Abstract
Cardiovascular disease (CVD) is the leading cause of mortality, disability, and healthcare costs, with a significant impact on the elderly and contributing to premature deaths across various age groups, including those below age 70. Despite decades of transformative discoveries and clinical efforts, the challenges of diagnosis, prevention, and treatment of CVD persist on a massive scale. This study aimed to unravel potential CVD-associated biomarkers and establish a machine learning model for the risk assessment of CVD. Untargeted metabolic assay with ultra-high performance liquid chromatography-tandem mass spectrometry and routine clinical biochemistry test were undertaken on the fasting venous blood specimens from 57 subjects. Four relevant clinical traits and 164 CVD-associated metabolites were identified, especially those related to glycerophospholipid metabolism and biosynthesis of unsaturated fatty acids. The machine learning model achieved from an integrated biomarker panel of palmitic amide, oleic acid, 138-pos (the 138th detected metabolomic feature in positive ion mode), phosphatidylcholine, linoleic acid, age, direct bilirubin, and inorganic phosphate, was able to improve the accuracy of CVD risk assessment up to a high satisfactory value of 0.91. The findings indicate that disorders in the metabolic processes of biological membranes and energy are significantly associated with increased risk of vascular damage in CVD patients. With machine learning methods, the pivotal metabolites and clinical biomarkers offer a promising potential for the efficient risk assessment and diagnosis of CVD.
Similar content being viewed by others
Introduction
Cardiovascular disease (CVD) refers to ischemic or hemorrhagic diseases that occur in the heart, brain and other tissues, due to arteriosclerosis, hypertension, hyperlipidemia1. The pathology is quite diverse, including diseases such as heart disease (HD), stroke, and others2,3. CVD is the leading cause of mortality, disability, and healthcare costs across various age groups, significantly affecting the elderly and also contributing to deaths in individuals below age 70. Despite decades of transformative discoveries and clinical efforts, the challenges of diagnosis, prevention, and treatment of CVD persist on a massive scale. The Global Burden of Disease has reported a rise in the prevalence of CVD from 271 million in 1990 to 523 million in 2019, and an increase in the CVD-related deaths from 12.1 million to 18.6 million accompanied during the same period1. Currently, the clinical diagnosis of CVD has been significantly advanced. For example, electrocardiogram testing, ultrasonography, and computed tomography are the commonly used techniques4. However, these techniques are mainly applicable for in-hospital use, due to the equipment size, cost, and technical requirements5. Many studies have shown that during the occurrence and progress of CVD, the body would undergo a series of early damage events such as oxidative stress, inflammatory responses, ion imbalances, and metabolic disturbances6,7,8. These disturbances contribute to the damage of endothelial cells, neurons, and glial cells. As a result, the circulatory and nervous systems are significantly affected, and a large number of DNA, protein, and metabolite molecules are changed9. Therefore, it is possible to detect and unravel these substances as potential biomarkers for the investigation and diagnosis of CVD.
Numerous omics-based studies have been conducted to analyze, identify, validate, and apply CVD biomarkers, including genomics, proteomics, and metabolomics10,11. Genome-wide association studies (GWAS) have identified pivotal loci associated with CVD12, which have a positive impact on disease risk prediction and prevention. For instance, a large-scale GWAS discerned a single nucleotide polymorphism at chromosome 6p21.1 related to large artery atherosclerotic stroke13. A meta-analysis concerning MRI-confirmed patients found five genetic loci linked to lacunar stroke14. These discoveries were beneficial to screening high-risk populations for CVD. Proteomics plays a critical role in exploring the composition and variation of cellular, tissue or organismal proteomes, elucidating the underlying mechanisms of physiological and pathological changes in organisms10. Proteomics has been applied to seek CVD biomarkers, such as growth differentiation factor 15, kidney injury molecule, and WAP 4-disulfide core domain protein 215. These CVD-related genes and proteins not only help understand the disease pathophysiology but also provide valuable targets for diagnosis.
Metabolomics is a burgeoning, powerful technology to describe the collection of endogenous small-molecule metabolites and lipids within an organism or the state of a biological system. It underscores to directly and accurately disclose the terminal state and phenotype of organisms, revealing metabolic changes across a series of events, such as alterations in genes, proteins, and other functional components16. A large diversity of metabolomes and potential metabolites as biomarkers not only help understand organisms or cell responding to stimuli at different stages but also provide deep insights into cell identification, disease occurrence and development17. Therefore, metabolomics has been applied to risk assessment and diagnosis of CVD. For instance, untargeted metabolomics in the Bogalusa Heart Study discovered serum metabolites and potential biological pathways associated with the pathogenesis of heart failure18. The study of coronary heart disease and stroke disclosed the relationship between stroke risk and linoleic acid (LA)19.
This study not only aimed to unravel the metabolic changes of CVD through analysing the fasting venous blood specimens of 57 population subjects, but to use pivotal metabolite and clinical trait biomarkers for assessing the CVD risk in conjunction with machine learning methods. Plasma samples were detected by means of ultra-high performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) technique for untargeted metabolomics analysis. Serum samples were measured on an automatic biochemical analyzer for routine clinical biochemistry test. The successful CVD screening model provided an alternative promising potential for the efficient risk assessment and diagnosis of CVD.
