5b−/− zebrafish was short, and the position of the cerebellum moved to the front and overlapped with that of the midbrain (Fig. 3A, B and C).
Abnormal morphology of the brain in paqr5b−/−. (A) Comparison of the morphology of dissected brains between paqr5b+/+ and paqr5b−/− fish. The scale bars in the upper panels are 200 μm. The scale bars in the lower panels are 100 μm. Sagittal sections (B) and transverse sections (C) of the heads of paqr5b+/+ and paqr5b−/− are shown. The olfactory bulb (OB), telencephalon (Tel), tectum opticum (TeO) and corpus cerebeli (CCe) are indicated. Scale bars are 200 μm in sagittal sections. The scale bars are 50 μm in transverse sections.
The internal structures of the head were observed by micro-CT (Fig. 4, Supplementary Movies 4). The results showed that in paqr5b−/− zebrafish, the cranial cavity was enlarged, and the size of the olfactory rosette (OR) was greatly reduced. The OR was barely connected to the olfactory bulb (OB) at the tip of the brain by nerve cells by an extremely thin tubular structure. Micro-CT images allow comparison of the morphology of the whole head of paqr5b−/− zebrafish with that of wild type (paqr5b+/+) siblings. This study focused on the OR because it was the most affected tissue by the paqr5b knockout mutation.
Observation of soft tissues inside the head region by micro-CT. Sagittal micro-CT images of the head region of adult paqr5b+/+ and paqr5b−/− are shown. The olfactory mucosa (OM), olfactory bulb (OB), telencephalon (Tel), tectum opticum (TeO) and corpus cerebeli (CCe) are indicated. Connections between the OM and OB are indicated by arrowheads. The scale bars in the upper panels are 500 μm. The scale bars in the lower panels are 100 μm.
The expression of Paqr5b was then examined by Western blot analysis using an anti-Paqr5b antibody, and the Paqr5b protein was lost in paqr5b−/− zebrafish, as predicted from the gene sequence (Fig. 5A). A rather broad protein band with a molecular weight of 55 kDa corresponding to the Paqr5b protein, presumed to be a glycoprotein, was detected in paqr5b+/+ zebrafish. The specificity of the anti-Paqr5b antibody was also confirmed by Western blot using recombinant Paqr5b protein expressed in yeast (Supplementary Fig. S3). Having confirmed the specificity of the antibody, the next step was to examine the expression sites of Paqr5b in the ORs of paqr5b+/+ and paqr5b−/− zebrafish by immunohistochemical staining using this antibody (Fig. 5B).
Expression of Paqr5b in olfactory mucosa (OM). (A) Expression analysis of Paqr5b in the OM. Proteins in the OM of paqr5b+/+ and paqr5b−/− zebrafish were detected by Coomassie Brilliant Blue R staining (CBBR). Paqr5b in the OM was detected with an anti-zebrafish Paqr5b antibody (a-Paqr5b). The arrowhead indicates a protein band corresponding to Paqr5b. (B) Immunohistochemical observation of Paqr5b in OM. Immunohistochemical staining results for Paqr5b in paqr5b+/+ and paqr5b−/− OMs. Sagittal sections of the OM of paqr5b+/+ and paqr5b−/− zebrafish were stained with an anti-zebrafish Paqr5b antibody (a-Paqr5b) and DAPI. Images of DAPI signals observed through a DAPI filter (DAPI) and images of immunostaining signals observed through the FITC filter. Merged images (Merge) of differential interference contrast (DIC) images and images of DAPI and anti-Paqr5b (a-Paqr5b) staining are shown. Enlarged images of DAPI and anti-Paqr5b-stained sections are indicated by red squares (lower panels). Neuronal cells (Crypt, Microvillus and Ciliated) that reacted with anti-Paqr5b in paqr5b+/+ are indicated by arrows. These OSNs are absent in paqr5b−/−. Nonspecific staining with the anti-Paqr5b antibody is indicated by a white arrowhead. The scale bars indicate 200 μm in the upper panels and 50 μm in the enlarged images. (C) Sagittal sections of paqr5b+/+ and paqr5b−/− were stained with hematoxylin, eosin and Alcian blue (H-E-A). The stained glycan layers are indicated by arrows. Scale bars are 200 μm in the upper panels and 50 μm in the lower panels.
