Abstract
Adeno-associated virus (AAV) vectors can mediate long-term expression of immunogenic transgenes in vivo through transduction of tolerogenic cells in the liver. Tissue-targeted AAV vectors allow transduction of non-hepatic cells, but this necessitates development of strategies to minimize transgene immunogenicity. Here, we first validated that AAV capsids with tissue-specific tropism and transgene promoters enabled expression of the immunogenic protein, firefly luciferase, in liver, muscle, or adipose tissue. Cellular immunity was detectable in animals where luciferase was expressed in muscle or adipose, but not liver tissue. With the objective of enhancing tolerance of transduced non-hepatic cells, AAV vectors were engineered to co-express luciferase plus the immune checkpoint protein, PD-L1. In animals where transduced cells expressed luciferase but not PD-L1, there was incremental depletion of transduced cells over time. By contrast, the bioluminescent signal increased incrementally over the study, and was significantly greater, in the muscle and adipose tissue of animals where PD-L1 was co-expressed with luciferase. Our data demonstrate that PD-L1 co-expression facilitates persistent, tissue-targeted expression of immunogenic transgenes without transducing tolerogenic hepatic cells. Our strategy of PD-L1 co-expression may provide a versatile platform for sustained expression of immunogenic transgenes in gene and cell therapies.
Similar content being viewed by others
Introduction
Immune response to therapeutic transgene products represents safety and efficacy barriers in gene and cell therapies1,2. For instance, in gene replacement therapies, the patients’ immune system will recognize a corrected wildtype protein as foreign, and similarly, patients’ treated with chimeric antigen receptor (CAR) T cell therapies may evolve a humoral and cellular immune responses to the CAR scFv or VHH domains, which are often of non-human origin2,3. Recombinant adeno associated virus (AAV) vectors are preferred gene delivery vehicles for systemic gene therapies due to their high in vivo transduction efficiency, tunable tissue tropisms and relatively low immunogenicity4,5. Notably, AAV vectors induce systemic immunological tolerance to transgene products through transduction of tolerogenic liver cells, such as hepatocytes, hepatic regulatory T and Kupffer cells6,7,8,9.
Liver toxicity is a major safety concern associated with high dose systemic AAV gene therapies10. To minimize liver toxicity and off-targeted transgene expression, AAV vectors with tissue-selective capsids and tissue-specific promoters have been employed to target extrahepatic cells. However, achieving tissue-selective transgene expression while avoiding the transduction of tolerogenic liver cells may heighten the risk of triggering an immune response to the AAV transgene product. Indeed, muscle-targeted AAV gene therapy approaches for Duchenne muscular dystrophy have demonstrated anti-dystrophin immune response in patients and animal models11,12,13. Similarly, AAV-mediated selective expression of an immunogenic transgene in beta cells led to a robust transgene-specific immune response and decreased beta cell mass in a mouse model14. Thus, it is critical to develop strategies to minimize transgene immunogenicity for tissue-targeted AAV therapies which are independent of tolerogenic liver cell transduction.
PD-L1 is a potent immune checkpoint regulator which binds the PD-1 receptor found on lymphocytes, macrophages, and dendritic cells15. Importantly, PD-1 receptor binding prevents cytotoxicity and inflammatory cytokine release from cytotoxic CD8+ T cells as well as natural killer cells16. PD-L1 is ubiquitously expressed in the liver in stellate, Kupffer, and dendritic cells17,18,19, and it is a key signaling factor that facilitates immune tolerance via the liver20. PD-L1 expression is also induced in hepatocytes in response to a novel or immunogenic antigen21. It is of note that AAV-mediated delivery of PD-L1 in joints was shown to block progression of arthritis in a mouse model without impacting systemic immune responses22. Similarly, beta cell-targeted expression of PD-L1 and CTLA4-Ig by AAV has been shown to block development of autoimmune-mediated diabetes in a mouse model23.
In this study, our aims were (i) to determine the influence of AAV-mediated, tissue-targeted expression of an immunogenic transgene on persistent transduction and immune response induction, and (ii) to develop a strategy to achieve persistent expression of an immunogenic transgene without transducing tolerogenic hepatic cells. We used a muscle-targeted AAV vector because of its known immunogenicity risk12,13. We also selected an adipose-targeted AAV vector, as the immune consequences of adipose-targeted transgene expression have not been well characterized, despite its metabolic importance and secretory potential24,25,26. We also included a liver-targeted AAV vector as its immune tolerance induction has been well established6,7. Firefly luciferase was selected as an immunogenic transgene because of its well-established immunogenicity and T cell epitope in C57BL/6 mice27,28,29, and because its expression in vivo can be easily quantified using bioluminescence imaging. We then developed a novel strategy that used co-expression of PD-L1 to mitigate immunogenicity and promote sustained transgene expression in adipose or muscle tissue without the involvement of tolerogenic hepatic cells.
Results
Screening study and assessment of liver, muscle and adipose tissue targeting
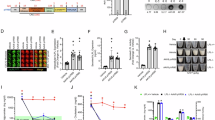
In screening studies, we assessed tissue targeting in mice using a combination of AAVs with differing tissue tropisms, tissue-specific promoters, and infusion of virus via different routes (Figs. 1 and 2). AAV8 and AAV9 were selected for their tropism for liver30, adipose31,32 or muscle33 tissue. Transgenes were driven by a chimeric human α anti-trypsin and human apolipoprotein E (AAT/APOE)34, Adiponectin35, or MHCK736 promoters, which are expressed in liver, adipose, or muscle cells, respectively. Schematics of the viral constructs used in screening assays are shown in Fig. 1A, with vector maps provided in Supplementary Fig. 1. Viral constructs expressed the immunogenic transgene firefly luciferase27,28,29, and with the goal of eliciting an immune response, AAV’s were delivered at a high dose of 6.8 × 1013 viral genomes (VG)/kg, or 1.7 × 1012 VG for an average 25 g mouse.
Screening of tissue-specific firefly luciferase expression, targeting liver and muscle tissues. (A) Screening assay vector construct design. (B) Liver targeting via AA8-AAT/APOE-Luc IV dosing, with bioluminescence imaging at weeks 1 and 3. (C) Skeletal and cardiac muscle targeting via AAV8-MHCK7-Luc IV or AAV9-MHCK7-Luc IV, with bioluminescence imaging at weeks 1 and 3. (D) Quantification of bioluminescence from mice treated with AA8-AAT/APOE-Luc IV at weeks 1 and 3. (E) Ex vivo tissues from animals treated with AA8-AAT/APOE-Luc IV at week 3. (F) Quantification of bioluminescence from mice treated with AAV8-MHCK7-Luc IV or AAV9-MHCK7-Luc IV at weeks 1 and 3. (G) Ex vivo tissues from animals treated with AAV8-MHCK7-Luc IV or AAV9-MHCK7-Luc IV at week 3. Firefly luciferase abbreviated as “Luc” in figure. Statistics, GraphPad Prism, two-way ANOVA with Tukey’s multiple comparison test, n = 5, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Screening of tissue specific firefly luciferase expression, targeting adipose tissue and assessing immune responses. (A) Adipose tissue targeting via AAV8-Adipose delivered via intravenous (IV), subcutaneously (SC), or intraperitoneal (IP) injection with bioluminescence imaging of mice at weeks 1 and 3. Control animals received AAV8-Adipose-Nil, containing no transgene, delivered via IV. (B) Quantification of bioluminescence imaging at weeks 1 and 3. (C) Quantification of bioluminescence from ex vivo tissues at week 3. (D) Example IFN-γ ELISPOT data used in caption E; complete raw IFN-γ ELISPOT data presented in Supplementary Fig. 2. (E) Quantification of IFN-γ ELISPOT used to detect a cellular immune response to the firefly luciferase transgene. (F) Assessment of humoral immune response through quantification of relative western blot signals for Anti-firefly luciferase IgG, relative to Gapdh. Firefly luciferase abbreviated as “Luc” in figure. Statistics, GraphPad Prism, A-D two-way ANOVA with Tukey’s multiple comparison test, F one-way ANOVA with Dunnett’s multiple comparison test, n = 5, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Liver was targeted using a combination of AAV8, the AAT/APOE promoter, and intravenous (IV) infusion (AAV8-AAT/APOE-Luc IV), resulting in a strong localized bioluminescence signal detectable in the abdominal regions of mice at weeks 1 and 3 (Fig. 1B). The bioluminescence signal was similar at weeks 1 and 3 (Fig. 1D). At week 3, animals were euthanized, and the heart, liver, quadriceps muscle, spleen, and adipose tissues isolated, and tissue bioluminescence signal assessed ex vivo. The bioluminescence signal from the liver was statistically greater than from any other tissue or organ in these animals (Fig. 1E). While the bioluminescence signal resulting from AAV8-AAT/APOE-Luc-IV transduction was largely restricted to the liver, the signal was so great that the absolute whole-body luminescence was orders of magnitude greater than that achieved using viral systems that targeted muscle or adipose tissues (Fig. 1D, versus Figs. 1F and 2B).