Materials and methods
Study design and participants
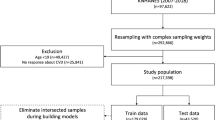
A case-control study was designed to investigate the metabolic changes and underlying biological processes in HD and ischemic stroke (IS) patients. Study participants were recruited in the Affiliated Hospital of Guizhou Medical University from February to April 2021. According to the diagnostic results by professional clinicians for each CVD, subjects were divided into the HD group including 17 patients, the IS group composed of 20 patients, and control group of 20 subjects. Especially, the control group meant the individuals precluding any observable symptoms or history of CVD. Demographic information, including sex and age, was recorded. For details of patient-related information, refer to Data S1 in the Supporting Information. The study was reviewed and approved by the Ethics Committee of Guizhou Medical University (Approval number: 202006).
Figure 1 summarizes the design of the study that was deployed in two phases. In the phase I, a routine biochemistry test was conducted with serum samples to acquire the levels of 36 biochemical indicators of each participant. These biochemical indicators together with sex and age information comprised a set of clinical traits which must be generally first checked for the health assessment of patient and initial diagnosis of CVD when the patient is hospitalized. The phase II was the main focus of this study, in which the untargeted metabolic analysis of plasma samples was carried out to unravel CVD-associated metabolites and profile the pivotal metabolic changes.
Blood specimens and sample preparation
Fasting venous blood specimens were collected from all individual subjects into 5 mL dry vacutainer tubes, and 5 mL vacutainer tubes containing ethylene diamine tetraacetic acid (EDTA) as a chelating agent. All the blood specimens were immediately centrifuged for 15 min at 1500 rpm and 4 °C to separate the serum and/or plasma from the other blood components. The serum was aliquoted from each dry tube and measured on an E602 automatic biochemical analyzer (Cobas, Switzerland) for a routine clinical biochemistry test. The quantitative levels of 33 biochemical traits were acquired (Table 1). The plasma was aliquoted from each EDTA tube and preserved at − 80 °C until further analysis.
Prior to the UHPLC-MS/MS detection, plasma samples were first thawed in ice water. Then, 100 µL of each plasma sample was pipetted into Eppendorf tube and resuspended with 400 µL of ice-cold 80% methanol by well vortex. The mixture was incubated on ice for 5 min and subsequently centrifuged at 15, 000 g and 4 °C for 20 min, yielding a supernatant. Supernatant underwent further dilution with ultrapure water until the methanol content reached 53% (v/v). The diluted supernatant was transferred to a new Eppendorf vial and centrifuged at 15,000 g and 4 °C for 20 min in preparation before loading to the UHPLC-MS/MS20. Quality control (QC) sample was prepared by pooling 5 µL of each obtained supernatant from all the individual 100 µL aliquots and mixing together into a total of 285 µL.
UHPLC-MS/MS assay
The UHPLC-MS/MS analysis was performed on a UHPLC system (Vanquish, Thermo Fisher Scientific) coupled with an Orbitrap Q Exactive™ HF mass spectrometer (Q Exactive, Thermo Fisher Scientific). Hypersil GOLD C18 column (100 mm × 2.1 mm, 1.9 μm, Thermo Scientific) was used for chromatographic separation at column temperature of 40 °C with a flow rate of 0.2 mL/min. The mobile phases A and B were respectively water containing 0.1% formic acid and methanol for the positive ion mode. While for the negative mode the mobile phases A and B were water containing 5 mmol/L ammonium acetate (pH 9.0) and methanol. The gradient eluting procedure was as follows: 2% B held for 1.5 min, then increased to 100% B in 12.0 min and held for 2.0 min, finally declined to 2% B at 14.1 min and held until 17.0 min21. Q Exactive™ HF mass spectrometer was operated in both positive and negative polarity modes with a spray voltage of 3, 500 V, capillary temperature of 320 °C, the flow rates of sheath gas and auxiliary gas were 35 and 10 in arbitrary unit, respectively. S-lens RF level was 60 and auxiliary gas heater temperature was 350 °C. The mass-to-charge ratio (m/z) range was 70.0‒1,050 with a resolution of 120,000. The data-dependent acquisition-based mass fragment (MS2) scans were automatically determined in terms of fragments for dissociation22. Three blank and six QC samples were successively run to balance the system before the analysis of plasma samples. QC sample was tested every ten plasma samples during the UHPLC analysis (Fig. S1 in the Supporting Information), which was used for the data quality assessment and variation monitoring purpose.
Metabolite annotation
The acquired UHPLC-MS/MS data were subjected to a variety of data processing with SIRIUS (version 5.5.7)23 (Fig. S2 in the Supporting Information). This processing encompassed peak extraction, baseline correction, signal deconvolution, and peak alignment, among others, to obtain relevant features in the form of peak table. The obtained peak table not only included qualitative information of retention time (tR), precursor ion (MS1), and MS2 of annotated metabolites but also the quantitative information of peak area, etc. The qualitative information of tR, MS1, and MS2 was utilized to identify every specific compound through comparison with those in known databases by searching and similarity scoring. Based on the annotating accuracy, compounds were identified at levels 1–424: (1) confidently identified compounds with spectral similarity to real chemical reference standards (level1), (2) putatively annotated compounds matching the mass characteristics with MS1 and MS2 in database such as Kyoto Encyclopedia of Genes and Genome (KEGG)25, Human Metabolome Database (HMDB)26, Chemical Entities of Biological Interest (ChEBI)27 and PubChem28 (level2), (3) putatively annotated compound classes with spectral and/or physicochemical similarity to known class compounds (level3), and (4) unknown compounds with accurate mass or molecular formula matching in database (KEGG, PubChem, HMDB) (level4). All the candidate compounds were ranked for the most probable structural match.