In the paqr5b+/+ zebrafish OR, a large number of cells were stained with the anti-Paqr5b antibody (Fig. 5B, left panels). Five types of olfactory sensory neurons (OSNs) were identified in zebrafish OR. Among them, three major types of OSNs were found with immunofluorescent signals. Ciliated OSNs have long dendrites with cilia containing rounded olfactory knobs and express the Paqr5b protein. Microvillus neurons are another type of olfactory sensory neuron with short dendrites and moderate Paqr5b expression. Crypt neurons, found in the apical region of the OR, contain globular, short cell bodies with few cilia, resulting in relatively weak Paqr5b signals. In paqr5b−/−, Paqr5b immunoreactivity was absent in lamellar cells, confirming the loss of paqr5b expression and antibody specificity for this protein (Fig. 5B, right panels). In contrast, the surface portion of the lamellae showed strong signals in both paqr5b+/+ and paqr5b−/− zebrafish. The olfactory epithelium was positively stained with Alcian blue, which stains glycans, suggesting that Paqr5b antibodies bind to the secretory mucoid covering the OR lamellae (Fig. 5C). It was demonstrated by immunofluorescence analysis that Paqr5b was expressed in neurons in the OR (Fig. 6). The signals representing Paqr5b and a neuron cell-specific protein, acetylated tubulin, overlapped well. In paqr5b−/−, acetylated tubulin immunoreactivity was absent in lamellar cells, confirming the loss of neurons in paqr5b−/− OR (Supplementary Fig. S4). These results demonstrated that Paqr5b is expressed in OSNs in OR. We also attempted to demonstrate the involvement of Paqr5b in the regulation of cell proliferation and apoptosis of neurons by immunostaining. We performed immunostaining with phosphorylated histone H3 (anti-phospho-H3) as a marker of cell proliferation and with cleaved-caspase3 (anti-Casp3) as a marker of apoptosis. However, no significant signals were observed in either staining (Supplementary Fig. S4). It was suggested that cell proliferation and apoptosis mainly occurred during development and could not be detected in adult tissues.
Paqr5b is expressed in neurons in the OR. Immunofluorescence staining of sections of OR from paqr5b+/+. Sections were double stained with anti-rabbit Paqr5b polyclonal antibodies (α-Paqr5b), anti-acetylated tubulin monoclonal antibody (α-acetylated tubulin) and DAPI. Merged images of both antibodies and DAPI are also shown (merge). Scale bars = 50 μm.
Histological observation of OR. HE-stained longitudinal sections of paqr5b+/+ and paqr5b−/− are shown. Total morphology of the OR (upper panels). Enlarged images of lamellae in OR (lower panels). Crypts, microvilli, ciliated olfactory sensory neurons (OSNs), basal lamina and supporting cells are indicated. The scale bars indicate 200 μm in the upper panels and 20 μm in the lower panels.
The results of the immunohistochemical staining indicated that Paqr5b is a protein expressed in the OSNs of the zebrafish olfactory epithelium. These findings suggest that these neurons are lost in paqr5b−/− zebrafish. This finding was also confirmed by histological analysis. HE-stained OR sections revealed that only the basal cells and supporting cells on both sides of the basal cells were present in the lamella, resulting in a thin lamella with a low overall cell number and a lack of neurons in paqr5b−/− (Fig. 7). The OR was very small in paqr5b−/− zebrafish due to this extreme decrease in cell number. These analyses indicate that paqr5b is an essential gene for neuronal differentiation in the OR.
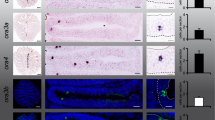
The expression of paqr5 mRNA in the OR was examined by in situ hybridization (ISH). ISH signals detected by the antisense probe were observed in the middle part of the olfactory epithelium, while signals were almost absent in the apical part of the olfactory epithelium (Fig. 8). In contrast, almost no signals were detected by the sense probe. The result suggests that differentiating neurons express paqr5b transcripts.
In situhybridization of OR. Fluorescence-stained in situ hybridization sections of paqr5b+/+ with antisense (Antisense) or sense (Sense) probes are shown. Enlarged images of sections are indicated by red squares (lower panels). Scale bars indicate 100 μm in the upper panels and 20 μm in the lower panels.
Discussion
The expression of seven paqr genes, potential pheromone targets, was examined in the ORs of zebrafish, of which paqr5b showed the highest expression6. The results of our expression analysis of paqr5b in the present study also support previous reports. Although the survival rate of the paqr5b−/− lines we produced this time was low, both the female and male homozygous mutants were fertile, and the body as a whole did not have any major problems. However, it showed a depressed morphology near the tip of the head. A micro-CT scan revealed severe abnormalities in the OR. Our current phenotypic analysis of paqr5b KO fish indicated that Paqr5b is essential for the formation of the OR. In the OR of the paqr5b−/− mutants, all OSNs appeared to be absent.