Skeletal and cardiac muscle were targeted through IV infusion of AAV8 or AAV9, with the firefly luciferase gene driven by the MHCK7 promoter (AAV8-MHCK7-Luc or AAV9-MHCK7-Luc). Figure 1C and F show whole body imaging and quantification of bioluminescence signal, demonstrating that the bioluminescence signal was similar for each viral condition at weeks 1 and 3. At week 3, both AAV8-MHCK7-Luc-and AAV9-MHCK7-Luc yielded significantly greater bioluminescence signal from the heart, quadricep muscle, and liver, relative to other tissues in the same animals (Fig. 1G). There was no significant difference between animal groups treated with AAV8 or AAV9 (Fig. 1F).
Prior to this study, immune responses to adipose tissue-specific transgene expression had not been well characterized. To target firefly luciferase transgene expression to adipose tissues, AAV8 with a modified Adiponectin promoter used (AAV8-Adipose-Luc), as well as an empty or Nil firefly luciferase control (AAV8-Adipose-Nil, Fig. 2A). Three routes of administration were compared for overall transduction efficiency and immunogenicity. Animals were dosed intravenously (IV) via tail vein injection, subcutaneously injection (SC) into the lower central back, or via intraperitoneal (IP) injection. Bioluminescence signal from animals was assessed at weeks 1 and 3, with tissues assessed ex vivo at termination. Based on day 7 IVIS imaging, IP delivered virus transduced adipose yielded the greatest signal, while SC delivered virus yielded the weakest signal (Fig. 2B). As expected, no firefly luciferase activity was apparent in the empty virus treated cohort (AAV8-Adipose-Nil). In contrast to what was observed in muscle and liver targeted groups, there was loss (albeit a statistically insignificant loss) of bioluminescence signal in IP and SC AAV8-Adipose-Luc treated animals between weeks 1 and 3. While greater bioluminescence signal was detected from all adipose tissues (and some off target signal from livers), statistical significance was only observed for visceral adipose relative to other tissues where AAV8-Adipose-Luc was delivered via IP (Fig. 2C). Failure to observe statistical significance for adipose tissues, relative to other tissues, in the IV and SC animal groups was likely due to the substantial loss in bioluminescence signal between weeks 1 and 3. SC animals had the weakest whole body bioluminescence signal at both weeks 1 and 3, and weakest visceral adipose, subcutaneous adipose, and brown adipose signals, relative to other injection routes. These data suggested that SC delivery is associated with either reduced transduction efficiency or heightened transgene immunogenicity.
IFN-γ ELISPOT was performed to assess the cellular immune response to firefly transgene expression (Fig. 2D). AAV8-AAT/APOE-Luc-IV targeting the liver and AAV8-Adipose-Nil (no transgene expression) yielded equivalent, and effectively no cellular immune response. By contrast, AAV8-Adipose-Luc-SC adipose targeted firefly luciferase expression did yield a significant cellular immune response. This study is the first to evaluate the cellular immune response to adipose specific transgene expression. The strong cellular immune response and loss of firefly luciferase activity observed for SC delivered AAV8-Adipose-Luc are indicative of cytotoxic lymphocyte mediated immune clearance of transduced cells. Similarly, AAV8 or AAV9 virus with MHCK7 driven firefly luciferase expression in the muscle elicited a significant cellular immune response (see IFN-γ ELISPOT data in Fig. 2D and data graphed in Fig. 2E). All routes of administration induced a humoral immune response to the firefly luciferase transgene as assessed via western blot quantification of anti-firefly luciferase IgG antibodies in the blood, albeit with only a statistically meaningful signal generated from AAV8-Adipose-Luc-IV and AAV8-MHCK7-Luc relative to the AAV8-Adipose-Nil control (Fig. 2F).
Using PD-L1 to mitigate the cellular immune response to muscle and adipose transgene expression
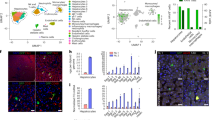
Firefly luciferase transgene expression in muscle and adipose tissue elicited a cellular immune response, while firefly luciferase expression in the liver did not. As our goal was to mitigate the cellular immune response and sustain transgene expression, we used highly immunogenic transduction strategies in the next portion of our study. IV delivery of AAV9, having muscle tropism33, and the MHCK736 muscle-specific promoter were selected as a clinically relevant and immunogenic model. SC delivery of AAV8, having adipose tissue tropism31,32, with a modified Adiponectin35 adipose-specific promoter, were selected as a highly immunogenic model; this combination resulted in significant and almost complete loss of transduced cells during the 3-week screening study (Fig. 2A,B). AAV expression constructs were designed to express the immunogenic peptide firefly luciferase under regulation of a modified Adiponectin or MHCK7 promoters, with or without a P2A linker followed by PD-L1 (Fig. 3A,B). PD-L1 expression by transduced cells was predicted to dampen T cell cytotoxicity and inflammatory cytokine release as depicted schematically in Fig. 3C.
Tissue targeting and induction of immune tolerance through co-expression of PD-L1. (A) Graphical abstract for tissue targeting. (B) AAV construct design for tissue specific expression and immune tolerance through the co-expression of PD-L1. (C) Graphical abstract for PD-L1 driven blockade of T-cell mediated immune clearance of transduced cells. Luciferase refers to firefly luciferase in figure. Captions were drawn using BioRender.
