Abstract
Stuccoes are very delicate decorative elements of Roman age. Very few of them survived almost intact to present days and, for this reason, they are of great interest to restorers and conservators. In this study, we combined metabarcoding and untargeted metabolomics to characterise the taxonomic and metabolic profiles of the microorganisms forming biofilms on the stuccoes located on the ceiling of the laconicum, a small thermal environment in the archaeological park of Baia (southern Italy). We found that some samples were dominated by bacteria while others by eukaryotes. Additionally, we observed high heterogeneity in the type and abundance of bacterial taxa, while the eukaryotic communities, except in one sample (at prevalence of fungi), were dominated by green algae. The metabolic profiles were comparable across samples, with lipids, lipid-like molecules and carbohydrates accounting for roughly the 50% of metabolites. In vitro and in vivo tests to remove biofilms on stuccoes using essential oils blends were successful at a 50% dilution for one hour and half. This integrative study advanced our knowledge on taxonomic and metabolic profiles of biofilms on ancient stuccoes and highlighted the potential impacts of these techniques in the field of cultural heritage conservation.
Similar content being viewed by others
Introduction
Stuccoes are traditional decorative elements for ceilings and vaults, but also vertical surfaces, in buildings and villas of the Roman age1,2. Before the discovery of Nero’s Domus Aurea in Rome in the 15th century, the only evidence of these artworks came from written accounts, such as like Pliny the Elder’s Naturalis Historia3. Roman stuccoes are frequently polychrome, but for many years were supposed to be white or monochromatic; however, likewise Greek statues and temples, we now know that the white coloration was generally due to the loss of pigments rather than a lack of them. No single recipe seemed to exist for their preparation4. Recent analyses of mineralogical composition of stuccoes from the Domus Aurea revealed the same elemental composition of plaster, i.e., the use of calcium hydroxide as binder and calcite as aggregates5. Due to the delicate carving techniques, very few stuccoes have survived intact or almost intact to present days and, in rarest cases, have preserved the original colours. Examples can be found in the abovementioned Domus Aurea in Rome6, in some tombs excavated in the area of Pozzuoli7,8, in the area of Baia9 and Pompeii, Herculaneum and Stabiae10,11. Furthermore, after being uncovered by archaeological excavations, as all decorative features, stuccoes are also exposed to deterioration by abiotic and biotic agents, which alter both their structure and aesthetics. For all these reasons, they are of particular conservation interest.
In recent years, the study of deterioration caused by biological agents (biodeterioration) has been fostered by the introduction of -omics techniques12. These techniques have enabled not only the taxonomic characterisation of microbial communities involved in the colonisation of substrata (metabarcoding and metagenomics) but also their metabolic activity (metatranscriptomics, metabolomics, proteomics). Some of these applications like metabarcoding are becoming routinary (e.g13,14,15,16). , while the others are still at their infancy. Understanding the type and activity of microorganisms is crucial for planning the best strategies for their removal and to prevent future colonisations. To date, no published studies have used -omics tools for the study of biodeterioration of ancient stuccoes.
In this study, we employed a multi-omics approach combining metabarcoding of two molecular markers (16S for bacterial communities and 18S for eukaryotic communities) and untargeted metabolomics to assess the taxonomic and metabolic profiles of the bacterial and eukaryotic community involved in the biodeterioration of Roman stuccoes in a thermal environment called laconicum in the archaeological site of Baia (Campania region, Italy). To the best of our knowledge, this is the first attempt to characterise both microorganism and metabolite diversity using -omics approaches in ancient stuccoes. Furthermore, we tested the efficacy of extracts of essential oils at different dilutions to remove the biological patinas from the stuccoes, utilising homemade tiles as test samples.
Materials and methods
Study site and sampling
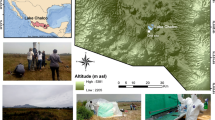
The laconicum is located on the terrace of the upper peristyle of the so called “Sosandra sector” in the archaeological park of Baia17 (Campania region, Italy). The room is characterised by an “L”-shaped plan, with a main rectangular body opening eastward, and a corridor that opens southward into an adjacent room, where the statue of the Venus Sosandra was found in 19539. The room is of small size, with a surface area of approximately 5 m2. It is a balneum, a private environment with a thermal function, specifically used for steam baths (laconicum). The internal wall structure of the laconicum is made of opus reticulatum flanked by opus vittatum, and the ceiling is decorated with stuccoes (Fig. 1A). The narrow corridor showcases a sequence of medallions arranged along the main north-south axis, each connected to its neighbours and to the edges of the ceiling through short, straight bands. The frames are decorated with delicate pearl mouldings, triple for the medallions and double for the connecting strips (Fig. 1B). The medallions have a diameter of 40 cm, while the mouldings are 6 cm wide (triple) and 3 cm wide (double). Among the figures depicted are animals such as lions, swans, and marine creatures and mythological characters like Cupid and Nereids (Fig. 1B). The stuccoes are made of dolomitic lime with calcium and magnesium carbonate as binders. The crystalline structure of the dolomite gives greater compactness and hardness to the lime, making it more resistant. The high quantity of dehydrated calcium sulphate on the surface of some of them can be attributed to the presence of saline efflorescence.
Biological patinas were sampled with sterile scalpels by a restoration student (Sara Scamardella) thanks to an agreement between the University of Studies Suor Orsola Benincasa and the archaeological park of Baia. Two samples (S1 and S2) were collected from the inner east side of the laconicum, while the other five from the ceiling decorated with stuccoes (S3-S7). Sample S2 was close to a saline efflorescence. The samples were placed into plastic tubes and stored at -20 °C until later analyses.
Metabarcoding analyses
The bacterial and eukaryotic components of biological patinas from the stuccoes were characterised through a metabarcoding approach, amplifying the V3-V4 region of 16S rRNA, and the V4 region of 18S rRNA, respectively. Total DNA was extracted using the DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. A qualitative and quantitative analysis of extracted DNA was carried out through visualisation on gel electrophoresis and with a Qubit v.4 fluorometer using the dsDNA HS Assay Kit (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA). Extracted DNA was stored at -20 °C until shipment for amplification and high-throughput sequencing. Metabarcoding analyses were carried out by Integrated Microbiome Resource (IMR; Halifax, Canada) at the condition specified on the company website (http://imr.bio/protocols) and using the primer sets (“Bacteria-specific “Illumina” V3-V4, B969F and BA1406R”18 and “Eukaryote-specific V4, E572F and E1009R”19) described in the same webpage. To monitor possible biological contaminations, an extraction blank (sample “ctrl”) was processed as negative control alongside the other samples till the sequencing step; in addition, PCR controls were carried out by the company.