Data processing and statistical analysis
To facilitate the subsequent statistical analysis of clinical traits and metabolic data, it was necessary to initially remove the traits with missing values exceeding 50%. Separately, individual data points exceeding 1.5 times the interquartile range were identified and removed as outliers. After these steps, the remaining missing values were imputed using the median value. Metabolites with a relative standard deviation greater than 25% in QC samples were filtered out. Data normalization was performed through median normalization, log10 transformation, and autoscaling.
Principal component analysis (PCA) was carried out to observe sample clustering and dispersion. One-way analysis of variance (ANOVA) was implemented to detect the difference among groups, and Fisher’s least significant difference (LSD) test was used for pairwise comparisons between groups. A threshold of p < 0.05 was set for determining significant changes in metabolites between groups or p < 0.01 for highly significant changes.
To identify differential features more accurately in the metabolomics data, age adjustment was performed. Only those features exhibiting significant differences (p < 0.05) in both age-adjusted analysis and ANOVA were selected for further identification. For controlling the overall false discovery rate, the p-values calculated from ANOVA and LSD were corrected by means of the Benjamin-Hochberg method, using the R command p. adjust in the stats package. Statistical analysis was conducted in the software R (version 4.2.3). PCA was carried out using MetaboAnalyst 5.029.
Metabolomics pathway analysis
Metabolomics pathway analysis was performed through running MBROLE 2.0 (http://csbg.cnb.csic.es/mbrole2)30 with the supported compound identifies of the significant metabolites. The species were restricted to Homo sapiens for library searching and the enrichment analysis involved KEGG, SMPDB (HMDB) and BioCyc Pathways, ChEBI roles, HMDB biofunctions, and KEGG roles. Pathway overrepresentation analysis was performed using a cumulative hypergeometric test. The metabolic pathways and biofunctions with p < 0.05 were considered specific to the CVD, HD, and IS patients.
CVD risk assessment with machine learning
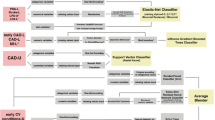
The recursive feature elimination (RFE) method was first implemented for selecting the clinical traits and metabolites in caret 6.0–92 package in R 4.3.1. RFE is a multi-step feature selection approach that has demonstrated better performance and synergistic effects in multivariate calibration compared to univariate statistical methods like the t-test and ANOVA31. Afterwards, the CVD risk was assessed with the applications of five sophisticated machine learning methods of linear discriminant analysis (LDA), partial least squares-discriminant analysis (PLS-DA), support vector machine (SVM), gradient boosting machine (GBM), and random forest (RF)32. The assessment models were individually established with the above-selected traits and metabolites.
Using stratified sampling, the 57 participants across three groups were randomly divided into a calibration set and a validation set at approximately a 4:1 ratio. That is, the calibration set included 16 control, 13 HD, and 16 IS subjects, while the independent validation set contained four control, four HD, and four IS subjects for each group, respectively.
The model performance was evaluated in terms of metrics: accuracy (ACC), sensitivity (SEN), specificity (SPE), Matthews correlation coefficient (MCC), F1 score, the receiver operating characteristic (ROC) curve, and the area under ROC curve (AUC). All these procedures were implemented in R 4.3.1 using the caret 6.0–92 package.
Results
Clinical traits of participants
The clinical traits of the subjects are available in Table 1, despite the limited source of subjects, which is a common problem in a population-based disease study. Their chi-square test and ANOVA revealed that eight traits out of 38 ones showed significant differences (p < 0.05) between groups and even extremely significant differences (p < 0.01). They were age, total bilirubin (TBIL), direct bilirubin (DBIL), indirect bilirubin (IBIL), aspartate aminotransferase/alanine aminotransferase (AST/ALT), inorganic phosphate (IP), magnesium (Mg) and anion gap (AG). Their boxplots in Fig. S3 in the Supporting Information disclosed the distribution and dispersion of in each group. For instance, the age, TBIL, DBIL and IBIL indicators of control group showed large variations.
The eight traits of three groups were submitted to a PCA procedure. Figure 2A shows the resultant scores. Clearly, it failed to distinguish the HD, IS and control subjects. Therefore, the RFE was executed to further refine these eight traits. Ultimately, the age, IP, AG and DBIL were spotted and subjected to five machine learning methods for establishing discrimination models (Table 2). It could be observed that LDA performed well on both the calibration and validation sets, and the resulting accuracy, sensitivity, specificity, Matthews correlation coefficient, F1 score, and AUC were fair. The values corresponding to the calibration set were respectively close to those of the validation set. Hence, the LDA method was subsequently used to distinguish between HD and IS groups, as well as between CVD and non-CVD participants. Figure 2B depicts the obtained ROC curves. As in the HD versus control case, the AUC value calculated for the calibration reached 0.923 and 0.875 for the validation, respectively. While in the IS versus control case, both the AUC values approached 1.0. This indicated a good efficiency in discriminating HD, IS and control groups, by using the trait panel of age, IP, AG and DBIL.