Co-expression of PD-L1 elevates and sustains transgene expression in the heart and skeletal muscle
AAV9-MHCK7-Luc was delivered at an IV dose of 4 × 1013 vg/kg, or 1.7 × 1012 vg for an average 25 g mouse. One group of mice received AAV9-MHCK7-Luc, while the other received AAV9-MHCK7-Luc-PD-L1. IVIS bioluminescent imagining was performed weekly for four weeks (day 7 to day 29), then animals sacrificed, and tissues imaged ex vivo (Fig. 4). Whole torso bioluminescence signal derived from AAV9-MHCK7-Luc transduced mice peaked on day 14, followed by a non-statistical tapering of signal until day 29, with 3 out of 5 animals yielding little bioluminescence signal by the end of the study (Fig. 4B). In striking contrast, animals transduced by AAV9-MHCK7-Luc-PD-L1 exhibited peak bioluminescence signal at day 14, with maintenance of this signal until day 29. Expression of PD-L1 was accompanied by quantifiably greater basal firefly luciferase signal in these animals at days 14, 21, and 29. Tissue-specific expression was confirmed via ex vivo imaging of excised tissues (Fig. 4C). Bioluminescence signal was confined primarily to the heart and skeletal muscle, with modest off-target bioluminescence signal in livers for AAV9-MHCK7-Luc-PD-L1 transduced animals. PD-L1 co-expression in animals transduced with AAV9-MHCK7-Luc-PD-L1 resulted in significantly greater bioluminescence signal from hearts (P < 0.05) and quadriceps muscle (P < 0.0001) relative to animals transduced with AAV9-MHCK7-Luc. The relative function and transduction efficiency of the AAV9 with or without the PD-L1 transgene was assessed on a cell line (C2C12 myoblast cells37) in vitro, and the addition of PD-L1 to the vector construct was found to not impact transduction efficiency nor luciferase expression (Supplementary Fig. 3). Based on these data we presumed that the increased in number of luciferase-expressing cells in animals transduced with AAV9-MHCK7-Luc-PD-L1 was due to greater maintenance of transduced cells rather than a greater initial transduction efficiency. When histology was performed on cardiac tissue strong PD-L1 staining was detected (Supplementary Fig. 4A). The combination of bioluminescence imaging and PD-L1 histology demonstrate that the cardiac tissue was expressing both PD-L1 and firefly luciferase. The capacity for both luciferase and PD-L1 expression to be driven by the MHCK7-Luc-PD-L1 construct was also validated by transfecting C2C12 myoblast cells. Transfected cells emitted bioluminescence when the culture medium was supplemented with luciferin, and these cells also demonstrated greater PD-L1 expression than non-transfected controls (Supplementary Fig. 5). Finally, body weight measurements collected over the study suggest that there was no incremental increase in toxicity due to co-expression of PD-L1, with animals transduced with AAV9-MHCK7-Luc or AAV9-MHCK7-Luc-PD-L1 gaining mass over the course of the study (Supplementary Fig. 6).
PD-L1 sustains transduction and muscle specific firefly luciferase expression. (A) In vivo IVIS imaging of the bioluminescence signal of animals dosed with either AAV9-MHCK7-Luc or AAV9-MHCK7-Luc-PD-L1 from day 1 to day 29. (B) Quantification of bioluminescence signal emanating from mouse torsos. (C) Quantification of bioluminescent signal from specific tissues/organs assessed ex vivo at day 29. Firefly luciferase abbreviated as “Luc” in figure. Statistics, GraphPad Prism, two-way ANOVA with Tukey’s multiple comparison test, n = 5, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
PD-L1 expression sustains transgene expression in adipose tissue
AAV8-Adipose-Luc or AAV8-Adipose-Luc-PD-L1 was delivered SC at a dose of 4 × 1013 vg/kg or 1.7 × 1012 vg for an average 25 g mouse (Fig. 5). Peak bioluminescence signal for animals dosed with AAV8-Adipose-Luc was at day 14, with signal tapering for 4 out of 5 animals by day 21 (Fig. 5A,B). Firefly luciferase bioluminescence signal increased from day 7 to day 28 (P < 0.05) for AAV8-Adipose-Luc-PD-L1, in contrast to the weakening signal observed in the AAV8-Adipose-Luc group (Fig. 5B). The signal from day 21 and day 28 was statistically greater in the Adipose-Luc-PD-L1 group compared to the AAV8-Adipose-Luc control group. At day 28, animals were sacrificed, and bioluminescence signal from tissues was assessed ex vivo (Fig. 5C). There was bioluminescence signal from the subcutaneous, visceral, and subscapular brown adipose tissues, as well as some off-target signal from livers. AAV8-Adipose-Luc-PD-L1 transduction resulted in significantly greater (P < 0.001) firefly luciferase bioluminescence signal from brown and visceral adipose tissues compared to animals transduced with AAV8-Adipose-Luc. When histology was performed on adipose tissue, PD-L1 staining was detected (Supplementary Fig. 4B). The combination of bioluminescence imaging and PD-L1 histology demonstrate that the adipose tissue was expressing both PD-L1 and firefly luciferase.
PD-L1 sustains transduction of adipose tissue-specific firefly luciferase expression. (A) In vivo IVIS imaging of bioluminescence signal from animals dosed with either AAV8-Adipose-Luc or AAV8-Adipose-Luc-PD-L1 from days 1 to 28. (B) Quantification of bioluminescence signal emanating from mouse torsos. (C) Quantification of bioluminescent signal from specific tissues/organs assessed ex vivo at day 28. Firefly luciferase abbreviated as “Luc” in figure. Statistics, GraphPad Prism, two-way ANOVA with Tukey’s multiple comparison test, n = 5, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
PD-L1 co-expression abrogates immune responses to the firefly luciferase transgene
As previously shown in Fig. 2E, AAV9-MHCK7-Luc again elicited a strong cellular immune response as quantified by IFN-γ ELISPOT analysis (Fig. 6A). While AAV9-MHCK7-Luc-PD-L1 led to a similar cellular immune response as animals dosed with AAV9-MHCK7-Luc, co-expression of PD-L1 led to greater and sustained firefly luciferase expression in muscle tissue. Again, as observed in the screening assay (Fig. 2D), AAV8-Adipose-Luc elicited a strong cellular immune response (Fig. 6B). In contrast to that observed with muscle targeting, PD-L1 co-expression (AAV8-Adipose-Luc-PD-L1) did not elicit a strong cellular immune response. Both AAV9-MHCK7-Luc-PD-L1 and AAV8-Adipose-Luc-PD-L1 dampened the humeral immune response relative to controls without PD-L1, but in neither instance was this dampening statistically significant (Fig. 6C,D).
PD-L1 co-expression abrogates immune responses to the firefly luciferase transgene. (A) Quantification of IFN-γ ELISPOT cellular immunity in AAV9-MHCK7 animal groups, relative to no stimulation or ovalbumin controls. (B) Quantification of IFN-γ ELISPOT cellular immunity for AAV8-Adipose animal groups, relative to no stimulation or ovalbumin controls. (C) Western Blotting to detect humoral immunity via anti-firefly luciferase antibodies, upper band: Anti-firefly luciferase IgG, lower band: Gapdh. (D) Quantitation of western blot densitometry, with raw data provided in Supplementary Figs. 7 and 8. Firefly luciferase abbreviated as “Luc” in figure. Statistics, GraphPad Prism, A and B, two-way ANOVA with Tukey’s multiple comparison test, D one-way ANOVA, Holm-Sidak’s multiple comparison test, n = 5, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
PD-L1 co-expression and muscle tissue immune cell infiltration
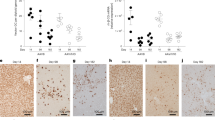
As expected, there was relatively little immune cell infiltration into cardiac and quadricep muscle in animals infused with the adipocyte targeting AAV8-Adipose-Luc or AAV8-Adipose-Luc-PD-L1 (Fig. 7A,B). By contrast, there was substantial infiltration of cardiac muscle by macrophages (F4/80) as well as CD4 and CD8 positive lymphocytes in animals infused with AAV9-MHCK7-Luc (Fig. 7A). This immune cell infiltration into cardiac tissue was almost entirely abrogated by co-expression of PD-L1 in animals infused with AAV9-MHCK7-Luc-PD-L1. Reduced immune cell infiltration within the cardiac tissue of AAV9-MHCK7-Luc-PD-L1 treated animals aligned with the sustained firefly luciferase signal observed in the cardiac tissue of these animals (Fig. 4C). There was relatively little immune cell infiltration into the quadricep muscle of animals treated with AAV9-MHCK7-Luc or AAV9-MHCK7-Luc-PD-L1. Given the trend towards a reduction in firefly luciferase signal after week 3 in animals infused with AAV9-MHCK7-Luc, we reasoned that immune cell infiltration likely peaked earlier in the study, likely leading to a steady decline in immune cell infiltration within the quadriceps by day 29. Immune cell infiltration in inguinal lymph nodes and H&E staining of liver tissue was similar in all animals across all groups (Supplementary Fig. 9).