Raw data (fastq format) were processed to generate amplicon sequence variants (ASVs) using the dada2 pipeline20 in R21. At the end of the pipeline, singletons were removed and, to account for differences in the number of ASVs across samples, data were normalised to the median value (20,206 for 16S dataset and 138 for 18S dataset) using the “rrarefy” function of the vegan R package22. Taxonomic assignation of eukaryotes was carried out using a BLAST approach23 against the PR2 database v4.14.024 (https://github.com/pr2database/pr2database/releases) and, for hits not assigned, against the nucleotide (nr) database. Five hits were stored for each query and the final assignment was determined using a lowest common ancestor (lca) approach to identify ASVs at the lowest taxonomic level when different matches occurred at the same similarity percentage. For the latter task, we used the galaxy-tool-lca script25 (https://github.com/naturalis/galaxy-tool-lca), which is partly based on MEGAN’s lca method26. Taxonomic assignation of bacterial ASVs was carried out using the naive Bayesian classifier method27 against the Silva reference database v138.128 (https://zenodo.org/record/4587955#.Ylqor9NBw2w); assignation at species level were determined by exact match (100% identity) between ASVs and sequenced reference strains always within dada2 using the “silva_species_assignment_v138.1” database.
Taxonomic composition of bacterial, eukaryotic, and combined communities was represented as barplots in RStudio29 using the phyloseq package30 and plotted with ggplot231. Venn diagrams were built using the file2meco32 and microeco33 R packages to assess whether communities with similar taxonomic composition at Domain rank (bacteria vs. eukaryotes) in the barplots were also similar at lower taxonomic levels (phylum and family). A principal component analysis (PCA) was carried out on the ClustVis webserver34 (https://biit.cs.ut.ee/clustvis/) to detect structure in our 16S, 18S, and 16S + 18S data.
Metabolomic analysis
Approximately 10 mg of pulverized sample material was weighed into a brown vial and exact mass was noted. A quantity of 0.5 mL of acetone: methanol (50:50) was added to each sample. Samples were mixed using a vortex at 1650 rpm for 3 min and then placed in a glass container filled with crushed ice; the container was kept in an ultrasonic bath for 30 min. The extract was filtered into a LC vial using a 0.2 μm filter, and the vial was stored in a freezer (-80 °C) until analysis. The extraction procedure was repeated two times or more for the same sample.
Extracted samples were analyzed by mass spectrometry using the Q-Orbitrap EXPLORIS 120 (Thermo Fisher Scientific, Foster City, CA, USA) and by Ultra-Performance Liquid Chromatography–Mass Spectrometry (UPLC-MS) analysis to get the metabolite profile, using a VANQUISH UPLC (Thermo Fisher Scientific, Foster City, CA, USA) coupled to a Q EXACTIVE mass spectrometer (Thermo Fisher Scientific, Foster City, CA, USA) equipped with an electrospray ionization (ESI) source in positive mode. A volume of 5 µL of sample was injected into a Hypersil GOLD™ C18 column (2.1 × 200 mm, 1.9 μm, Thermo Fisher Scientific, Foster City, CA, USA). Mobile phase was A: water + 0.1% TFA and B: Acetonitrile + 0.1% TFA. Gradient settings were: 0 min 5%B, 10 min 70%B, 11 min 70–95%B, isocratic for 1 min. Total flow was 0.35 ml min-1, column temperature was 40 °C. Chromatographic data were also recorded using a Photodiode array detector operating with a frequency of 12.5 Hz.
Metabolites were assigned to functional groups using ClassyFire35 (http://classyfire.wishartlab.com) after conversion of chemical names to several formats (ChEBI, KEGG, and InChI codes) in the Chemical Translation Service (CTS; http://cts.fiehnlab.ucdavis.edu/batch). Metabolic profiles across samples were showed as barplots of functional groups using the packages phyloseq30 and ggplot231. A heatmap was plotted to visualise the abundance patterns of selected metabolites across samples; we selected metabolites that occurred at least in two samples and with abundance > 1%. A PCA was also carried out as per metabarcoding data to detect possible structure in our samples. To link the metabolites to specific metabolic pathways, we used the standard KEGG compound names (C codes) previously retrieved in CTS as input for MetaboAnalyst v6.036 (https://www.metaboanalyst.ca/MetaboAnalyst/Secure/utils/NameMapView.xhtml), and then we used the KEGG37,38 mapper search tool (https://www.genome.jp/kegg/mapper/search.html).
Metabarcoding-metabomolomics associations
A PermANOVA analysis with the adonis function of the vegan package22 was carried out to detect significant associations between abundance of microbial (bacterial and eukaryotic) communities and metabolite concentrations.
Removal of biological patinas with essential oils
For the removal of biological patinas, we used ESSENZIO (IBIX Biocare, Lugo, Italy), a biodegradable and biocompatible product based on a blend of essential oils (mainly extracts of Origanum vulgare and Thymus vulgaris). Before in vivo removal of patinas, the product was first tested on homemade tiles of slaked lime and marble powder contaminated with biofilms taken in situ and grown in vitro to simulate the surface of the stuccoes. Based on the results obtained by Cennamo et al.39, the mixture was tested at different dilutions in demineralized water (10%, 20% and 50%) and after different application times (30 min, 1 h, 1 h and 30 min, 2 h) on specimens prepared in the laboratory (data not shown). The effectiveness of the treatment was evaluated through a visual comparison with clean specimens. Once the most suitable times and concentrations have been identified, the treatment was extended to the entire portion of the stucco decorations where biological patinas were observed.
Results
Metabarcoding analyses
The Illumina sequencing of V3-V4 16S region generated 215,759 raw reads distributed across seven samples. The clean, annotated dataset contained 82,352 sequences corresponding to 265 ASVs in four samples (S1, S2, S3, and S6); after the normalization procedure, 73,039 sequences and 265 ASVs remained (Table S1). The raw eukaryotic dataset based on the 18S–V4 region included 34,150 sequences across seven samples. The clean, annotated dataset contained 23,245 sequences corresponding to 23 ASVs in three samples (S4, S5, and S7); after normalization, 552 sequences and 18 ASVs remained (Table S2). Details regarding the number of sequences discarded in each pre-processing step per sample are provided in Table S3. The negative control (sample “ctrl”) resulted in no sequences for 16S marker and 21 sequences assigned to Cladosporium and 24 to humans for 18S marker; since human sequences were discarded as non-target and the fungal ones were not found in any of our samples, we excluded a possible role of contaminants in our diversity estimates.