Since OSNs originate from a population of multipotent basal cells19,37,38, Paqr5b is presumed to be involved in the induction of neuronal differentiation from basal cells. However, in differentiated neurons, Paqr5b is thought to function as a receptor for odorants. In particular, Paqr5b is a notable receptor for progestin, a pheromone in fish. However, it is presumed that paqr5b is involved in the reception of a wide range of substances, as it was observed by immunostaining in almost all OSNs. However, following the ‘one receptor–one neuron’ rule39,40, it is unlikely that Paqr5b functions as a receptor. Since it has been shown that progestin is necessary for dendritic development in cerebellar neurons41,42,43, it is highly possible that Paqr5b is not a receptor for odorant substances but is responsible for inducing neuronal differentiation and maintaining neuronal activity via progesterone stimulation. The role of Paqr5b as a progestin receptor for neuron cell differentiation or a receptor for odorants should be investigated in the future. Zebrafish paqr5b is expressed throughout the embryo during the early stages of development, with particularly high expression levels in the head (Thisse, B., Thisse, C. (2004) Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission (http://zfin.org)). Thus, it is predicted that paqr5b is more widely responsible for brain development. The paqr5b−/− lines showed abnormalities in whole-brain structure during early development and in adults. The whole-brain structure is small, especially the OB, which was poorly developed and connected to the OR by a thread-like structure. The cerebellum covers the posterior part of the midbrain. Although the effects of the brain structure abnormalities on the mutant zebrafish are currently unknown, paqr5b−/− zebrafish were not very active compared to wild-type (paqr5b+/+) zebrafish (Supplementary Movies 5; wild-type (paqr5b+/+) and 6; paqr5b−/−), and their lack of neurons in the OR is expected to prevent them from sensing odors; however, their foraging behavior does not appear to be affected. It is thought that they visually target their prey, but this needs to be verified. Additionally, the increase in the size of the egg membranes of paqr5b−/− was slightly abrogated, and the egg membranes were soft and tended to be broken (Fig. 1C). These pleiotropic aberrant traits may suggest that paqr5b has broad functions. However, redundancy among paqr subtypes results in the absence of severe abnormalities.
In our study, the Western blot analysis results showed that Paqr5b was expressed in the zebrafish olfactory epithelium. An approximately 55 kDa band was detected in the olfactory epithelium of paqr5b antibody-treated zebrafish. We previously demonstrated that paqr5b (mPRγ) was expressed in the oocytes of goldfish8, but no effect on oocyte maturation was observed in morpholino knockdown experiments. mPRγ was presumed to be involved in processes other than the induction of oocyte maturation, and no severe abnormalities were observed in female or male paqr5b−/− zebrafish with regard to reproductive functions. mPR subtypes have been found to be expressed in the ovary and testis, and it is presumed that redundancy with these subtype groups results in the absence of abnormalities. Our current study provided evidence that Paqr5b is expressed in the OR and is responsible for the formation of OSNs in the OR. This report provides the first evidence of the distinct physiological role of Paqr5b (mPRγ).
Methods
Ethics statement
All the methods in this study were performed in accordance with the ARRIVE guidelines 44. The use of zebrafish and the experimental protocol were approved (approval no. 2019 F-5, 2020 F-4, 2021 F-2, 2022 F-3, 2023 F-9) by the Institutional Ethics Committee of Shizuoka University, Japan. All the methods were carried out in accordance with relevant Institutional guidelines and regulations.
Zebrafish
Zebrafish were nurtured and developed under standard laboratory conditions45. Larva were kept in Petri dishes in an incubator set at 28.5 °C until they started to swim on the surface of the water (4–5 days), after which they were moved to a plastic or glass tank with an electric heater and maintained at 28.5 °C under a 14 h light:10 h dark cycle. During the larval period, the zebrafish were fed Paramecium spp. for approximately 1 month. Adult zebrafish were fed a diet consisting of live brine shrimp in the morning and instant food (Tetra Guppy, Tetra GmbH, Melle, Germany) in the evening. The original strains of roy (mpv17a9/a9) and albino (golden-2; brsb2/b2) were generously donated by Dr. K. Kawakami and the AB wild type was donated by Dr. N. Sakai at the National Institute for Genetics, Mishima, Japan.
Genome editing
The crRNA for the paqr5b gene and tracrRNA were purchased from Horizon Discovery Ltd. The Cas9 nuclease (Guide-it™ Recombinant Cas9) was purchased from Takara (Shiga Japan). Equal volumes of crRNA (10 µM), Cas9 (500 ng/ml), and tracrRNA (20 µM) were mixed, and approximately 4 nl of the mixture was microinjected into a 1-cell-stage embryo by using an IM300 microinjector (Narishige Scientific Instrument Lab, Tokyo, Japan) under a binocular microscope. The edited larvae (F0) were incubated in Petri dishes at 28.5 °C for 5 days and were released to a culture tank under suitable conditions after they started to swim. The genotype of each fish was analyzed through a heteroduplex mobility assay (HMA) and DNA sequencing as previously described36.