PD-L1 co-expression and immune cell infiltration into muscle tissue. (A) CD8 cell, CD4 cell, and macrophage infiltration into cardiac tissue of animals treated with AAV8-Adipose-Luc, AAV8-Adipose-Luc-PD-L1, AAV9-MHCK7-Luc, or AAV9-MHCK7-Luc-PD-L1. (B) CD8 cell, CD4 cell, and macrophage infiltration into quadricep muscle tissue of animals. Firefly luciferase abbreviated as “Luc” in figure.
PD-L1 co-expression and adipose tissue immune cell infiltration
Firefly luciferase signal in animals transduced with AAV8-Adipose-Luc peaked on day 14 and was largely undetectable by day 28 when animals were sacrificed, and immune cell infiltration quantified. At day 28, CD8 and CD4 cell infiltration appeared to be modestly greater in the adipose tissue of animals treated with AAV8-Adipose-Luc-PD-L1 relative to animals treated with AAV8-Adipose-Luc (Fig. 8). This pattern of modestly greater CD8/4 cell infiltration was observed in inguinal adipose tissue (Fig. 8A), subscapular brown adipose surrounded by white adipose tissue (Fig. 8B), and in the viseral adipose tissue (Supplementary Fig. 9). Likely, immune cell infiltration peaked in the adipose tissue of animals treated with AAV8-Adipose-Luc early in the study, aligning with peak firefly luciferase signal at day 14 (Fig. 5A,B), followed by a decline in immune cell infiltration until the day 28 harvest. Thus, despite co-expression of PD-L1, the sustained expression of luciferase at the day 28 endpoint would have led to greater immune cell infiltration in these animals than in the AAV8-Adipose-Luc-treated animals where the firefly luciferase signal had been largely nullified at this time point. For both quadricep muscle and adipose tissue, in animals where PD-L1 was not co-expressed, immune cell infiltration likely peaked with maximal firefly luciferase expression and then declined by the study endpoint(s).
PD-L1 co-expression and immune cell infiltration into adipose tissue. (A) CD8 cell, CD4 cell, and macrophage infiltration into inguinal adipose of animals treated with AAV8-Adipose-Luc, AAV8-Adipose-Luc-PD-L1, AAV9-MHCK7-Luc, or AAV9-MHCK7-Luc-PD-L1. (B) CD8 cell, CD4 cell, and macrophage infiltration into subscapular brown adipose tissue surrounded by white adipose tissue of animals. Firefly luciferase abbreviated as “Luc” in figure.
Discussion
The continued evolution of gene therapies will require integration of strategies that mitigate immune response to transgenes. Herein, we explored the utility of PD-L1 co-expression to mitigate immune-mediated clearing of transduced muscle and adipose cells. We first compared the immunogenicity and stability of transgene expression in muscle and adipose tissue, using liver tissue as a control. AAV8-AAT/APOE-Luc yielded stable targeted firefly luciferase expression in the liver, with negligible cellular immunity, consistent with previous reports38,39. By contrast, using AAV8-MHCK7-Luc or AAV9-MHCK7-Luc to target firefly luciferase expression in the muscle did elicit cellular immunity, which parallels human studies where transgene expression in the muscle is immunogenic11,12, likely necessitating immune modulation for long-term retention of transduced muscle cells. Targeted firefly luciferase transgene expression in adipose tissue (AAV8-Adipose-Luc SC) also elicited cellular immunity, but with more rapid clearing of transgene-expressing cells than observed when muscle tissue was targeted.
Adipose tissue contains a variety of immune cells types40 and is known to secrete inflammatory cytokines41,42, but it is also similar to the liver in that portions of the tissue are connected directly to portal circulation43. Adipose transgene expression proved to be immunogenic, and perhaps due to the greater immune cell infiltration in adipose tissue, the clearance of transduced cells occurred more rapidly than in skeletal muscle, especially following SC delivery of AAV. It may seem surprising that SC delivery of AAV would enable efficient transduction of tissues distal to the injection site, and to our knowledge no group has previously transduction of adipose tissue in mice using SC delivery of AAV. However, other reports suggest that effective transduction of distal tissues can be achieved with SC delivered AAV. For example, Wang et al. used SC delivery of AAV9 to efficiently transduce skeletal (60%) and cardiac muscle (65%), introducing human Tafazzin (hTAZ) into 1-day old TAZ KO mice44. Inagaki and colleagues compared AAV8 and AAV9 infused into C57BL/6 male or female mice via portal vein (PV), tail vein (TV), intraperitoneal (IP) or SC routes45. While SC administration of AAV8 or AAV9 resulted in reduced liver transduction compared to PV, TV, or IP at most time points, stable liver transduction was achieved at rates 30-to-50% of the levels obtained with PV, TV, or IP. Thus, while it appears that no group has previously reported on SC AAV delivery and adipose tissue transduction, the two cases described above suggest that high levels of transduction of tissues distal from the site of SC AAV infusion could reasonably be expected. These screening data sets suggested that adipose- and muscle-targeted transgene expression were ideal environments in which to evaluate the merits of PD-L1 co-expression.
When PD-L1 was co-expressed in muscle tissue (AAV9-MHCK7-Luc-PD-L1) of mice the bioluminescence signal increased substantially from day 7 to day 14 and then was sustained over the study. By contrast, the much weaker bioluminescence signal in the control group (AAV9-MHCK7-Luc) peaked at day 14 and then tapered over the study. There was no detectable difference in cellular or humoral immunity between the AAV9-MHCK7-Luc-PD-L1 and AAV9-MHCK7-Luc groups at day 29 endpoint. At this endpoint there was greater immune cell infiltration into the cardiac tissue of the AAV9-MHCK7-Luc treated animals, but little immune cell infiltration observed in the skeletal (quadricep) muscle of either animal group. When PD-L1 was co-expressed in the adipose tissue (AAV8-Adipose-Luc-PL-D1) of mice the bioluminescence signal increased until day 14, and was then sustained over the study period. By contrast, in control animals (AAV8-Adipose-Luc) the bioluminescence signal achieved a substantially reduced maximum at day 14, after which it began to taper. There was significantly greater cellular immunity in the AAV8-Adipose-Luc relative to the AAV8-Adipose-Luc-PD-L1 treated animals, but no difference in humoral immunity. By contrast, histological sections of adipose tissue collected at the day 28 endpoint suggested modestly greater CD8 and CD4 cell infiltration in animals treated with AAV8-Adipose-Luc-PD-L1. Thus, in both the muscle and adipose tissue targeted studies the immunological protection enabled by PD-L1 co-expression was visually appreciable through the substantially greater and sustained bioluminescence signal in these animal groups. However, the measured immunological responses assessed at study endpoints provided less compelling evidence for the benefit of PD-L1 co-expression. This observation likely reflects the fact that most of the firefly luciferase expressing cells had been cleared from the AAV9-MHCK7-Luc or AAV8-Adipose-Luc treated animals well before the study endpoint. We reason that peak immunological response in AAV9-MHCK7-Luc or AAV8-Adipose-Luc treated mice would have corresponded with peak firefly luciferase expression at mid study, and that as luciferase expression tapered so would have the immunological response in these animals. In previous work we used AAV to transduce firefly luciferase in mouse beta cells, and tracked bioluminescence signal, as well as immune cell infiltration within the pancreas, over time14. Waves of immune cell infiltration spiked with peak firefly luciferase expression, and then dwindled as luciferase expression declined. It is likely that a similar pattern permeated through the current study; in animals without PD-L1 co-expression maximal immunological response likely occurred with peak firefly luciferase expression, and then tapered by the study endpoint.