In four out of seven samples, microbial community was constituted almost exclusively by bacteria, while in the remaining three samples, by eukaryotes (Fig. 2A); regarding samples from stuccoes, three out of five (S4, S5, and S7) were dominated by eukaryotes. Bacterial taxa were absent in samples S4, S5, and S7, while eukaryotic taxa occurred in all but S2 sample. The 95.1% of bacterial ASVs assigned to phylum level were shared by all samples (Fig. 2B), while only the 59.6% of ASVs at family level was shared (Fig. 2C). Regarding eukaryotes, only the 33.8% of ASVs attributed to phylum level was shared, and the remaining 66.2% of ASVs were exclusive to sample S7 (Fig. 2D). At family rank, no ASVs were shared, with the 67.6% of them exclusive to sample S7, the 29.9% to sample S5 and only 2.5% to sample S4 (Fig. 2E). A total of 14 bacterial families with abundance > 5% of total sequences were identified (Fig. 2F). Euzebyaceae was the only family present in all taxa and at notable abundance; members of Nitriliruptoraceae were present only in three samples (S1, S3, and S6). Not considering unclassified bacteria and families collapsed at abundance < 5%, bacterial community of most samples was represented by 6–8 families; the only exception was sample S2, with only three families (Euzebyaceae, Termosynechoccaceae, and Trueperaceae), of which Termosynechoccaceae included roughly the half of ASVs. With the only exception of Euzebyaceae, Pseudonocardiaceae, and Rhizobiaceae, all the other families were represented by single genera or unclassifiable sequences at lower taxonomic levels (Table S1). Most of the latter were in the family Nitriliruptoraceae, while among cyanobacteria we observed some taxa particularly abundant for which assignation was not achieved even at family level (Table S1). Only five assignments at species level were obtained (Aliihoeflea aestuarii, Halomonas chromatireducens, Nocardiopsis exhalans, Pelagibacterium lentulum, and Streptomyces sodiiphilus), plus two ambiguities (Brevundimonas bacteroides/B. variabilis and Nocardiopsis exhalans/N. valliformis).
Taxonomic composition of microbial communities at different ranks. (A) Taxonomic composition as bacteria vs. eukaryotes across the seven samples analysed; (B) Venn plot of bacterial ASVs assigned at Phylum level; (C) Venn plot of bacterial ASVs assigned at Family level; (D) Venn plot of eukaryotic ASVs assigned at Phylum level; (E) Venn plot of eukaryotic ASVs assigned at Family level; (F) barplots of abundance of bacterial ASVs annotated at Family level; (G) barplots of abundance of eukaryotic ASVs annotated at Phylum level.
The eukaryotic component was dominant in samples S4, S5 and S7 (Fig. 2A) but overall, taxonomically limited, accounting almost exclusively for chlorophytes and fungi (Fig. 2G). Among the former ones, we identified Picocystis salinarum R.A.Lewin, and Ctenocladus circinnatus Borzì, and assigned other ASVs to the genera Picochlorum W.J.Henley & al., and Pseudostichococcus L.Moewus (Table S2); for fungi, the only attribution at genus level was for Cyphellophora de Vries (Eurotiomycetes), while all the other ASVs belonged to the classes Dothideomycetes and Sordariomycetes. Such taxa were not equally distributed across samples, with Picocystis salinarum occurring only in samples S1 and S3, Ctenocladus in samples S1 and S6, Picochlorum in sample S7, and Pseudostichococcus in samples S5 and S6. Similarly, ASVs assigned to fungi, despite particularly abundant in sample S7, were exclusive of each sample and never shared (Table S2). The PCA analysis based on the bacterial dataset (Supplementary Figure S1a) separated along the first axis (46.5% of variance) sample S6 from S2, S3 and S1, the latter further separated from the others on the second axis by 33.7% of variance. Similarly, in the PCA analysis based on eukaryotic data (Supplementary Figure S1b), sample S6 was separated by all the others on PC1 (43.3% of variance), and S1 by the remnant samples by PC2 (32%); eukaryotic communities of samples S3, S4, and S5 were closely related. The same pattern of distinctiveness of samples S6 and S1 along PC1 and PC2, respectively was observed in the combined dataset (Supplementary Figure S1c). The PCA based on metabolomic data (Supplementary Figure S1d) showed a different pattern, with samples S1, S4, and S6 separated from the others on the first axis (57.3% of variance), and S1 and S2 from the others on the second axis (17.3% of variance).
Metabolomic analyses and metabarcoding-metabolomics associations
Employing mass spectrometry, we identified and annotated 162 metabolites across six powdered material samples (Table S4). Sample S7 was excluded from the analysis due to insufficient source material, which led to a failed extraction. Almost a quarter of metabolites belonged to lipids and lipid-like (fatty acids) molecules (23.5%) and carbohydrates (21.6%), followed by organic acids and derivatives (13.6%) and amino acids, peptides, and analogues (12.3%) (Fig. 3A); a list of the other classes of compounds is available in Table S4. According to the heatmap based on the most abundant metabolites (Fig. 3B), the sample S2 showed a different metabolic profile in respect to the other, with lactic acid constituting around the 20% of all metabolites, and followed by 2-Hydroxyisocaproic acid (~ 7%). For the other samples, we observed a clustering that was compatible with the distance among samples (see also Fig. 1A). Regarding metabolites, Samples S3 and S4 were characterized for 10–15% by sorbose and lactic acid, while samples S1, S5 and S6 showed highest abundances of the former.
KEGG compound codes (C) were obtained for 129 metabolites (Table S4) and attributed to the following pathways: “metabolic pathways” (85 compounds), biosynthesis of secondary metabolites (38), microbial metabolism (29), biosynthesis of aminoacids (18), and carbon and protein metabolism (10 and 17, respectively) (Tables S5 and S6).
No correlation was found between microbial and metabolite abundances (p > 0.05) according to the adonis test.