Metabolite detection and annotation
A total of 4819 metabolic features were detected from the original UHPLC-MS/MS data of the HD, IS and control groups within the analysis time of 12.0 min, including 2532 features in the positive mode and 2287 features in the negative mode (Fig. S4 in the Supporting Information). A complete list of the features was available in the Supplementary Data 1. After data preprocessing, 848 features were generated for metabolite annotation (Fig. 3A), as detailed in peak table of the Supplementary Data 2. Metabolite annotation was completed through searching in known databases, comparing the experimentally obtained tR, MS1, and MS2 to those standard compounds in libraries, and scoring the similarities within MS-Finder33, mzCloud34, SIRIUS23, Metfrag35, and MetaboAnalyst29.
Close inspection of each annotated metabolite revealed that lipids and lipid-like molecules had a maximal proportion of 33% in the metabolite profiles, and followed by organic acids and derivatives (24%), unannotated metabolites (14%), and organ heterocyclic compounds (8%). Figure 3B summarizes the classes of annotated metabolites. Although PCA was tried to visualize these metabolic profiles, the subjects in the control, HD, IS, and QC groups failed to discriminate from one another on the score scatterings (Fig. 3C). In contrast, seven QC samples were clustered within a small area, and their Cronbach’s alpha score was 0.89, quite higher than those of the control, HD, and IS samples (Table S1 in the Supporting Information). This at least implied a satisfactory stability of the UHPLC-MS/MS measurements and reliability of data manipulation.
Metabolic pathway of CVD and control groups
By ANOVA of the metabolite profiles associating metabolic pathways with CVD, 165 differential metabolites were identified between the CVD and control groups (Fig. 4A), accounting for 19.34% of the 848 features. The correlation between the age and metabolites was also analysed (Fig. 4B), through conducting a significant test with covariate adjustment on age. Finally, 164 differential metabolites were determined in the intersection of ANOVA and linear age-adjusted correlation, with significant differences of p < 0.05. Their heat map (Fig. 4C) covered a variety of categories of metabolites (Fig. 4D) annotated at four levels (Fig. 4E). Among them, 42 lipid and lipid-like molecules held the highest percent of 25.61%. The Supplementary Data 3 tabulated these metabolites.
In order to understand the metabolome of CVD, both pathway enrichment and biofunction analysis were performed with an imposed p < 0.05. The result was that nine metabolic pathways were identified and presented in Fig. S5A in the Supporting Information. These pathways were primarily associated with energy metabolism and membrane components. Glycerophospholipid metabolism had a minimum p-value of 7.88 × 10–6. The biofunction with the smallest p-value of 1.00 × 10–8 was energy source and membrane component. Figure 5 illustrates these metabolic pathways in the CVD patients. Particular attention was given to glycerophospholipid metabolism and biosynthesis of unsaturated fatty acids, which involved nine metabolites of choline (C00114), lysophosphatidylcholine (LPC, C04230), phosphatidylcholine (PC, C00157), oleic acid (C00712), 1-acyl-sn-glycero-3-phosphoethanolamine (C04438), 2-acyl-sn-glycero-3-phosphoethanolamine (C05973), LA (C01595), eicosadienoic acid (C16525), and docosahexaenoic acid (DHA) (C06429). These two pathways were critically related through PC conversion to LA, accounting for the major metabolome of CVD different from that of the control group, to a certain extent.
Significant metabolic pathways accounting for the difference between CVD patients and control group. “Matched metabolites” refer to the differential metabolites identified through comparative analysis between CVD and control groups. “Unmatched metabolites” are those involved in the main pathways yet not detected during the analysis.
Metabolic alteration in HD and IS
Given the differences in symptoms and clinical diagnoses between HD and IS patients, pairwise comparisons were made on the HD versus control, IS versus control, and IS versus HD groups, respectively. The p-values corresponding to each metabolite feature were computed and then subjected to logarithmic transformation with a base of 10. As a consequence, 132 differential metabolites were identified and classified into three main groups. Nine metabolites of them were specifically associated with HD, four of which belonged to benzene series, accounting for 44.44%. This metabolite alteration involved four perturbed metabolic pathways and one disturbed biofunction of protein synthesis (Table S2 and Fig. S5B in the Supporting Information). The phenylacetate and phenylalanine metabolisms were closely related to the protein synthesis and amino acids biosynthesis.
There were 107 metabolites specific to the IS metabolic alterations, mainly lipid and lipid-like molecules (25%), and organic acids and derivatives (25%). Seven important pathways and six disordered biofunctions (Table S2 and Fig. S5C in the Supporting Information) explained that the glycerophospholipid metabolism was mostly correlated, followed by the α-linolenic acid and LA metabolism, biosynthesis of unsaturated fatty acids, and LA metabolism, respectively. The disturbed biofunctions were mainly membrane component and energy source.