Our data highlight the increased risk of transgene immunogenicity associated with extrahepatic transgene expression enabled by tissue-targeted AAV vectors. We demonstrated the feasibility of using PD-L1 co-expression to evade cellular immunity and prevent cytotoxic lymphocyte mediated clearance of transduced extrahepatic cells. While our work is not the first to utilize PD-L1 co-expression to mitigate transgene immunogenicity46, it is the first to show the sufficiency of PD-L1 co-expression to prevent immune clearance of transduced cells without requiring systemic immune suppression. A previous study utilized a system of two vectors with non-specific promoters to express PD-L1 and the immunogenic secreted peptide, ovalbumin, but immune evasion was only achieved in conjunction with systemic immunosuppression with CTLA4-Ig46. By using a single vector, we ensured that all transduced cells expressed both luciferase and PD-L1, thus protecting the cells from cytotoxic clearance.
As firefly luciferase is not a secreted protein the focus of this study was on cellular rather than humoral immunity. Anti-firefly luciferase IgG antibodies were generated in all treatment groups; there was a trend in their reduction with PD-L1 co-expression, but this was not statistically significant. Hepatic transgene expression has been shown to induce immune tolerance in several mouse studies and this tolerance included a reduction in antibody titers20,47. This reduction in antibody titer does not occur immediately, and perhaps if our study were extended we would have also observed a reduction in luciferase antibodies.
Limitations associated with our report include the use of only male mice, the relatively brief 4-week studies, and risks associated with life-long PD-L1 expression in the targeted tissues. Mouse gender influences AAV transduction efficiency, but not necessarily in a uniform and predictable manner. Davidoff and colleagues demonstrated that AAV2 transduced mouse livers 5-13-fold more efficiently in male mice than in female mice48. Similarly, Piechnik et al., reported significantly greater transduction of heart, kidney, lung, spleen, and bone tissues in male versus female mice when using AAV849. Alternatively, Maguire and colleagues reported on the more efficient targeting female mouse brains with AAV950. There also appears to be an interplay between animal gender and study duration; Guenzel and colleagues followed the transfection of multiple tissues in mice over 1.5 years51. They observed that luciferase and alpha subunit of propionyl-CoA carboxylase (PCCA) remained elevated in cardiac tissue in both male and female mice, but that transgene expression in liver and skeletal muscle of female, but not male, mice waned. These data point to the complex role gender may play on therapeutic outcome, suggesting that putative AAV therapies should be tested in both male and female animals, and over extended time periods. Finally, while PD-L1 co-expression does enable sustained transgene expression in our model, there are risks and potential limitations with this approach. PD-L1 plays a role in naturally occurring liver-mediated tolerance20, and is able to deactivate cytotoxic lymphocytes16. These same features introduce the risk that PD-L1 co-expression may enable the evolution of an aggressive cancer52. It may be possible to manage such cancers with PD-L1 inhibitors or PD-L1 antibody therapies, but nevertheless this risk must be integrated into cost-benefit analysis. There is the added complexity that PD-L1 may not be the best solution when working beyond murine models. Basal PD-1 expression level is lower in human immune cells than in mice53, and further work is needed to assess this approach in a humanized mouse or primate model. Further, there is the possibility that PD-L1 is insufficient to protect transduced cells in humans. If PD-L1 was found to be insufficient, there are alternative immunomodulatory proteins that could be utilized, including HLA-E54, CTLA-455 or LAG-356. Through further iteration and refinement, it is likely that a co-expression system could be established that overcomes limitations associated with transgene immunogenicity.
In conclusion, we have shown that AAV-mediated gene therapy targeting the adipose tissue elicits strong cellular and humoral immune responses to the transgene product. Through the design of a novel PD-L1 co-expression system we were able to mitigate cellular immune responses in a murine model enabling sustain transgene expression in both muscle and adipose tissue. Our system demonstrates the ability to induce immune tolerance without systemic immune suppression or involvement of the liver, providing a platform strategy that could have utility in both exploratory and clinical gene therapy settings.
Materials and methods
AAV vector production
AAV vectors were produced as described previously57, using a 3-plasmid transfection system in adherent HEK293 cells. Transgene cassettes were synthesized and inserted into expression constructs flanked by AAV2 ITR’s by GenScript (Piscataway, NJ). In screening studies, four different transgene cassettes were used as shown graphically in Fig. 1A. These cassettes included luciferase driven by the MHCK7 promoter (MHCK7-Luc), luciferase driven by the AAT/APOE promoter (AAT/APOE-Luc), luciferase driven by the Adiponectin promoter (Adipose-Luc), and an empty or Nil transgene driven by the Adiponectin promoter (Adiponectin-Nil). The Adiponectin promoter comprises of 226 bp of human Adiponectin promoter enhancers and 523 bp of murine Adiponectin enhancers/promoter sequences, followed by 510 bp of human-beta-globin splicing unit. AAV vector maps are provided in Supplementary Fig. 1. In the PD-L1 co-expression studies, four different transgene cassettes and AAV capsid combinations were used as shown graphically in Fig. 3B. These included AAV8 with a cassette containing luciferase driven by the Adiponectin promoter (AAV8-Adiponectin-Luc), AAV8 with a cassette containing luciferase, P2A, and PD-L1 driven by a modified Adiponectin promoter (AAV8-Adiponectin-Luc-PDL1), AAV9 with a cassette containing luciferase driven by the MHCK7 promoter (AAV9-MHCK7-Luc), and AAV9 with a cassette containing luciferase, P2A and PD-L1 driven by the MHCK7 promoter (AAV9-MHCK7-Luc-PD-L1). The cassette sizes were as follows: MHCK7-Luc (6998 bp, 3881 bp between ITRs), MHCK7-Luc-PD-L1 (7943 bp, 4826 bp between ITRs), Adiponectin-Luc (6956 bp, 3839 bp between ITRs), and Adiponectin-Luc-PD-L1 (7901 bp, 4784 bp between ITRs).
Viral particles were harvested from cell lysate for AAV9 and isolated from both the media and lysate for AAV8 pseudo-typed vectors. AAV particles were purified using a single round of iodixanol gradient ultracentrifugation, followed by buffer exchange on a Vivaspin 20 column (100 kDa PES, Sartorious) to remove iodixanol, and finally passed through a 0.22 μm filter.
Co-expression of luciferase and PD-L1
Co-expression of firefly luciferase and PD-L1 from the MHCK7-Luc-PD-L1 plasmid was validated by transfecting C2C12 myoblast cells37. C2C12 cells were cultured in DMEM plus 10% FBS. The transfection solution was formulated from 200 µL of Opti-MEM, 12 uL of Lipofectamine 3000 reagent (ThermoFisher Cat# L3000001) combined with 200 µL Opti-MEM, 6 µg DNA and 8 uL of Lipofectamine P3000 reagent, and this was used to transfect C2C12 cells in two wells of a 6-well plate. After two hours, 3 mL of DMEM plus 10% FBS was added to each well. On the following day, 20 µL of IVISbrite D-Luciferin Ultra Bioluminescent Substrate was added to each well, and bioluminescence quantified on a IVIS SpectrumBL High-Throughput In Vivo Optical Imaging System (PerkinElmer Cat# CLS137325). Cells were then harvested with TrypLE™ Express Enzyme (ThermoFisher), stained with an anti-PD-L1 antibody (Mouse B7-H1/PD-L1 APC-conjugated Antibody, RnDSystems Cat# FAB1019A) in PBS plus 1% bovine serum albumin (ThermoFisher, Cat# 26170035). Cells were characterized on a MACSQuant Analyzer flow cytometer (Miltenyi Biotec) and data analyzed using FlowJo software (Version 10.9.0).