RNA extraction and qPCR
Total RNA was extracted from the ORs from paqr5b+/+ and paqr5b−/− fish using the ISOGEN-LS RNA extraction reagent (Nippon Gene, Tokyo, Japan) according to the manufacturer’s protocol. For cDNA synthesis, 1 µg of each RNA and Illustra Ready-To-Go RT–PCR beads (GE Healthcare Life Science, IL, USA) were used according to the manufacturer’s instructions. Quantitative PCR was conducted to investigate the expression level of paqr5b in each sample. The 20 µl reaction mixture contained 1 µl of each of the 10 µM forward primer (5’-TGCCTACCCGTACCTGTTTG-3’) and reverse primer (5’-CTCTCGGGTAAGTGGGTTGC-3’) solutions, 10 µl of SYBR green PCR master mix, and 5 µl of 10-fold-diluted cDNA. qPCR was performed using a LightCycler 480 (Roche Applied Science, Mannheim, Germany) with the following program: 95 °C for 5 min; 45 cycles of 95 °C for 15 s, 56 °C for 15 s, and 72 °C for 20 s; and then 65 °C and 95 °C for 20 s each for the final melting curve analysis. Each sample was analyzed 3 times as replicates. The mRNA expression level was normalized to the expression level of a common reference gene, elongation factor 1α (EF1α).
Mutant line generation and phenotype observation
Heterozygous mutant paqr5b (paqr5b+/−) zebrafish were generated by pairing F0 mutant zebrafish and wild-type zebrafish. This paqr5b+/− strain was inbred to produce a homozygous mutant (paqr5b−/−) in the F2 generation. F2 paqr5b−/− were outbred with wild-type or albino strains. Additionally, the wild-type siblings (paqr5b+/+) were outbred with the wild-type or albino strains. Homozygous outbred wild-type or albino paqr5b+/+ and paqr5b−/− strains were obtained from the F4 generation. Phenotypic observation started from the F5 generation produced from inbred paqr5b+/+ or paqr5b−/− zebrafish.
Stereoscopic observation
A stereomicroscope (Olympus SZX12) and a microscope camera (Olympus DP70) were used for observation and imaging. Paqr5b+/+ and paqr5b−/− embryos were observed and photographed under a microscope.
Micro-CT imaging
Wild-type paqr5b+/+ and mutant paqr5b−/− adult zebrafish were fixed with 4% paraformaldehyde and stored in 70% ethanol. For whole head skeleton observation, the samples were observed directly (Fig. 2C). The heads of the zebrafish were scanned using an X-ray micro-CT device (ScanXmate-CF110TSH320/460, Comscantechno Co., Ltd., Japan) at a tube voltage peak of 40 kVp and a tube current of 70 µA. The sample was rotated 360 degrees in steps of 0.24 degrees, generating 1500 projection images of 2064 × 1548 pixels. The micro-CT data were reconstructed at an isotropic resolution of 5.0 μm for the observation of skeletons. For observation of soft tissues, the samples were stained with 25% Lugol staining solution for 4 days (Fig. 4). The heads of the zebrafish were scanned using an X-ray micro-CT device (ScanX- mate-E090S105, Comscantechno Co., Ltd., Japan) at a tube voltage peak of 85 kVp and a tube current of 90 µA. The sample was rotated 360 degrees in steps of 0.2 degrees, generating 1800 projection images of 992 × 992 pixels. To obtain high-resolution data for the entire head, the position of the sample was shifted, and the images were taken in two stages. The micro-CT data were reconstructed at an isotropic resolution of 2.85 μm. Finally, the two datasets were combined into a single set of data for the entire head, which was used for image analysis. We conducted micro-CT on three replicates each from paqr5b+/+ and paqr5b−/− zebrafish (Fig. 4). Three-dimensional tomographic images were obtained using OsiriX MD software (version 13, Pixmeo, SARL, Switzerland) and Imaris software (version 9.1, Carl Zeiss Microscopy Co., Ltd., Japan). Supplementary Movies were edited using Adobe Premiere Pro CC (Adobe Systems Co., Ltd., Japan).
Production of the anti-Paqr5b polyclonal antibody
Polyclonal antibodies against three synthetic 13-mer peptides derived from the C-terminal domain (no. 336–347 amino acids) of zebrafish paqr5b (YSSNPKNTANKE) conjugated to keyhole limpet hemocyanin were generated by a commercial vendor (Eurofins Genomics, Tokyo, Japan) in rabbits.