Removal of biofilms
The conditions that were found to be optimal for removing the biological patinas on the mortar test specimens were dilution of ESSENZIO at 50% in demineralized water and application time of an hour and a half (Fig. 4A, point 3). In all the other tiles, dark spots are visible as indicative of the presence of microorganisms. After this trial, the above-mentioned treatment was applied in-situ on all the stuccoes under restoration; an example of treatment is provided in Fig. 4B and C, and 4D. Since the biofilm layers were rather thin and not uniformly distributed, it was not necessary to use a support that would allow for a longer exposure time. At last, the surface was rinsed and cleaned with scalpel and swab. After 11 months from the treatment, no biofilms have been observed yet. An example of stucco decoration before and after removal of biofilms with essential oils is provided in Supplementary Figure S2.
Removal of biological patinas with ESSENZIO. (A) Test treatment with contact times of half an hour (1), one hour (2), one hour and half, and two hours (4); (B) stucco’s area before treatment; (C) stucco’s area during the application of the biocidal product, and before being covered with film; (D) result after treatment.
Discussion
The mechanisms of biodeterioration of cultural heritage have been studied for many years. From the first, culture-based approaches, which often favour the detection of few, fast-growing opportunistic species over other, less abundant ones40,41,42, the introduction of culture-independent methods has revealed an astonishing taxonomic diversity of microorganisms colonising monuments and historical manufacts43,44,45. However, such pioneer techniques as DGGE (Denaturing Gradient Gel Electrophoresis) and ARISA (Automated Ribosomal Intergenic Spacer Analysis), to cite just a few, did not provided a taxonomic information about the members of a given microbial community, and were often coupled to culture-based approaches46,47. The introduction of Next Generation Sequencing (NGS) approaches and their affordability of in terms of data processing, and reduced costs over years, has empowered the knowledge of microorganisms causing biodeterioration of cultural heritage in terms of taxonomic, physiological and metabolic diversity48,49,50. Among the various techniques, DNA metabarcoding has attracted great interest in the last few years, by allowing the simultaneous amplification and sequencing of short fragments of previously established markers as 16S, 18S, 23S, and ITS2 for the characterization of prokaryotes, eukaryotes, algae and fungi (e.g.,14,51,52). These studies have contributed to enrich the “taxonomic library” of microorganisms associated to different cultural heritage items as common stone monuments53,54, man-made artefacts55, and wall- and traditional paintings56 to peculiar materials as ceramics57. Stuccoes fall within the latter group, being very fragile materials that rarely survive intact or almost intact to present days. Available literature on the taxonomic diversity of microorganisms forming biofilms on such materials is from the vault of a XVIII century Italian church58, and Mayan stucco masks from Guatemala59 or buildings from Mexico60. In all of these studies, no similarities in microbial communities of stuccoes were observed: fungi (Aspergillus, Chaetomium, Sarocladium, and Stachybotrys) were dominant in the Italian church, while cyanobacteria (especially Gloecapsopsis, Pseudoanabaena, and Rhabdoderma in the former, and Gloeocapsa, Synechocystis-like, and Xenococcus in the latter) were the dominant taxa. In both Mayan samples, as also reported by Garcia de Miguel et al.61, eukaryotic algae except for Chlorella were absent from most samples, and this trend was explained with their exposition to direct sunlight, which was likely responsible for desiccation. The laconicum object of this study is, on the contrary, a small, semi-confined environment, with rather uniform values of temperature, light and humidity across the year. High humidity (75–95%) is also favoured by its exposition close to a hill and phenomena of capillary rise of water from the ground and infiltration from several sides. This environment is also constantly protected from direct sunlight and excessive ventilation. This could be the reason why we found several eukaryotic algae like Ctenocladus, Picochlorum, Picocystis salinarum, and Pseudostichococcus, in addition to different genera of cyanobacteria (Leptolyngbya, Loriellopsis, Nodosilinea, and Nodularia,) and fungi (Cyphellophora). In addition to climatic factors, the diversity of taxa found on stuccoes could be explained with the porosity and water retention capacity of this material stuccoes, which is known to present high bioreceptivity62. It should be taken into consideration that most of the biological patina here found developed both in direct contact with the surface of the stuccos and on the thin limestone encrustations that were present above them. Regarding bacterial colonisation of stuccoes data are even more limited, but Agarossi et al.63, found Nocardia and Streptomyces as the most abundant genera in the subterranean Neo-Phytagorean basilica of Porta Maggiore in Rome (1st century AD). We found several bacterial genera (excluding cyanobacteria) particularly abundant like Chelativorans, Longispora, Nitrolancea, Phytoactinopolyspora, and Pseudonocardia, but none of them was typical of stuccoes, but also found on adjacent, non-stucco samples. Nonetheless, microbial colonization is also driven by the spatially different micro-environments, including both exposure and physical-chemical characteristic of stuccoes. For instance, Halomonas chromatireducens, a species of halophilic bacteria, was only found in sample S1, which is in proximity to saline incrustations, while Chelativorans, a genus of Gram-negative, strictly aerobic bacteria generally isolated from nutrient-poor environments64 was abundant in all samples. In sample S1, the one close to saline incrustations, we also found bacteria typical of marine or tidal environments as Oceanicaulis, Aliihoeflea aestuarii, and Pelagibacterium lentulum: their occurrence could be due to the influence of nearby sea sprays (the distance of the laconicum from the sea is of just 200 m), as also explained for other samples collected in Baia15 and for some eukaryotic species mentioned above, typical of saline environments. Salt-tolerant bacteria and archaea have been reported as colonisers of stone monuments, especially in porous building materials subjected to rainwater and rising damp that contain soluble salts65,66,67. We did not find any archaeal sequences in our dataset, and we cannot exclude, beside a real absence, a bias due to primer choice. According to the company’s protocol (https://imr.bio/protocols.html), the primer pair for the V3-V4 region18 of 16S should have a moderate coverage (0–90%) at amplifying archaea. Further studies using archaeal-specific primers are needed to assess their contribution in the biodeterioration of cultural heritage sites, especially the ones interested by saline incrustations.
Regarding the biodeterioration potential of the microorganisms here detected, some of them were already known to be involved in aesthetic (e.g., discolouration) and structural (e.g., corrosion, deterioration and decay) damages to cultural heritage items. Among the most abundant bacterial genera identified, we report Actinomycetospora68, Egibacter69, Loriellopsis70, Pseudonocardia68,71,72, Rubrobacter68,70,73, Streptomyces71, and Truepera74. Although none of these studies focused on the biodeterioration of stuccoes, it is likely that these microorganisms could have a similar impact on this substrate. In addition, other abundant bacteria, such as Chelativorans, have been indirectly associated with wood decay75, or biopolymers degradation in marine environments, as in the case of Oceanocaulis76. For eukaryotes, the available literature on the biodeterioration is mostly focused on algae and fungi, especially from stone monuments77,78,79. Regarding stuccoes, most of studies have investigated the role of fungi, reporting Aspergillus80 and Chaetomium, Penicillium, Sarocladium, and Stachybotrys58. Among these, in our study we only found Sarocladium in the stucco sample S6. Other studies (see above) reported only on the presence of microorganisms, therefore allowing no inferences on their role in biodegradative phenomena. About algae, the genus Ctenocladus here found was already reported81 for stuccoes, despite the direct role in biodeterioration was not assessed. For the genera Picochlorum, Picocystis, and Pseudostichococcus, we report their occurrence for the first time on stuccoes and we did not find any study implying their involvement in the deterioration of cultural heritage.