Quantitative PCR and primers
DNA was isolated from snap frozen tissues using the QIAmp DNA mini kit (Qiagen Cat. Number # 51306) as per the manufacturer’s protocol. RNA was isolated from frozen tissues by homogenization followed by TRIzol extraction (ThermoFisher Cat# 15596026) as per the manufacturer’s instructions. AAV titration and genome copy number was assessed by quantitative polymerase chain reaction (qPCR) using a Quantstudio 7 system (Applied Biosystems) and TaqMan™ Fast Universal PCR Master Mix (Applied Biosystems Cat# 4352042) reaction buffer with a standard curve consisting of linearized AAV expression plasmid. Detection was performed with a primer probe set specific to the AAV2 ITR Forward GGAACCCCTAGTGATGGAGTT, Probe CACTCCCTCTCTGCGCGCTCG, and Reverse CGGCCTCAGTGAGCGA.
Gene expression was assessed through RT-qPCR using the Quantstudio 7 system and TaqMan™ RNA-to-CT™ 1-Step Kit (ThermoFisher Cat. # 4392653). Codon optimized mouse CD274: Forward GAGTGCAGATTCCCTGTAGAAC, Probe TACACCACTAACGCAAGCAGGTCC, and Reverse CTCTCCTGCCACAAACTGAA. Firefly Luciferase: Forward CTTCGAGGAGGAGCTATTCTTG, Probe TGCTGGTGCCCACACTATTTAGCT, and Reverse GTCGTACTTGTCGATGAGAGTG. Eukaryotic 18 S rRNA was used as an endogenous control (VIC™/TAMRA™ probe, ThermoFisher Cat# 4310893E).
Animal studies
All animal experiments were approved by the Institutional Animal Care and Use Committee of AstraZeneca (Gaithersburg, Maryland) and carried out according to established guidelines. Animals were housed in standard laboratory conditions with 5 animals per cage, fed ad libitum on standard chow, with a 12-hour dark and light cycle. Six-week-old C57/Bl6 male mice were purchased from the Jackson Laboratory and were quarantined for one week prior to use in studies. Mice were infused with AAV at a dose of 4 × 1013 vg/kg or 1.7 × 1012 vg for an average 25 g mouse via intravenous (IV), intraperitoneal (IP), or subcutaneous (SC) injection. We aimed to achieve a high rate of transduction and luciferase expression, which would be immunogenic, and then assess if co-expression of PD-L1 would dampen immune cell-mediated elimination of the transduced cells. One of our target tissues was cardiac and skeletal muscle, which could be viewed as being specifically relevant to AAV therapies for Duchenne muscular dystrophy. Our AAV dose was similar to previous Duchenne muscular dystrophy studies58,59,60,61, indicating the relevance of this viral dose for transducing muscle tissue. Similar doses (4 × 1013 vg/kg) have also been used in non-Duchenne muscular dystrophy studies62,63,64. In our studies, mouse body weight was recorded weekly. Periodic IVIS imaging was performed. Animals were euthanized by carbon dioxide asphyxiation. Tissues and serum were collected with terminal imaging between 20 and 29 days after virus delivery.
IVIS imaging
Animals were anesthetized with 2.5% isoflurane for 3 min before intraperitoneal injection of 150 µL of IVISbrite D-Luciferin Ultra Bioluminescent Substrate in RediJect Solution (XenoLight Cat# 770505). Animals were placed into an IVIS SpectrumBL High-Throughput In Vivo Optical Imaging System, remaining on flurane, and imaged for luminescence 6 minutes after substrate dosing, using automatic exposure times. For periodic in vivo imaging animals were monitored throughout recovery and returned to standard housing. For live animals, image scaling for the Adipose or MHCK7 promoters was set at 3 × 105–3 × 106 or 1 × 106–1 × 107, respectively. Whole body bioluminescence radiance quantification was performed capturing 6.5 × 4.0 regions of interest, and then intensity rescaled to 5 × 105–2 × 107 to facilitate comparison between groups. For terminal ex vivo imaging, animals were euthanized immediately after whole body imaging by carbon dioxide asphyxiation, and organs harvested. Radiance was quantified in circular regions of interest specified for and fully encompassing each individual tissue, sized 3.0 × 3.0 cm for liver, 2.0 × 2.0 cm for brown adipose, 2.0 × 2.0 cm for inguinal adipose, 1.5 × 1.5 cm for spleen, and 1.5 × 1.5 cm for the heart. Images were compiled and luminescence was quantified using Living Image Software Version 4.8.0 (PerkinElmer Cat# 128113).
IFN-γ ELISPOT
T cell activation was assessed on isolated splenocytes using the Mouse IFN-gamma ELISpot Kit (R&D Systems Cat# EL485) as per the manufacturer’s protocol. Excised whole spleens were immediately placed in chilled RPMI 1640 media (ThermoFisher Cat# 11875093) until processing. Spleens were disrupted using a soft plastic pestle, passed through a 40 μm filter, and washed with chilled HyClone HBSS(1x) from (ThermoFisher Cat# SH30588.01). Erythrocytes were eliminated with a one-minute incubation in ACK lysis buffer (Gibco Cat# A10492-01). Splenocytes were resuspended in warmed RPMI 1640 media (ThermoFisher Cat # 11875093), containing 5% fetal bovine serum (ThermoFisher Cat # 10082147), and 1X Penicillin-Streptomycin (ThermoFisher Cat# 15070063). Splenocytes were counted before adding 500,000 cells to each well. Wells were left unstimulated (US) or treated with 3 micrograms of purified stimulation peptide. Purified peptides for cell stimulation were synthesized by GenScript for ovalbumin, amino acid sequence: ISQAVHAAHAEINEAGR65, and firefly luciferase amino acid sequence: LMYRFEEEL29. Cells were incubated for 18 h at 37 °C before proceeding with the ELISA portion of the assay. After completion of the ELISA kit protocol, plates were permitted to dry for at least 48 h before imaging and quantification using a CTL-Immunospot S7 Universal Analyzer.
Western blotting to detect serum anti-firefly luciferase IgG
For Western Blots, 20 mg of cell lysate protein from HEK293 transfected with a CMV-firefly luciferase construct (LUC) or left un-transfected (UT) was run on alternating lanes of polyacrylamide gels. Novex™ Sharp Pre-stained Protein Standard (ThermoFisher Cat# LC5800), was used as a marker of molecular weight. Protein was transferred to a nitrocellulose membrane using an Invitrogen Iblot system and blocked with StartingBlock TBS Blocking Buffer (ThermoFisher Cat# 37542). Each blot section was probed with 1:2000 mouse anti-gapdh (Cell Signaling Cat# D4C6R), and 1:500 mouse serum in multiplex overnight at 4 C. Blots were washed 3X with TBST buffer before probing with Anti-mouse IgG, (HRP-linked Antibody CST Cat# 7076) for chemiluminescent imaging or for near infrared imaging with IRDye® 800CW Donkey anti-Mouse IgG Secondary Antibody (Li-Cor Cat# 926-32212). Blots were washed 4X with TBST before imaging. Near IR imaging was performed and quantified using a Li-Cor Odyssesy DLx system. Chemiluminescent imaging was performed using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoFisher Cat#34579) and was quantified with Image J software as previously described66.