Western blot analysis
Ten ORs were dissected from five paqr5b+/+ and paqr5b−/− zebrafish. ORs were homogenized with 20 µl of SDS sample buffer solution. Ten microliters of sample was added to each lane of an SDS‒PAGE gel (12%) and subjected to electrophoresis at 30 mA for 70 min. The separated proteins were stained with CBBR or electroblotted onto an ImmobilonR-P Hydrophobic PVDF Transfer Membrane (0.45 μm pore size, Merck) at 15 V and 400 mA for 45 min on an ATTO Western blotting semidry system. Nonspecific binding sites were blocked by incubation with 5% skim milk powder in TBS containing 0.1% Tween-20 (TTBS) at 4 °C overnight. The next day, the membrane was washed with 10 ml of 1× TTBS for 5 min 3 times. The blot was then incubated with zebrafish paqr5b antiserum (1:1000) diluted in TBS for 2 h at 4 °C. After washing with TTBS 3 times, the blot was incubated with a goat anti-rabbit IgG (H + l) HRP-conjugated secondary antibody (1:2000) in TBS for 2 h at 4 °C. The blot was then rinsed with 10 ml of 1× TTBS for 5 min 3 times. Then, the membrane was incubated with 400 µL of a mixture of chemiluminescent reagent from a Western Lightning Plus-ECL kit. The blot was imaged using a Fujifilm LAS 4000 mini (ImageQuant™ LAS 4000 mini, version 1.3), and the acquired images were analyzed with ImageQuant TL software.
Immunohistochemistry
Eight-month-old zebrafish were used for analysis. The head was fixed with 4% paraformaldehyde overnight or for 24 h at 4 °C. The fixed samples were then dehydrated through an ethanol gradient (70%, 90%, 100% ethanol) for 30 min 3 times and washed with LEMOSOL for 30 min 2 times, after which paraffin-embedded sections were prepared. Sagittal Sect. (5 μm thick) were prepared, and the sections were cleared with xylene and hydrated with a graded ethanol series (100%, 90%, 70%, and ethanol). Slides containing samples were treated with blocking solutions containing 5% skim milk in 3% bovine serum albumin (BSA)/1× Tris-buffered saline (TBST, pH 7.4) and kept at 4 °C for 1 h. The sections were thoroughly washed twice with 1× TTBS buffer solution for 10 min. The sections were then incubated with a zebrafish paqr5b monoclonal antibody (1:50) and anti-acetylated a-tubulin (1:50, Sigma-Aldrich T6793) at 4 °C overnight. The next day, the sections were fully immersed in 1× TTBS buffer solution twice for 10 min and incubated with anti-rabbit IgG (H + L) secondary Alexa Fluor 488 conjugate (1:500) (Cell Signaling Technology, MA, USA), anti-mouse IgG (H + L), and F(ab’)2 fragment (Alexa Fluor 555 conjugate) (1:500, Cell Signaling) for 1 h at room temperature. For DAPI staining (1:1000), the samples were rinsed for 30 min at 4 °C for 1 h. After two 10 min washes with the same buffer solution, the samples were mounted with ProlongTM Glass Antifade Mountant and NucBlue™ (Invitrogen by Thermo Fisher Scientific). Immunostained sections were observed using a Carl Zeiss LSM-700 microscope. The fluorescence signal was detected by laser excitation of the fluorophores at 405 and 488 nm, and detection was performed with internal photomultipliers.
Histology
Zebrafish heads were fixed with Bouin’s solution (15:5:1 = picric acid: formaldehyde: acetic acid) for 2 days. The fixed samples were then dehydrated through a graded ethanol series (70%, 90%, 100% ethanol) and washed with 100% LEMOSOL. Paraffin-embedded sections were prepared with sagittal slices of 5 μm thickness and kept on a heater at 65 °C overnight for two days. Slides were deparaffinized with 100% xylene and rehydrated with 100%, 90%, and 70% ethanol and ultrapure water. Hematoxylin and eosin were used for staining. Mounting was performed with a Mount Quick (Daido Sangyo Co., Ltd., Japan). Arsenic blue staining was performed for additional staining of the mucosa. Images of the sections were taken with an Olympus fluorescence microscope (Olympus, BX60, Japan).