Metabolomic studies are still at their infancy in the field of cultural heritage (e.g.,82,83,84), but are gaining increasing importance due to their capabilities at providing qualitative and quantitative data on small molecules that are part of the metabolic pathways. Specifically, untargeted metabolomics is particularly useful to search for biomarkers likely involved in the biodeterioration of CH, by collecting data from hundreds to thousands of metabolites that belong to various classes of chemicals in a single analysis12. According to the studies reviewed in12, pathways like biosynthesis and degradation of aminoacids, ubiquinone and other terpenoid-quinone biosynthesis, pigments biosynthesis and degradation were shared among different CH objects. In our study, most of metabolites belonged to biosynthesis of secondary metabolites, microbial metabolism, biosynthesis of aminoacids, and carbon and protein metabolism, indicating active metabolic processes. In some cases, it has been possible to link the presence of specific metabolites to particular taxa, e.g. chlorophyll a to photosynthetic eukaryotes and cyanobacteria, and diatoxanthin to Picocystis salinarum. Diatoxanthin is a pigment typical of heterokonts (especially diatoms) and, outside this group, to date it has been only found in this species; because we found no diatom sequences in our samples and P. salinarum was particularly abundant in some of them, we are confident in the inferred attribution of such metabolite to this species. However, our correlation analysis between abundance of taxa and metabolites provided weak signals. Finding statistically significant correlations could be difficult in CH studies for several reasons. One factor responsible for this could be the bias in the sampling strategy of metabolomics but also metagenomics studies. Indeed, historical objects are sampled in non-invasive ways, which result in collecting very small amounts of superficial material from small portions that could not be representative of the entire surface12. For instance, there is the risk that the communities living in the deeper layers of the biofilm, which are often at contact with the CH, are not sampled to avoid damages to the object. In addition, it should be considered that a statistically robust sampling could not be often achieved because some manufacts are unique (impossibility to compare biofilms from similar substrata) or, on the same manufact, the number of biofilms that can be collected is limited (different degree of biodeterioration). Another affecting factor could be the choice of the barcoding marker, with broad-spectrum markers unable of capturing fine taxonomic resolution that could be, instead, relevant to link some organisms to specific metabolites. For instance, the universal 18S-V4 region could have underestimated the diversity of some fungi groups, which are better detected with ITS region marker, by amplifying other eukaryotic taxa85. However, the nature of detected metabolites could be responsible for such weak associations. Indeed, most of metabolites found here and in other studies on different substrata (e.g.,86) belong to generic pathways and cannot be associated to specific communities.
The use of green biocides such as essential oil extracts represents a valid alternative to synthetic biocides, allowing a restoration to be carried out using products with low toxicity, easy to handle and environmentally sustainable. Indeed, in the last few years there was an increase in the use of phyto-derivatives like liquorice leaf extract and essential oils as safer and eco-friendly alternative to chemicals to be used as natural biocides for the restoration of cultural heritage87,88,89. Despite the wide spectrum of efficacy of essential oils across different taxa and domains, it was found that Gram negative bacteria were generally more resistant than Gram positive bacteria, and that some had major effects on fungi than others90. In the field of cultural heritage, attention has been paid on extract oils from the family Lamiaceae, especially from Thymus and Origanum, because they have proven to be effective on different microorganisms and for their availability in commerce as ready-to-use blends91,92,93. An example of the application of these essential oils concerns the restoration of some artworks from the Catholic cemetery for foreigners in Rome94, where up to three applications were repeated into a hydroalcoholic solution (70% ethanol and 30% water), with a concentration of 5%. In other cases, as the removal of biofilms under the tiles of the floor mosaic of Leda’s House in the archaeological park of Solunto in Sicily95 and on the sculpture “The Silvano” in the archaeological museum of Florence96, applications of a thymus blend at 15% and a bush application in 2% demineralized water respectively, were proven to be effective. Regarding the specific application of ESSENZIO by IBIX Biocare, successful restoration interventions were achieved for the mosaics located in the room XIX of the Insula of the Muses in Ostia Antica97 and a mosaic fountain in Ravenna98. From the in-depth analysis of the different application methods tested in the studies described above, we identified the most important parameters to take into consideration for the application of essential oils and we were able to remove the biofilms on the surfaces of the laconicum. The treatment with ESSENZIO at 50% in demineralized water with an application exposure of one hour and half was successful, and no biofilms were observed after eleven months from the restoration.
Conclusions
Genetic and biochemical high-throughput techniques (-omic tools) have opened new paths toward the study of microorganisms forming biofilms on cultural heritage and their metabolic activity that is responsible of biodeterioration phenomena. Despite the potential of such -omic tools, which reflects in increasing literature on metabarcoding studies of CH objects, the number of studies using metabolomics and integrating both approaches is still small. In this study, we reported on the taxonomic and metabolite diversity of microorganisms forming biofilms on ancient stuccoes. At best of our knowledge, this is the first wide-spectrum intervention on ancient Roman stuccoes (besides the work by Bruno et al.87 on catacombs in Rome), as well as the first study integrating -omics approaches as metabarcoding and metabolomics on such fragile cultural heritage object. We confirmed that metabarcoding is a powerful technique to quickly and thoroughly characterise the taxonomic diversity of microorganisms in complex matrices as biofilms in respect to classical, culture-based approaches, also in fragile and delicate materials as stuccoes. In addition, we demonstrated that it is possible to extract and isolate numerous metabolites from very low amount of material and that such untargeted-metabolomics analyses are indicative of metabolic pathways active in the biofilms. However, we also argued that the paucity of biological material collected from stuccoes or cultural heritage items in general, as well as the scattered distribution of biofilms in the study system could affect the detection of statistically significant correlations between abundance of taxa and metabolites. Last, we have proven that a treatment based on essential oils from thyme and oregano effectively removes both bacterial and eukaryotic biofilms from stuccoes, thus confirming its utility in restoration of cultural heritage. To promote the adoption of such integrative approaches, future efforts should focus on the establishment of standardised strategies for sampling, data pre-processing, and statistical analyses of cultural heritage objects. Additionally, it is important not to overlook the value of culture-based methods in enriching taxonomic and metabolomics reference libraries, which are essential for any -omic study.