Histology
Tissues were harvested and fixed for 24 hours in paraformaldehyde before transfer to 70% ethanol for at least 24 hours prior to embedding in paraffin blocks. Tissues sectioning and staining was performed by Histoserv Inc. (Germantown, Maryland). Tissue slices 5–6 μm thick were collected onto Superfrost Plus Microscope Slides (Fisher Scientific). When tissue did not contained fat, 5 μm slices were collected, and when tissue did contain fat, 6 μm slices were collected.
For immunohistochemistry, slides were first deparaffinized in xylene and then hydrated through graded alcohols. Next, slides were subject to antigen retrieval in a Tris/EDTA buffer (pH 9) for 20–30 min. Antigen retrieval was performed for 20 minutes for F4/80 and CD4, and 30 minutes for CD8. Slides were then blocked against endogenous peroxidases. Following blocking, slides were incubated with the primary rabbit antibodies: anti-F4/80 (Cell Signaling Technology, Cat#70076, 1:250), anti-CD8a (Cell Signaling Technology, Cat# 98941, 1:250), anti-CD4 (Abcam Cat# ab183685, 1:3000), and anti-PD-L1 (Cell Signaling Technology, Cat# 64988). Primary antibodies were diluted in Agilent Antibody Diluent and Background Reducing reagent (Cat# S3022). Primary antibodies were detected with a goat-anti-rabbit horseradish peroxidase secondary (neat) and visualized with chromogen DAB (3, 3’-diaminobenzidine). Finally, slides were counterstained with hematoxylin, dehydrated in graded alcohols, cleared in xylene, and mounted with permanent mounting media. All rinses were performed using either distilled water or TBST (0.05% Tween 20 in Tris-buffered saline).
Statistical analysis
Statistical analysis was performed in Graphpad Prism. Statistical tests are described in figure captions.
Data availability
Data is available as a supplementary File.
References
Ertl, H. C. J. T cell-mediated immune responses to AAV and AAV vectors. Front. Immunol.12, 666666 (2021).
Singh, N. et al. Mechanisms of resistance to CAR T cell therapies. Semin. Cancer Biol.65, 91–98 (2020).
Khan, A. N. et al. Immunogenicity of CAR-T cell therapeutics: Evidence, mechanism and mitigation. Front. Immunol.13, 886546 (2022).
Srivastava, A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 21, 75–80 (2016).
Verdera, H. C., Kuranda, K. & Mingozzi, F. AAV vector immunogenicity in humans: A long journey to successful gene transfer. Mol. Ther. 28(3), 723–746 (2020).
Hoffman, B. E. et al. Nonredundant roles of IL-10 and TGF-beta in suppression of immune responses to hepatic AAV-factor IX gene transfer. Mol. Ther. 19(7), 1263–1272 (2011).
Breous, E., Somanathan, S., Vandenberghe, L. H. & Wilson, J. M. Hepatic regulatory T cells and Kupffer cells are crucial mediators of systemic T cell tolerance to antigens targeting murine liver. Hepatology 50(2), 612–621 (2009).
Dobrzynski, E. et al. Induction of antigen-specific CD4+ T-cell anergy and deletion by in vivo viral gene transfer. Blood 104(4), 969–977 (2004).
Keeler, G. D. et al. Gene therapy-induced antigen-specific tregs inhibit neuro-inflammation and reverse disease in a mouse model of multiple sclerosis. Mol. Ther. 26(1), 173–183 (2018).
Asokan, A. & Shen, S. Redirecting AAV vectors to extrahepatic tissues. Mol. Ther. 31(12), 3371–3375 (2023).
Anthony, K. et al. T cell responses to dystrophin in a natural history study of Duchenne muscular dystrophy. Hum. Gene Ther. 34(9–10), 439–448 (2023).
Chung Liang, L., Sulaiman, N. & Yazid, M. D. A decade of progress in gene targeted therapeutic strategies in Duchenne muscular dystrophy: A systematic review. Front. Bioeng. Biotechnol. 10, 833833 (2022).
Li, N. et al. The effect of immunomodulatory treatments on anti-dystrophin immune response after AAV gene therapy in dystrophin deficient mdx mice. J. Neuromuscul. Dis. 8(s2), S325-s340 (2021).
Tonne, J. M. et al. Beta cell regeneration after single-round immunological destruction in a mouse model. Diabetologia 58(2), 313–323 (2015).
Zhang, H. et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 40(1), 184 (2021).
Francisco, L. M., Sage, P. T. & Sharpe, A. H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 236, 219–242 (2010).
Agina, H. A. et al. Hepatic expression of programmed death-1 (PD-1) and its ligand, PD-L1, in children with autoimmune hepatitis: Relation to treatment response. Clin. Exp. Hepatol. 5(3), 256–264 (2019).
Weiskirchen, R. & Tacke, F. Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology. Hepatobiliary Surg. Nutr. 3(6), 344–363 (2014).
Heymann, F. & Tacke, F. Immunology in the liver–from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 13(2), 88–110 (2016).
Kumar, S. R. P., Hoffman, B. E., Terhorst, C., de Jong, Y. P. & Herzog, R. W. The balance between CD8(+) T cell-mediated clearance of AAV-encoded antigen in the liver and tolerance is dependent on the vector dose. Mol. Ther. 25(4), 880–891 (2017).
Sun, X. et al. Transcriptional switch of hepatocytes initiates macrophage recruitment and T-cell suppression in endotoxemia. J. Hepatol. 77(2), 436–452 (2022).
Li, W. et al. Intra-articular delivery of AAV vectors encoding PD-L1 attenuates joint inflammation and tissue damage in a mouse model of rheumatoid arthritis. Front. Immunol. 14, 1116084 (2023).
El Khatib, M. M. et al. beta-Cell-targeted blockage of PD1 and CTLA4 pathways prevents development of autoimmune diabetes and acute allogeneic islets rejection. Gene Ther. 22(5), 430–438 (2015).
Jimenez, V. et al. FGF21 gene therapy as treatment for obesity and insulin resistance. 10(8) (2018).
O’Neill, S. M. et al. Targeting adipose tissue via systemic gene therapy. Gene Ther. 21(7), 653–661 (2014).
Bates, R., Huang, W. & Cao, L. Adipose tissue: An emerging target for adeno-associated viral vectors. Mol. Ther. Methods Clin. Dev. 19, 236–249 (2020).
Podetz-Pedersen, K. M., Vezys, V., Somia, N. V., Russell, S. J. & McIvor, R. S. Cellular immune response against firefly luciferase after sleeping beauty-mediated gene transfer in vivo. Hum. Gene Ther. 25(11), 955–965 (2014).
Jeon, Y. H. et al. Immune response to firefly luciferase as a naked DNA. Cancer Biol. Ther. 6(5), 781–786 (2007).
Limberis, M. P., Bell, C. L. & Wilson, J. M. Identification of the murine firefly luciferase-specific CD8 T-cell epitopes. Gene Ther. 16(3), 441–447 (2009).
Vilà, L. et al. AAV8-mediated Sirt1 gene transfer to the liver prevents high carbohydrate diet-induced nonalcoholic fatty liver disease. Mol. Ther. Methods Clin. Dev. 1, 14039 (2014).