In situ hybridization histochemistry
DIG-labeled antisense and sense probes for zebrafish paqr5b mRNA were generated by a commercial vendor (Tokushima Molecular Pathology Institute, Inc, Tokushima, Japan). Freshly collected OR samples were fixed in 4% w/v paraformaldehyde in PBS (pH 7.4) overnight at 4 °C, followed by immersed in 30% w/v sucrose in PBS at 4 °C. The fixed OR was transferred to an embedding chamber and embedded with Tissue-tek O.C.T. compound. The embedding chamber was dipped into liquid N2. Samples were cut into 10 μm thick sections on a cryostat microtome (CryoStar NX70, Thermo Fisher Scientific, MA, USA) at -20 °C. Free-floating sections washed for 5 min in diethylpyrocarbonate-treated phosphate-buffered saline (DEPC-PBS) were pretreated with 0.2 N HCl for 20 min, washed twice for 5 min in DEPC-PBS, and then acetylated in 0.1 M triethanolamine–HCl (pH 8.0) containing 0.25% acetic anhydride for 10 min. Before the hybridization step, sections were washed again twice for 5 min with DEPC-PBS. All pretreatments were performed at 4 °C. Following the pretreatment, sections were preincubated in hybridization buffer (50% Formamide, 2X SSC, 1 µg/µl tRNA, 1 µg/µl Salmon sperm DNA, 1 µg/µl BSA, 10% Dextran sulfate ) at 55 °C for 1 h and then hybridized with DIG-labeled cRNA probes (800 ng/ml; denatured at 95 °C for 5 min and cooled at 4 °C for 5 min shortly before use) in the same buffer at 64 °C for 16 h. After hybridization, the sections were washed with 2XSSC (300 mM NaCl, and 30 mM sodium citrate, pH 7.0) containing 50% formamide at 55 °C for 1 h, rinsed in wash buffer (500 mM NaCl, 10 mM Tris–HCl, pH 8.0, and 1 mM EDTA, pH 8.0) for 10 min and then incubated with RNase A (20 mg/ml; Sigma–Aldrich, St. Louis, MO) in wash buffer at 37 °C for 30 min. After being rinsed in wash buffer again for 10 min, they were soaked in 2XSSC containing 50% formamide and 0.2XSSC containing 50% formamide at 55 °C for 30 min each. Bound DIG probes were detected with Anti-Digoxigenin-POD, Fab fragments (Roche) followed by fluorescence detection by Styramide™ Signal Amplification (PSA™) system (AAT Bioquest, CA USA). The samples were mounted with ProlongTM Glass Antifade Mountant and NucBlue™ (Invitrogen by Thermo Fisher Scientific). Sections were observed using a Carl Zeiss LSM-700 microscope.
Statistical analysis
Summary data are presented as the mean ± S.D. Student’s t test was used to determine the statistical significance of the differences in the data using GraphPad Prism (San Diego, CA, USA). The data were considered significant at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****).
Reporting summary
Further information on the research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided in this paper.
References
Tang, Y. T. et al. PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J. Mol. Evol. 61, 372–380. https://doi.org/10.1007/s00239-004-0375-2 (2005).
Thomas, P. et al. Steroid and G protein binding characteristics of the seatrout and human progestin membrane receptor alpha subtypes and their evolutionary origins. Endocrinology. 148, 705–718. https://doi.org/10.1210/en.2006-0974 (2007).
Zhu, Y., Bond, J. & Thomas, P. Identification, classification, and partial characterization of genes in humans and other vertebrates homologous to a fish membrane progestin receptor. Proc. Natl. Acad. Sci. USA. 100, 2237–2242. https://doi.org/10.1073/pnas.0436133100 (2003).
Wu, X. J., Liu, D. T., Chen, S., Hong, W. & Zhu, Y. Impaired oocyte maturation and ovulation in membrane progestin receptor (mPR) knockouts in zebrafish. Mol. Cell. Endocrinol. 511, 110856. https://doi.org/10.1016/j.mce.2020.110856 (2020).
Pang, Y., Dong, J. & Thomas, P. Characterization, neurosteroid binding and brain distribution of human membrane progesterone receptors delta and epsilon (mPRdelta and mPRepsilon) and mPRdelta involvement in neurosteroid inhibition of apoptosis. Endocrinology. 154, 283–295. https://doi.org/10.1210/en.2012-1772 (2013).
Saraiva, L. R. et al. Molecular and neuronal homology between the olfactory systems of zebrafish and mouse. Sci. Rep. 5, 11487. https://doi.org/10.1038/srep11487 (2015).
Tokumoto, M., Nagahama, Y., Thomas, P. & Tokumoto, T. Cloning and identification of a membrane progestin receptor in goldfish ovaries and evidence it is an intermediary in oocyte meiotic maturation. Gen. Comp. Endocrinol. 145, 101–108. https://doi.org/10.1016/j.ygcen.2005.07.002 (2006).
Tokumoto, T. et al. Characterization of multiple membrane progestin receptor (mPR) subtypes from the goldfish ovary and their roles in the induction of oocyte maturation. Gen. Comp. Endocrinol. 177, 168–176. https://doi.org/10.1016/j.ygcen.2012.03.005 (2012).