Both the IS and HD groups had 16 significant differential metabolites in common, comprising the majority (50%) of lipid and lipid-like molecules. Eight perturbed pathways and four biofunctions were identified to alter significantly (Table S2 and Fig. S5D in the Supporting Information).
To examine the differential expression of these 132 metabolites in plasma of the IS, HD, and control subjects, the Mfuzz package36 was run to cluster them. It turned out that eight clusters revealed the distinct formation of significant metabolite features at different expression levels (i.e., clusters 1–8, Table S3 and Figs. S6‒14 in the Supporting Information). For instance, cluster 7 was composed of 15 metabolites, showing a good agreement that the lowest levels were expressed in the control group, and the expressions increased to middle levels in the HD group, while the IS group had nearly the highest expressions. The top five metabolites with large membership values from 0.64 to 0.42 corresponded to (9Z,12Z,15Z)-octadecatrien-1-ol, oleic acid, linoleamide, arachidyl linoleate, and oleamide, respectively.
Assessment of CVD risk with metabolic biomarkers and machine learning
The machine learning methods of LDA, PLS-DA, SVM, GBM and RF were employed to address the issue of discrimination and risk assessment of CVD. First, three crucial metabolites of 1594-pos (palmitic amide), 1698-pos (oleic acid), 138-pos (138th feature in the positive ion mode, unannotated), were identified from the above-mentioned 164 features through recursive feature selection (Fig. 6A). When these three metabolites were individually fed to the five methods, the resulting LDA model showed the best performance (Table S4 in the Supporting Information). However, the corresponding accuracies, sensitivities and F1 scores were just above 0.80 for both the calibration and validation. This model seemed not good enough for the CVD risk assessment. Therefore, an ultimate biomarker panel was constructed, not only including palmitic amide, oleic acid, 138-pos, but also PC and LA that linked the glycerophospholipid metabolism to the biosynthesis of unsaturated fatty acids, and the clinical traits of age, DBIL, IP (Fig. 6B). Once again, this panel was used to obtain another five models (Table 3). The accuracy, sensitivity, specificity, F1 score, and AUC values resulted from the LDA model were all larger than 0.90, which attained to a desire of the risk assessment. The MCC values of the validation and calibration sets were quite similar, both larger than 0.86. With this panel the LDA model was able to discriminate any group of the HD patients, IS patients, and control subjects from the two remaining. Figure 6C depicts the ROC curves, of which all the AUC values were equal or close to 1.00. This indicated a good efficiency in the CVD discrimination.
Machine learning prediction results based on selected metabolic biomarkers for disease differentiation. (A) Recursive feature selection from metabolite. (B) Expression level distributions of eight traits in biomarker panel (metabolites in the first five box plots, clinical traits in the last three box plots) used for machine learning. (C) ROC for risk assessment of CVD using the biomarker panel.
Discussion
The clinical traits of the HD, IS, and control groups showed significant differences in age, IP, AG, and DBIL indices. It is known that aging poses a large risk factor on many physiological processes, leading to an increasing prevalence of CVD. In this study, the average age was around 66 years for the HD patients, 65 years for the IS patients, in contrast to 48.5 years for the control subjects (Fig. S3 in the Supporting Information). IP indirectly participates in the vascular injury process, whereby an increase in the phosphate level in the body accelerates the deposition of calcium and phosphate in the form of hydroxyapatite in blood vessels, leading to the vascular calcification and increasing the risk of CVD. Serum AG is commonly used to measure the types and causes of metabolic acidosis. Acidosis can induce vasodilation, reducing both peripheral resistance and systolic blood pressure, and thus causing a decline of cardiac output. In addition, metabolic acidosis is a common complication of congestive heart failure. DBIL has antioxidant and anti-inflammatory attributes. An alteration in bilirubin concentration within the physiological range may imply the presence of disruption in redox homeostasis and liver impairment within the enrolled participants. This study shows that there was a significant difference between the IS and control groups (Fig. S3 in the Supporting Information). These four traits suggest that there are associations between CVD and the pathological changes involving oxidative stress, inflammatory responses, ion imbalances, and metabolic disturbances. Yet, the pathophysiological mechanisms directly linking them to CVD remain unclear and lack sufficient specificity. Therefore, there is a need to further understand the pathological and physiological mechanisms of CVD, along with the exploration of potential biomarkers for its diagnosis.