Data availability
Raw Illumina reads are available in fastq format in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA1135078. All other information not included in the main text is provided as Supplementary Information alongside this manuscript.
References
Wadsworth, E. L. Stucco reliefs of the First and Second Centuries still extant in Rome. Mem. Am. Acad. Rome. 4, 9–102 (1924).
Ling, R. Stucco Decoration in Pre-augustan Italy. Pap Br. Sch. Rome. 40, 11–57 (1972).
Gapper, C. What is ‘Stucco’? English interpretations of an Italian term. Archit. Hist. 42, 333–343 (1999).
Gapper, C., Orton, J. & Plaster Stucco and Stuccoes. J. Archit. Conserv. 17, 22–27 (2011).
Clementi, C. et al. Non-invasive and micro-destructive investigation of the Domus Aurea wall painting decorations. Anal. Bioanal Chem. 401, 1815–1826 (2011).
Ball, L. & Domus Aurea doi: (2020). https://doi.org/10.1093/acrefore/9780199381135.013.2283
Ling, R. Some roman stucco reliefs from Pozzuoli now in the British Museum. Pap Br. Sch. Rome. 34, 24–33 (1966).
Ling, R. Roman paintings and Stucco reliefs in the Victoria and Albert Museum. Pap Br. Sch. Rome. 49, 46–58 (1981).
Ling, R. Stucco decorations at Baia. Pap Br. Sch. Rome. 45, 24–51 (1977).
Mielsch, H. Römische Stuckreliefs (Kerle, Heidelberg, 1975).
Robertson, M. K. Schefold, Vergessenes Pompeji: unveröffentliche Bilder römischer Wanddekorationen in geschichtlicher Folge herausgegeben. Bern and München: Francke Verlag, 1962. Pp. 218; 180 plates (16 in colour). Sw. fr. 75. J. Rom. Stud 54, 225–226 (1962).
Gutarowska, B. The use of -omics tools for assessing biodeterioration of cultural heritage: a review. J. Cult. Herit. 45, 351–361 (2020).
Chimienti, G. et al. Profile of microbial communities on carbonate stones of the medieval church of San Leonardo Di Siponto (Italy) by Illumina-based deep sequencing. Appl. Microbiol. Biotechnol. 100, 8537–8548 (2016).
Alaoui-Sosse, B. et al. Assessment of microbial communities colonizing the Azé prehistoric cave. J. Cult. Herit. 59, 1–9 (2023).
De Luca, D., Piredda, R., Trojsi, G. & Cennamo, P. Close but different: metabarcoding analyses reveal different microbial communities in ancient roman nymphaea. Int. Biodeterior. Biodegradation. 181, 105619 (2023).
Petraretti, M. et al. Deterioration-associated microbiome of a modern photographic artwork: the case of Skull and crossbones by Robert Mapplethorpe. Herit. Sci. 12, 172 (2024).
Amalfitano, P., Camodeca, G. & Medri, M. I Campi Flegrei: un itinerario archeologico (1990).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1–e1 (2013).
Comeau, A. M., Li, W. K. W., Tremblay, J. É., Carmack, E. C. & Lovejoy, C. Arctic Ocean Microbial Community Structure before and after the 2007 Record Sea Ice Minimum. PLoS One. 6, e27492 (2011).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581–583 (2016).
R Core Team. R: A language and environment for statistical computing (2020).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.4-3 (2022).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Guillou, L. et al. The Protist Ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013).
Beentjes, K. K. et al. Increased performance of DNA metabarcoding of macroinvertebrates by taxonomic sorting. PLoS One. 14, e0226527 (2019).
Huson, D. H., Auch, A. F., Qi, J. & Schuster, S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naïve bayesian classifier for Rapid assignment of rRNA sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
McLaren, M. R. & Callahan, B. J. Silva 138.1 prokaryotic SSU taxonomic training data formatted for DADA2. (2021). https://doi.org/10.5281/ZENODO.4587955
RStudio Team. RStudio: Integrated Development for R (2020).
McMurdie, P. J. & Holmes, S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 8, e61217–e61217 (2013).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, Berlin, 2016).
Liu, C. et al. Microbial habitat specificity largely affects microbial co-occurrence patterns and functional profiles in wetland soils. Geoderma. 418, 115866 (2022).
Liu, C., Cui, Y., Li, X. & Yao, M. Microeco: an R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 97, fiaa255 (2021).
Metsalu, T. & Vilo, J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566–W570 (2015).
Djoumbou Feunang, Y. et al. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 8, 61 (2016).
Xia, J., Psychogios, N., Young, N. & Wishart, D. S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660 (2009).
Kanehisa, M. & Goto, S. K. E. G. G. Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Cennamo, P. et al. UV-C irradiation and essential-oils-based product as Tools to Reduce Biodeteriorates on the Wall paints of the archeological site of Baia (Italy). Coatings 13, 1034 (2023).
Albertano, P. Epilithic algal communities in hypogean environments. G Bot. Ital. 127, 386–392 (1993).
Ortega-Calvo, J. J., Ariño, X., Hernandez-Marine, M. & Saiz-Jimenez, C. Factors affecting the weathering and colonization of monuments by phototrophic microorganisms. Sci. Total Environ. 167, 329–341 (1995).
Gorbushina, A. A. & Petersen, K. Distribution of microorganisms on ancient wall paintings as related to associated faunal elements. Int. Biodeterior. Biodegradation. 46, 277–284 (2000).
Gurtner, C. et al. Comparative analyses of the bacterial diversity on two different biodeteriorated wall paintings by DGGE and 16S rDNA sequence analysis. Int. Biodeterior. Biodegradation. 46, 229–239 (2000).
Gonzalez, J. M. & Saiz-Jimenez, C. Microbial diversity in biodeteriorated monuments as studied by denaturing gradient gel electrophoresis. J. Sep. Sci. 27, 174–180 (2004).
Landy, E. T., Mitchell, J. I., Hotchkiss, S. & Eaton, R. A. Bacterial diversity associated with archaeological waterlogged wood: ribosomal RNA clone libraries and denaturing gradient gel electrophoresis (DGGE). Int. Biodeterior. Biodegradation. 61, 106–116 (2008).