Kazeto, Y., Goto-Kazeto, R., Thomas, P. & Trant, J. M. Molecular characterization of three forms of putative membrane-bound progestin receptors and their tissue-distribution in channel catfish, Ictalurus punctatus. J. Mol. Endocrinol. 34, 781–791. https://doi.org/10.1677/jme.1.01721 (2005).
Mourot, B., Nguyen, T., Fostier, A. & Bobe, J. Two unrelated putative membrane-bound progestin receptors, progesterone membrane receptor component 1 (PGMRC1) and membrane progestin receptor (mPR) beta, are expressed in the rainbow trout oocyte and exhibit similar ovarian expression patterns. Reprod. Biol. Endocrinol. 4, 6. https://doi.org/10.1186/1477-7827-4-6 (2006).
Morini, M. et al. Nuclear and membrane progestin receptors in the European eel: Characterization and expression in vivo through spermatogenesis. Comp. Biochem. Physiol. Mol. Integr. Physiol. 207, 79–92. https://doi.org/10.1016/j.cbpa.2017.02.009 (2017).
Firestein, S. How the olfactory system makes sense of scents. Nature. 413, 211–218. https://doi.org/10.1038/35093026 (2001).
Kermen, F., Franco, L. M., Wyatt, C. & Yaksi, E. Neural circuits mediating olfactory-driven behavior in fish. Front. Neural Circuits. 7, 62. https://doi.org/10.3389/fncir.2013.00062 (2013).
Tierney, K. B. et al. Olfactory toxicity in fishes. Aquat. Toxicol. 96, 2–26. https://doi.org/10.1016/j.aquatox.2009.09.019 (2010).
Howe, K. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 496, 498–503. https://doi.org/10.1038/nature12111 (2013).
Sakai, C., Ijaz, S. & Hoffman, E. J. Zebrafish models of neurodevelopmental disorders: Past, present, and future. Front. Mol. Neurosci. 11, 294. https://doi.org/10.3389/fnmol.2018.00294 (2018).
Ganzen, L., Venkatraman, P., Pang, C. P., Leung, Y. F. & Zhang, M. Utilizing zebrafish visual behaviors in Drug Screening for Retinal Degeneration. Int. J. Mol. Sci. 18https://doi.org/10.3390/ijms18061185 (2017).
Døving, K. B., Dubois-Dauphin, M., Holley, A. & Jourdan, F. Functional anatomy of the olfactory organ of fish and the ciliary mechanism of water transport. Acta Zool. 58, 245–255 (1977).
Hansen, A. & Zielinski, B. S. Diversity in the olfactory epithelium of bony fishes: Development, lamellar arrangement, sensory neuron cell types and transduction components. J. Neurocytol. 34, 183–208. https://doi.org/10.1007/s11068-005-8353-1 (2005).
Hara, T. J. Chemoreception in fishes (1982).
Byrd, C. A. & Brunjes, P. C. Organization of the olfactory system in the adult zebrafish: Histological, immunohistochemical, and quantitative analysis. J. Comp. Neurol. 358, 247–259. https://doi.org/10.1002/cne.903580207 (1995).
Hansen, A. & Zeiske, E. The peripheral olfactory organ of the zebrafish, Danio rerio: An ultrastructural study. Chem. Senses. 23, 39–48. https://doi.org/10.1093/chemse/23.1.39 (1998).
Villamayor, P. R. et al. A comprehensive structural, lectin and immunohistochemical characterization of the zebrafish olfactory system. Sci. Rep. 11, 8865. https://doi.org/10.1038/s41598-021-88317-1 (2021).
Ahuja, G. et al. Zebrafish crypt neurons project to a single, identified mediodorsal glomerulus. Sci. Rep. 3 (2063). https://doi.org/10.1038/srep02063 (2013).
Ahuja, G. et al. Kappe neurons, a novel population of olfactory sensory neurons. Sci. Rep. 4, 4037. https://doi.org/10.1038/srep04037 (2014).
Friedrich, R. W., Jacobson, G. A. & Zhu, P. Circuit neuroscience in zebrafish. Curr. Biol. 20, R371–381. https://doi.org/10.1016/j.cub.2010.02.039 (2010).
Baier, H. & Korsching, S. Olfactory glomeruli in the zebrafish form an invariant pattern and are identifiable across animals. J. Neurosci. 14, 219–230. https://doi.org/10.1523/JNEUROSCI.14-01-00219.1994 (1994).
Friedrich, R. W., Genoud, C. & Wanner, A. A. Analyzing the structure and function of neuronal circuits in zebrafish. Front. Neural Circuits. 7, 71. https://doi.org/10.3389/fncir.2013.00071 (2013).