The deployment of metabolomic analysis of the HD and IS groups revealed a prevalence of metabolite disturbances in metabolic pathways in CVD patients. The significant differences in the plasma metabolic profiles of CVD patients primarily lie in lipids, such as PC, LPC, LA, and oleic acid. The pathways involved in these metabolites are mainly related to biological membrane and energy metabolisms. As known, the main structural component of biological membrane in various human tissues and cells is glycerophospholipids, which provide the stability, permeability, and fluidity for biological membrane. The fluidity and asymmetry of biological membranes are the basis for effective energy and cellular signal transduction. PC and LPC are intermediates in glycerophospholipid metabolism and serve as the backbone of neural membranes. Compared with the control group, the expression level of PC was significantly decreased in the IS group, whereas the expressions of LPC, LA, oleic acid, eicosadienoic acid and DHA were significantly raised in their individual levels. This might be due to the fact that brain injuries caused by hypoxia, ischemia or trauma activate N-methyl-D-aspartate receptors, resulting in the influx of extracellular calcium ions into cells. This process mediates the activation of phospholipase A2 (PLA2). PLA2 catalyzes the hydrolysis of PC into LPC and free fatty acids of LA, DHA, and oleic acid37,38, which in turn generate free radicals, thus enhancing ischemic injury. LPC has significant biological effects, including inflammatory reactions, increasing oxidative stress, interfering with vascular endothelial function, and damaging endothelial dependent diastolic function. Rather higher levels of LPC in the IS group (see LysoPC in Fig. S12 in the Supporting Information) were consistent with the previous study that there was an excessive presence of LPC in carotid plaques in patients with transient ischemic attack and stroke37. An elevation of LA expression elicits a surge in the oxidized LA metabolites, which spurs the formation of foam cells in macrophages and culminates in endothelial dysfunction that is a pivotal pathophysiological hallmark of IS39. Additionally, LA and DHA can regulate tumor necrosis factor-alpha induced senescence and dysfunction in endothelial cells40. These mechanisms suggest that unsaturated fatty acids and their receptors could indeed act as mediators in activating oxidative stress and inflammation pathways, not only in vascular endothelial cells but also potentially in cardiac tissues of CVD patients, thereby exacerbating disease progression.
LA and oleic acid can cross the blood-brain barrier to provide energy to the brain, which metabolizes about 20% of total energy consumption of the body41. During the ischemia and hypoxia in neuronal cells of brain tissue, the energy deficiency occurs and engenders an inability to maintain normal ion balance in neurons42. To regulate the energy deficiency caused by IS, the brain initiates the degradation of fatty acids through negative feedback mechanisms to maintain the energy balance and preserve the homeostasis of body. The increase in LA and oleic acid in the IS group makes more fatty acids degraded to maintain energy homeostasis43,44.
Taking into consideration the fact that the heart derives approximately 60–80% of energy from the fatty acid metabolism to support both systolic and diastolic functions, the HD group showed a notable rise in the expression levels of choline and oleic acid. This rise may be attributed to the hydrolysis of cardiac PC mediated by PLA2. In the context of short-term ischemia followed by reperfusion, phospholipase D is also activated, which catalyzes the hydrolysis of the phosphodiester bond in PC, leading to the formation of phosphatidic acid and free choline. Interestingly, choline along with other bioactive lipids like phosphatidic acid and lysophosphatidic acid, plays some roles in platelet aggregation, endothelial cell migration, and fibrinogen binding to platelet glycoprotein IIb/IIIa receptors. Choline can be detected in the core region of atherosclerotic plaques45. A significant rise of the expression level of choline in the IS and HD groups is linked to a risk escalation of CVD (C00114 in Fig. 5), with respect to the higher susceptibility to strokes and possibility of their recurrence46,47.
The recommendations for current CVD diagnostics in clinical practice guidelines are predominantly based on clinical symptoms, electrocardiogram testing, ultrasound examination, and computed tomography scans, with subsequent risk assessment of patients. However, early pathological events and metabolic changes have already occurred in the body before the manifestation of clinical symptoms in CVD. Hence, developing an early screening method based on metabolic biomarkers would be beneficial to the CVD risk assessment.
The LDA model achieved from the biomarker panel composed of palmitic amide, oleic acid, 138-pos, PC, LA, age, DBIL, and IP showed a good identification of IS subjects, with the AUC value of 1.00 of ROC curves. Satisfactory outcomes were not only attained in discriminating the HD patients from the control group, but also differentiating between the HD and IS subjects. Such diagnostic models enable the risk assessment and early detection of individuals with CVD before the manifestation of clinical symptoms, thus promoting timely specified medical examination in the cases at high CVD risk.
The primary strength of this study lies in its population-based experiment, comprehensive metabolite detection, and the integration of metabolomic biomarkers and clinical traits for the CVD diagnosis with machine learning methods. The accuracy of the CVD diagnosis by the LDA model obtained from palmitic amide, oleic acid, 138-pos, PC, LA, age, DBIL, and IP was improved at least 10.1% from that only using palmitic amide, oleic acid, and 138-pos metabolites. Moreover, compared to the LDA model with four clinical traits of age, DBIL, IP, and AG, the accuracy enhancement was more than 28.1%. However, there are several limitations that might affect the practical application of the CVD risk assessment model. First, only 57 clinical samples were available. Although this sample size basically met a minimum requirement for three groups when a test power of 75% and effect size of 0.40 (Cohen’s f) were specified at a significance level of 0.05, it is still relatively small and may limit the generalizability of the findings. Additionally, the recruitment process may not have fully excluded participants with confounding factors such as varying ages or other underlying conditions, which could influence the robustness of the results. Second, the untargeted metabolomic analysis led to a number of differential metabolites that predominantly reflect the changes shortly occurring around the onset of CVD. Metabolites pertaining to specific CVD subtypes might not have been detected due to low abundance, and thus necessitating a consideration or integration of specific techniques for targeted sample enrichment or detection. To address these issues and corroborate the likelihood of the present findings in the clinical use, the future study will engage in the following aspects: (i) increase the sample size and implement more stringent inclusion and exclusion criteria to minimize confounding variables such as age differences and comorbid conditions; and (ii) validate and upgrade the CVD risk assessment model in a quantitative form by using the above pivotal metabolite and clinical trait biomarkers and assessing the accuracy of CVD diagnosis. This is not only to confirm the relevance and reliability of these biomarkers for CVD identification, but also help screen high-risk patients in a timely and appropriate means.