Ghosh, S., Bagheri, B., Morgan, H. H., Divol, B. & Setati, M. E. Assessment of wine microbial diversity using ARISA and cultivation-based methods. Ann. Microbiol. 65, 1833–1840 (2015).
De Luca, D., Caputo, P., Perfetto, T. & Cennamo, P. Characterisation of Environmental Biofilms Colonising Wall paintings of the Fornelle Cave in the Archaeological Site of Cales. Int. J. Environ. Res. Public Health 18, 8048 (2021).
Perito, B. & Cavalieri, D. Innovative metagenomic approaches for detection of microbial communities involved in biodeteriorattion of cultural heritage. IOP Conf. Ser. Mater. Sci. Eng. 364, 12074 (2018).
Marvasi, M., Cavalieri, D., Mastromei, G., Casaccia, A. & Perito, B. Omics technologies for an in-depth investigation of biodeterioration of cultural heritage. Int. Biodeterior. Biodegradation. 144, 104736 (2019).
Vilanova, C. & Porcar, M. Art-omics: multi-omics meet archaeology and art conservation. Microb. Biotechnol. 13, 435–441 (2020).
Pfendler, S. et al. Biofilm biodiversity in French and Swiss show caves using the metabarcoding approach: first data. Sci. Total Environ. 615, 1207–1217 (2018).
Cennamo, P. & De Luca, D. A metabarcoding approach for the study of biodeterioration of ancient wall paintings in an Italian cave. J. Phys. Conf. Ser. 2204, 012011 (2022).
Ortega-Morales, O. et al. Deterioration and microbial colonization of cultural heritage stone buildings in polluted and unpolluted tropical and subtropical climates: a meta-analysis. Int. Biodeterior. Biodegradation. 143, 104734 (2019).
Paiva, D. S. et al. Uncovering the Fungal Diversity Colonizing Limestone Walls of a Forgotten Monument in the Central Region of Portugal by High-Throughput sequencing and culture-based methods. Appl. Sci. 12, 10650 (2022).
Timoncini, A. et al. Insight on bacteria communities in outdoor bronze and marble artefacts in a changing environment. Sci. Total Environ. 850, 157804 (2022).
Vieto, S. et al. Biodeterioration and cellulolytic activity by fungi isolated from a nineteenth-century painting at the National Theatre of Costa Rica. Fungal Biol. 126, 101–112 (2022).
Pinna, D. et al. Damaging and protective interactions of lichens and biofilms on ceramic dolia and sculptures of the International Museum of Ceramics, Faenza, Italy. Sci. Total Environ. 877, 162607 (2023).
Favero-Longo, S. E. et al. Biocide efficacy and consolidant effect on the mycoflora of historical stuccos in indoor environment. J. Cult. Herit. 34, 33–42 (2018).
Ortega-Morales, B. O. et al. Implications of colonizing biofilms and microclimate on west stucco masks at North Acropolis, Tikal, Guatemala. Herit. Sci. 1, 32 (2013).
Ortega-Morales, O., Guezennec, J., Hernández-Duque, G., Gaylarde, C. C. & Gaylarde, P. M. Phototrophic biofilms on ancient mayan buildings in Yucatan, Mexico. Curr. Microbiol. 40, 81–85 (2000).
De Miguel, J. M. G., Sanchez-Castillo, L., Ortega-Calvo, J. J., Gil, J. A. & Saiz-Jimenez, C. Deterioration of building materials from the Great Jaguar Pyramid at Tikal, Guatemala. Build. Environ. 30, 591–598 (1995).
Sanchez-Moral, S. et al. Deterioration of building materials in roman catacombs: the influence of visitors. Sci. Total Environ. 349, 260–276 (2005).
Agarossi, G., Ferrari, R. & Monte, M. D. Microbial biodeterioration in the hypogea: the subterranean neo-pythagorean basilica of Porta Maggiore in Rome. in Vth Int. Congress Deterioration Conserv. Stone 2, 597–605 (1985).
Kalwasińska, A. et al. Microbial communities associated with the anthropogenic, highly alkaline environment of a saline soda lime, Poland. Antonie Van Leeuwenhoek. 110, 945–962 (2017).
Saiz-Jimenez, C. & Laiz, L. Occurrence of halotolerant/halophilic bacterial communities in deteriorated monuments. Int. Biodeterior. Biodegradation. 46, 319–326 (2000).
Adamiak, J., Otlewska, A. & Gutarowska, B. Halophilic microbial communities in deteriorated buildings. World J. Microbiol. Biotechnol. 31, 1489–1499 (2015).
Lepinay, C. et al. Bacterial diversity associated with saline efflorescences damaging the walls of a French decorated prehistoric cave registered as a World Cultural Heritage Site. Int. Biodeterior. Biodegradation. 130, 55–64 (2018).
He, D. et al. Insights into the bacterial and fungal communities and microbiome that causes a microbe outbreak on ancient wall paintings in the Maijishan Grottoes. Int. Biodeterior. Biodegradation. 163, 105250 (2021).
Silva, I. et al. Microbial induced stone discoloration in alcobaça monastery: a comprehensive study. J. Cult. Herit. 67, 248–257 (2024).
Li, Q., Zhang, B., Yang, X. & Ge, Q. Deterioration-Associated Microbiome of Stone monuments: structure, variation, and Assembly. Appl. Environ. Microbiol. 84, e02680–e02617 (2018).
Abdulla, H., May, E., Bahgat, M. & Dewedar, A. Characterisation of actinomycetes isolated from ancient stone and their potential for deterioration. Pol. J. Microbiol. 57, 213–220 (2008).
Diaz-Herraiz, M. et al. The Actinobacterial colonization of etruscan paintings. Sci. Rep. 3, 1440 (2013).
Schröer, L., De Kock, T., Cnudde, V. & Boon, N. Differential colonization of microbial communities inhabiting Lede stone in the urban and rural environment. Sci. Total Environ. 733, 139339 (2020).
Dyda, M. et al. Diversity of Biodeteriorative bacterial and fungal consortia in Winter and Summer on historical sandstone of the Northern Pergola, Museum of King John III’s palace at Wilanow, Poland. Appl. Sci. 11, 620 (2021).
Beccaccioli, M. et al. The neolithic site La Marmotta: DNA metabarcoding to identify the microbial deterioration of waterlogged archeological wood. Front. Microbiol. 14, 1129983 (2023).