Byrd, C. A. et al. Ontogeny of odorant receptor gene expression in zebrafish, Danio rerio. J Neurobiol 29, 445–458. https://doi.org/10.1002/(SICI)1097-4695(199604)29:4<445::AID-NEU3>3.0.CO;2-8 (1996).
Korsching, S. I. et al. Olfaction in zebrafish: What does a tiny teleost tell us? Semin Cell. Dev. Biol. 8, 181–187. https://doi.org/10.1006/scdb.1996.0136 (1997).
Hildebrand, J. G. & Shepherd, G. M. Mechanisms of olfactory discrimination: Converging evidence for common principles across phyla. Annu. Rev. Neurosci. 20, 595–631. https://doi.org/10.1146/annurev.neuro.20.1.595 (1997).
Miyasaka, N. et al. Functional development of the olfactory system in zebrafish. Mech. Dev. 130, 336–346. https://doi.org/10.1016/j.mod.2012.09.001 (2013).
Friedrich, R. W. & Laurent, G. Dynamic optimization of odor representations by slow temporal patterning of mitral cell activity. Science. 291, 889–894. https://doi.org/10.1126/science.291.5505.889 (2001).
Braubach, O. R., Fine, A. & Croll, R. P. Distribution and functional organization of glomeruli in the olfactory bulbs of zebrafish (Danio rerio). J. Comp. Neurol. 520 (Spc2311), 2317–2339. https://doi.org/10.1002/cne.23075 (2012).
Pozzuto, J. M., Fuller, C. L. & Byrd-Jacobs, C. A. Deafferentation-induced alterations in mitral cell dendritic morphology in the adult zebrafish olfactory bulb. J. Bioenerg Biomembr. 51, 29–40. https://doi.org/10.1007/s10863-018-9772-x (2019).
Pachoensuk, T. et al. Zebrafish stm is involved in the development of otoliths and of the fertilization envelope. Reprod. Fertil. 2, 7–16. https://doi.org/10.1530/RAF-20-0040 (2021).
Marui, T. & Caprio, J. H. in Teleost Gustation171–198 (& TJ, 1992).
Ma, E. Y., Heffern, K., Cheresh, J. & Gallagher, E. P. Differential copper-induced death and regeneration of olfactory sensory neuron populations and neurobehavioral function in larval zebrafish. Neurotoxicology. 69, 141–151. https://doi.org/10.1016/j.neuro.2018.10.002 (2018).
Malnic, B., Hirono, J., Sato, T. & Buck, L. B. Combinatorial receptor codes for odors. Cell. 96, 713–723. https://doi.org/10.1016/s0092-8674(00)80581-4 (1999).
Serizawa, S. et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 302, 2088–2094. https://doi.org/10.1126/science.1089122 (2003).
Sakamoto, H., Ukena, K. & Tsutsui, K. Effects of progesterone synthesized de novo in the developing Purkinje cell on its dendritic growth and synaptogenesis. J. Neurosci. 21, 6221–6232 (2001).
Tsutsui, K. Neurosteroids in the Purkinje cell: Biosynthesis, mode of action and functional significance. Mol. Neurobiol. 37, 116–125 (2008).
Haraguchi, S. et al. Light-at-night exposure affects brain development through pineal allopregnanolone-dependent mechanisms. Elife 8. https://doi.org/10.7554/eLife.45306.
du Sert, N. P. et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. Plos Biology 18. https://doi.org/10.1371/journal.pbio.3000411 (2019).
Westerfield, M. The Zebrafish Book: A Guide for the laboratory Use of Zebrafish (Danio rerio). (Univ. of Oregon Press, 1995).
Acknowledgements
This study was supported by Grants-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science and JSPS KAKENHI Grant Number 23K05830 (to TT).
Author information
Authors and Affiliations
Contributions
U.H.M. maintained the zebrafish strains and performed microinjection, histological analysis, immunofluorescence staining, Western blotting, analyzed the data and drafted the manuscript. A.M. conducted the micro-CT. H.A. and U.H.M. performed the Western blotting. MMR performed microinjection to establish the mutant strain. T.T. participated in the study design, supervised the study and wrote the paper. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 3
Supplementary Material 4
Supplementary Material 5
Supplementary Material 6
Supplementary Material 7
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mustary, U.H., Maeno, A., Rahaman, M.M. et al. Membrane progesterone receptor γ (paqr5b) is essential for the formation of neurons in the zebrafish olfactory rosette. Sci Rep 14, 24354 (2024). https://doi.org/10.1038/s41598-024-74674-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74674-0
Keywords
This article is cited by
-
Loss of responses to odorants and pheromones in mPRγ (paqr5b)-knockout zebrafish
Fish Physiology and Biochemistry (2025)