Liu, X. et al. Adipose tissue insulin receptor knockdown via a new primate-derived hybrid recombinant AAV serotype. Mol. Ther. Methods Clin. Dev. 1, 8 (2014).
Wu, S. C. & Lin, C. H. Direct adeno-associated viruses injection of murine adipose tissue. Bio Protoc. 13(10), e4674 (2023).
Shen, S. et al. Functional analysis of the putative integrin recognition motif on adeno-associated virus 9. J. Biol. Chem. 290(3), 1496–504 (2015).
Okuyama, T. et al. Liver-directed gene therapy: A retroviral vector with a complete LTR and the ApoE enhancer-alpha 1-antitrypsin promoter dramatically increases expression of human alpha 1-antitrypsin in vivo. Hum. Gene Ther. 7(5), 637–645 (1996).
Wang, Z. V., Deng, Y., Wang, Q. A., Sun, K. & Scherer, P. E. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology 151(6), 2933–2939 (2010).
Salva, M. Z. et al. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol. Ther. 15(2), 320–329 (2007).
Yaffe, D. & Saxel, O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270(5639), 725–727 (1977).
Wang, L. et al. Systematic evaluation of AAV vectors for liver directed gene transfer in murine models. Mol. Ther. 18(1), 118–125 (2010).
Liu, C. et al. FGF21 protects against hepatic lipotoxicity and macrophage activation to attenuate fibrogenesis in nonalcoholic steatohepatitis. Elife 12 (2023).
Bapat, S. P., Liang, Y. & Zheng, Y. Characterization of immune cells from adipose tissue. Curr. Protoc. Immunol. 126(1), e86 (2019).
Kawai, T., Autieri, M. V. & Scalia, R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. Cell Physiol. 320(3), C375-c391 (2021).
Coppack, S. W. Pro-inflammatory cytokines and adipose tissue. Proc. Nutr. Soc. 60(3), 349–356 (2001).
Ibrahim, M. M. Subcutaneous and visceral adipose tissue: Structural and functional differences. Obes. Rev. 11(1), 11–18 (2010).
Wang, S. et al. AAV gene therapy prevents and reverses heart failure in a murine knockout model of Barth syndrome. Circ. Res. 126(8), 1024–1039 (2020).
Inagaki, K. et al. Robust systemic transduction with AAV9 vectors in mice: Efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 14(1), 45–53 (2006).
Adriouch, S. et al. Improved immunological tolerance following combination therapy with CTLA-4/Ig and AAV-mediated PD-L1/2 muscle gene transfer. Front. Microbiol. 2, 199 (2011).
Ziegler, R. J. et al. AAV2 vector harboring a liver-restricted promoter facilitates sustained expression of therapeutic levels of alpha-galactosidase A and the induction of immune tolerance in Fabry mice. Mol. Ther. 9(2), 231–240 (2004).
Davidoff, A. M., Ng, C. Y., Zhou, J., Spence, Y. & Nathwani, A. C. Sex significantly influences transduction of murine liver by recombinant adeno-associated viral vectors through an androgen-dependent pathway. Blood 102(2), 480–488 (2003).
Piechnik, M. et al. Sex difference leads to differential gene expression patterns and therapeutic efficacy in mucopolysaccharidosis IVA murine model receiving AAV8 gene therapy. Int. J. Mol. Sci. 23(20) (2022).
Maguire, C. A. et al. Mouse gender influences brain transduction by intravascularly administered AAV9. Mol. Ther. 21(8), 1470–1471 (2013).
Guenzel, A. J., Collard, R., Kraus, J. P., Matern, D. & Barry, M. A. Long-term sex-biased correction of circulating propionic acidemia disease markers by adeno-associated virus vectors. Hum. Gene Ther. 26(3), 153–160 (2015).
Wang, X., Teng, F., Kong, L. & Yu, J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. 9, 5023–5039 (2016).
Butte, M. J., Pena-Cruz, V., Kim, M. J., Freeman, G. J. & Sharpe, A. H. Interaction of human PD-L1 and B7–1. Mol. Immunol. 45(13), 3567–3572 (2008).
Khan, M., Arooj, S. & Wang, H. NK cell-based immune checkpoint inhibition. Front. Immunol. 11, 167 (2020).
Hosseini, A., Gharibi, T., Marofi, F., Babaloo, Z. & Baradaran, B. CTLA-4: From mechanism to autoimmune therapy. Int. Immunopharmacol. 80, 106221 (2020).
Maruhashi, T. & Sugiura, D. LAG-3: From molecular functions to clinical applications. 8(2) (2020).
Kurasawa, J. H. et al. Chemically defined, high-density insect cell-based expression system for scalable AAV vector production. Mol. Ther. Methods Clin. Dev. 19, 330–340 (2020).
Albini, S. et al. Assessment of therapeutic potential of a dual AAV approach for duchenne muscular dystrophy. Int. J. Mol. Sci. 24(14) (2023).
Potter, R. A. et al. Dose-escalation study of systemically delivered rAAVrh74.MHCK7.micro-dystrophin in the mdx mouse model of duchenne muscular dystrophy. Hum. Gene Ther. 32(78), 375–389 (2021).
Hakim, C. H. et al. Micro-dystrophin AAV vectors made by transient transfection and herpesvirus system are equally potent in treating mdx mouse muscle disease. Mol. Ther. Methods Clin. Dev. 18, 664–678 (2020).
Zhou, K. et al. Sildenafil increases AAV9 transduction after a systemic administration and enhances AAV9-dystrophin therapeutic effect in mdx mice. Gene Ther. 31(1–2), 19–30 (2024).
Zhang, L. et al. Young mice administered adult doses of AAV5-hFVIII-SQ achieve therapeutic factor VIII expression into adulthood. Mol. Ther. Methods Clin. Dev. 26, 519–531 (2022).
Li, H. et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood 117(12), 3311–3319 (2011).
Sun, J. et al. Adeno-associated virus-mediated expression of activated factor V (FVa) for hemophilia phenotypic correction. Front. Med. (Lausanne) 9, 880763 (2022).
Sun, L. Z. et al. Comparison between ovalbumin and ovalbumin peptide 323–339 responses in allergic mice: Humoral and cellular aspects. Scand. J. Immunol. 71(5), 329–335 (2010).
Ohgane, K. & Yoshioka, H. Quantification of Gel Bands By an Image J Macro, Band/Peak Quantification Tool v1 (2019).
Acknowledgements
This work was funded by AstraZeneca. The authors would like to thank the AstraZeneca Histology Core for their assistance.
Author information
Authors and Affiliations
Contributions
T.B.M. conceived the concept, designed and performed experiments, analyzed and interpreted the data, and wrote the manuscript. A.P. designed and performed experiments and interpreted the data. P.J.H., C.B., and J.H.K. performed experiments and interpreted data. M.R.D. analyzed and interpreted the data, wrote and edited the revised manuscript. Y.I. designed and interpreted experiments, and wrote and edited the revised manuscript. All authors approved the manuscript.
Corresponding authors
Ethics declarations
Ethics declarations
All animal experiments were approved by the Institutional Animal Care and Use Committee of AstraZeneca (Gaithersburg, Maryland) and performed as per the approved guidelines. Animal studies complied with the ARRIVE guidelines.
Competing interests
The studies reported here were completed at AstraZeneca, and P.J.H, C.B., J.H.K, Y.I. and M.R.D. are employed by AstraZeneca PLC.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
McMurphy, T.B., Park, A., Heizer, P.J. et al. AAV-mediated co-expression of an immunogenic transgene plus PD-L1 enables sustained expression through immunological evasion. Sci Rep 14, 28853 (2024). https://doi.org/10.1038/s41598-024-75698-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-75698-2