Conclusions
This study has deployed a population-based experiment and untargeted metabolomic analysis of CVD for risk assessment. Five pivotal metabolites of palmitic amide, oleic acid, 138-pos, PC, and LA were identified in CVD patients as potential biomarkers, specifically those involved in phospholipid metabolism and biosynthesis of unsaturated fatty acids. These candidate biomarkers are closely related to oxidative stress and inflammation in the body. Their corresponding dysregulation and metabolic pathways provided a deep insight into the pathogenesis of CVD from a metabolic perspective. Using these biomarkers and three clinical traits of age, DBIL, IP, the risk of CVD was accurately evaluated, and both IS and HD patients were identified in an attempt to screen high-risk patients at an early stage, facilitating timely and appropriate clinical examination.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Roth, G. A. et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: Update from the GBD 2019 study. J. Am. Coll. Cardiol. 76, 2982–3021 (2020).
Feigin, V. L. et al. Global, regional, and national burden of stroke and its risk factors, 1990–2019: A systematic analysis for the global burden of disease study 2019. Lancet Neurol. 20, 795–820 (2021).
Tsao, C. W. et al. Heart disease and stroke statistics—2023 update: A report from the American heart association. Circulation. 147, e93–e621 (2023).
Jensen, R. V., Hjortbak, M. V. & Bøtker, H. E. Ischemic heart disease: An update. Semin Nucl. Med. 50, 195–207 (2020).
Di Biase, L., Bonura, A., Caminiti, M. L., Pecoraro, P. M. & Di Lazzaro, V. Neurophysiology tools to lower the stroke onset to treatment time during the golden hour: Microwaves, bioelectrical impedance and near infrared spectroscopy. Ann. Med. 54, 2658–2671 (2022).
Xu, M., Liu, P. P. & Li, H. Innate immune signaling and its role in metabolic and cardiovascular diseases. Physiol. Rev. 99, 893–948 (2019).
Cai, J. J., Xu, M., Zhang, X. J. & Li, H. L. Innate immune signaling in nonalcoholic fatty liver disease and cardiovascular diseases. Annu. Rev. Pathol. 14, 153–184 (2019).
Piccirillo, F. et al. Changes of the coronary arteries and cardiac microvasculature with aging: Implications for translational research and clinical practice. Mech. Ageing Dev. 184, 111161 (2019).
Severino, P. et al. Ischemic heart disease pathophysiology paradigms overview: From plaque activation to microvascular dysfunction. Int. J. Mol. Sci. 21, 8118 (2020).
Doran, S. et al. Multi-omics approaches for revealing the complexity of cardiovascular disease. Brief. Bioinform. 22, bbab061 (2021).
Fangma, Y. J., Liu, M. T., Liao, J., Chen, Z. & Zheng, Y. R. Dissecting the brain with spatially resolved multi-omics. J. Pharm. Anal. 13, 694–710 (2023).
Pulit, S. L. et al. Loci associated with ischaemic stroke and its subtypes (SiGN): A genome-wide association study. Lancet Neurol. 15, 174–184 (2016).
Holliday, E. G. et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat. Genet. 44, 1147–1151 (2012).
Traylor, M. et al. Genetic basis of lacunar stroke: A pooled analysis of individual patient data and genome-wide association studies. Lancet Neurol. 20, 351–361 (2021).
Lind, L. et al. Large-scale plasma protein profiling of incident myocardial infarction, ischemic stroke, and heart failure. J. Am. Heart Assoc. 10, e023330 (2021).
Chen, Y., Li, E. M. & Xu, L. Y. Guide to metabolomics analysis: A bioinformatics workflow. Metabolites. 12, 357 (2022).
Harshfield, E. L. et al. Metabolomic profiling in small vessel disease identifies multiple associations with disease severity. Brain. 145, 2461–2471 (2022).
Razavi, A. C. et al. Novel findings from a metabolomics study of left ventricular diastolic function: The Bogalusa Heart Study. J. Am. Heart Assoc. 9, e015118 (2020).
Borges, M. C. et al. Circulating fatty acids and risk of coronary heart disease and stroke: Individual participant data meta-analysis in up to 16 126 participants. J. Am. Heart Assoc. 9, e013131 (2020).
Pezzatti, J. et al. Implementation of liquid chromatography-high resolution mass spectrometry methods for untargeted metabolomic analyses of biological samples: A tutorial. Anal. Chim. Acta. 1105, 28–44 (2020).
Tabrez, S., Shait Mohammed, M. R., Jabir, N. R. & Khan, M. I. Identification of novel cardiovascular disease associated metabolites using untargeted metabolomics. Biol. Chem. 402, 749–757 (2021).
Zhang, J. et al. Identification of biomarkers for risk assessment of arsenicosis based on untargeted metabolomics and machine learning algorithms. Sci. Total Environ. 870, 161861 (2023).