Hu, S. et al. Degradation of chlorothalonil via thiolation and nitrile hydration by marine strains isolated from the surface seawater of the Northwestern Pacific. Int. Biodeterior. Biodegradation. 154, 105049 (2020).
Nowicka-Krawczyk, P., Komar, M. & Gutarowska, B. Towards understanding the link between the deterioration of building materials and the nature of aerophytic green algae. Sci. Total Environ. 802, 149856 (2022).
Komar, M. et al. Biodeterioration potential of algae on building materials - model study. Int. Biodeterior. Biodegradation. 180, 105593 (2023).
Gadd, G. M., Fomina, M. & Pinzari, F. Fungal biodeterioration and preservation of cultural heritage, artwork, and historical artifacts: extremophily and adaptation. Microbiol. Mol. Biol. Rev. 88, e00200–e00222 (2024).
Afifi, H. A. M., Mansour, M. M. A., Hassan, A. G. A. I. & Salem, M. Z. M. Biodeterioration effects of three aspergillus species on stucco supported on a wooden panel modeled from Sultan Al-Ashraf Qaytbay Mausoleum, Egypt. Sci. Rep. 13, 15241 (2023).
Ariño, X., Hernandez-Marine, M. & Saiz-Jimenez, C. Ctenocladus circinnatus (Chlorophyta) in stuccos from archaeological sites of southern Spain. Phycologia. 35, 183–189 (1996).
Gutarowska, B. et al. Metabolomic and high-throughput sequencing analysis—modern approach for the assessment of biodeterioration of materials from historic buildings. Front. Microbiol. 6, 979 (2015).
Szulc, J. et al. Metabolomics and metagenomics analysis of 18th century archaeological silk. Int. Biodeterior. Biodegradation. 156, 105120 (2021).
LI, X., Liu, S. I. W., Zhou, Q., LI, Q. & Y. & Insight into natural ageing of historic baltic amber objects by untargeted metabolomics approach. J. Cult. Herit. 67, 470–478 (2024).
Banos, S. et al. A comprehensive fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms. BMC Microbiol. 18, 190 (2018).
Checcucci, A., Borruso, L., Petrocchi, D. & Perito, B. Diversity and metabolic profile of the microbial communities inhabiting the darkened white marble of Florence Cathedral. Int. Biodeterior. Biodegradation. 171, 105420 (2022).
Bruno, L. et al. Biodeterioration of Roman hypogea: the case study of the catacombs of SS. Marcellino and Pietro (Rome, Italy). Ann. Microbiol. 69, 1023–1032 (2019).
Palla, F., Bruno, M., Mercurio, F., Tantillo, A. & Rotolo, V. Essential oils as natural biocides in Conservation of Cultural Heritage. Molecules 25, 730 (2020).
Rugnini, L. et al. Biocidal Activity of Phyto-Derivative products used on Phototrophic biofilms growing on Stone surfaces of the Domus Aurea in Rome (Italy). Appl. Sci. 10, 6584 (2020).
Chao, S. C., Young, D. G. & Oberg, C. J. Screening for inhibitory activity of essential oils on selected Bacteria, Fungi and viruses. J. Essent. Oil Res. 12, 639–649 (2000).
Sparacello, S. et al. Thymus vulgaris essential oil and Hydro-Alcoholic Solutions to counteract Wooden Artwork Microbial colonization. Appl. Sci. 11, 8704 (2021).
Bosco, F., Mollea, C., Demichela, M. & Fissore, D. Application of essential oils to control the Biodeteriogenic Microorganisms in Archives and libraries. Heritage. 5, 2181–2195 (2022).
Russo, R. & Palla, F. Plant Essential Oils as Biocides in Sustainable Strategies for the Conservation of Cultural Heritage. Sustainability 15, 8522 (2023).
Bartolini, M. & Pietrini, A. M. La disinfezione delle patine biologiche sui manufatti lapidei: biocidi chimici e naturali a confronto. Boll. ICR 33, 39–49 (2016).
Rotolo, V. S., Caro, M. L., De, Giordano, A. & Palla, F. Solunto archaeological park in Sicily: life under mosaic tesserae. Fl. Medit. 28, 233-245 (2018).
Spada, M. & Sorella, F. Studio E Verifica delle proprietà biocide degli oli essenziali. Il restauro della statua di Silvano Del Museo Archeologico Nazionale Di Firenze. Riv dell’Opificio Delle Pietre Dure E Lab. di Restauro Di Firenze. 32, 100–116 (2020).
Macchia, A. et al. In situ application of Anti-fouling Solutions on a Mosaic of the Archaeological Park of Ostia Antica. Materials 15, 5671 (2022).
Coccıa, C., Carra, I. M., Gandını, M. B., Perpıgnanı, P. & Zambruno, S. The Parco Della Pace in Ravenna: the restoration of the Mosaic Fountain Le Chaos Et La Source De Vie by Claude Rahir Modern approaches to the Conservation and Restoration of Contemporary Works of art exposed outdoors TT - Ravenna‘daki Parco Della Pa. J. Mosaic Res. 16, 447–458. https://doi.org/10.26658/jmr.1377317 (2023).
Acknowledgements
The authors acknowledge Dr. Enrico Gallocchio, archaeological officer of the Archaeological Park of Baia for the permissions at accessing the area to conduct the activities of this research.
Funding
This research was funded by the European Union - NextGenerationEU Project “PE 0000020 CHANGES” - CUP B53C22003780006, NRP Mission 4 Component 2 Investment 1.3.
Author information
Authors and Affiliations
Contributions
P.C. conceived and designed the experiments; P.C. obtained the funding for the study; P.C. and S.S. were involved in the sampling activity; D.D.L. and R.P. carried out the experiments on metabarcoding; J.T. and M.L. produced the metabolomics data; M.M.C. and S.S. were involved in the restoration of stuccoes; D.D.L. and R.P. analysed the data; D.D.L. prepared the original draft of the manuscript; D.D.L., J.T., M.M.C., O.D.C., P.C., and R.P. were involved in review and editing of the manuscript. All authors have read and agreed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
This article does not include any studies of human participants or animals by the authors of this investigation.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
De Luca, D., Piredda, R., Scamardella, S. et al. Taxonomic and metabolic characterisation of biofilms colonising Roman stuccoes at Baia’s thermal baths and restoration strategies. Sci Rep 14, 26290 (2024). https://doi.org/10.1038/s41598-024-76637-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-76637-x