Dührkop, K. et al. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods. 16, 299–302 (2019).
Sumner, L. W. A. et al. Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics. 3, 211–221 (2007).
Kanehisa, M. et al. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Wishart, D. S. et al. HMDB 5.0: The human metabolome database for 2022. Nucleic Acids Res. 50, D622–D631 (2022).
Hastings, J. et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 44, D1214–D1219 (2016).
Kim, S. et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109 (2019).
Pang, Z. Q. et al. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761 (2022).
López-Ibáñez, J., Pazos, F. & Chagoyen, M. MBROLE 2.0-functional enrichment of chemical compounds. Nucleic Acids Res. 44, W201–W204 (2016).
Zhang, J., Cui, X. Y., Cai, W. S. & Shao, X. G. A variable importance criterion for variable selection in near-infrared spectral analysis. Sci. China Chem. 62, 271–279 (2019).
Ward, A. et al. Machine learning and atherosclerotic cardiovascular disease risk prediction in a multi-ethnic population. NPJ Digit. Med. 3, 125 (2020).
Tsugawa, H. et al. Hydrogen rearrangement rules: Computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal. Chem. 88, 7946–7958 (2016).
mzCloud. https://www.mzcloud.org (accessed on December 8 2022).
Ruttkies, C., Schymanski, E. L., Wolf, S., Hollender, J. & Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 8, 1–16 (2016).
Kumar, L. & Futschik, M. E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation. 2, 5–7 (2007).
Mannheim, D. et al. Enhanced expression of Lp-PLA2 and lysophosphatidylcholine in symptomatic carotid atherosclerotic plaques. Stroke. 39, 1448–1455 (2008).
Belayev, L., Khoutorova, L., Atkins, K. D. & Bazan, N. G. Robust docosahexaenoic acid-mediated neuroprotection in a rat model of transient, focal cerebral ischemia. Stroke. 40, 3121–3126 (2009).
Wang, L. et al. Triglyceride-rich lipoprotein lipolysis releases neutral and oxidized FFAs that induce endothelial cell inflammation. J. Lipid Res. 50, 204–213 (2009).
Toborek, M. et al. Linoleic acid and TNF-alpha cross-amplify oxidative injury and dysfunction of endothelial cells. J. Lipid Res. 37, 123–135 (1996).
Ebert, D., Haller, R. G. & Walton, M. E. Energy contribution of octanoate to intact rat brain metabolism measured by 13 C nuclear magnetic resonance spectroscopy. J. Neurosci. 23, 5928–5935 (2003).
Murphy, T. H., Li, P., Betts, K. & Liu, R. Two-photon imaging of stroke onset in vivo reveals that NMDA-receptor independent ischemic depolarization is the major cause of rapid reversible damage to dendrites and spines. J. Neurosci. 28, 1756–1772 (2008).
Wang, X. et al. Changes of metabolites in acute ischemic stroke and its subtypes. Front. Neurosci. 14, 580929 (2021).
Schwartz, M. W., Woods, S. C., Porte, D. Jr, Seeley, R. J. & Baskin, D. G. Central nervous system control of food intake. Nature. 404, 661–671 (2000).
Rother, E. et al. Subtype-selective antagonists of lysophosphatidic acid receptors inhibit platelet activation triggered by the lipid core of atherosclerotic plaques. Circulation. 108, 741–747 (2003).
Guasch-Ferré, M. et al. Plasma metabolites from choline pathway and risk of cardiovascular disease in the PREDIMED (prevention with mediterranean diet) study. J. Am. Heart Assoc. 6, e006524 (2017).
Haghikia, A. et al. Gut microbiota–dependent trimethylamine N-oxide predicts risk of cardiovascular events in patients with stroke and is related to proinflammatory monocytes. Arterioscl Throm Vas. 38, 2225–2235 (2018).
Acknowledgements
The authors acknowledged the Affiliated Hospital of Guizhou Medical University for specimen collection and data acquirement from the participants in hospital.
Institutional review board statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Guizhou Medical University (Approval number: 202006).
Funding
This research was funded by the National Natural Science Foundation of China (Grant nos. 82360700, 22004022 and 21864008), the Guizhou Provincial Science and Technology Projects (Grant nos. ZK[2021]045 and [2018]1130), the Excellent Young Talents Plan of Guizhou Medical University ([2021]104).
Author information
Authors and Affiliations
Contributions
X. Z.: Data curation, Formal analysis, Writing—original draft. X. S. and H. Z.: Formal analysis. F. X.: Writing—review & editing. B. L.: Conceptualization, Supervision, Writing—review & editing, Funding acquisition. J. Z.: Resources, Methodology, Writing—review & editing, Supervision, Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, X., Sun, X., Zhao, H. et al. Biomarker identification and risk assessment of cardiovascular disease based on untargeted metabolomics and machine learning. Sci Rep 14, 25755 (2024). https://doi.org/10.1038/s41598-024-77352-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-77352-3
Keywords
This article is cited by
-
Glycerophospholipid and Sphingosine- 1-phosphate Metabolism in Cardiovascular Disease: Mechanisms and Therapeutic Potential
Journal of Cardiovascular Translational Research (2025)