Abstract
Pancreatic ductal adenocarcinoma (PDAC) has high mortality and rising incidence rates. Recent data indicate that the gut microbiome and associated metabolites may play a role in the development of PDAC. To complement and inform observational studies, we investigated associations of genetically predicted abundances of individual gut bacteria and genetically predicted circulating concentrations of microbiome-associated metabolites with PDAC using Mendelian randomisation (MR). Gut microbiome-associated metabolites were identified through a comprehensive search of Pubmed, Exposome Explorer and Human Metabolome Database. Single Nucleotide Polymorphisms (SNPs) associated by Genome-Wide Association Studies (GWAS) with circulating levels of 109 of these metabolites were collated from Pubmed and the GWAS catalogue. SNPs for 119 taxonomically defined gut genera were selected from a meta-analysis performed by the MiBioGen consortium. Two-sample MR was conducted using GWAS summary statistics from the Pancreatic Cancer Cohort Consortium (PanScan) and the Pancreatic Cancer Case-Control Consortium (PanC4), including a total of 8,769 cases and 7,055 controls. Inverse variance-weighted MR analyses were performed along with sensitivity analyses to assess potential violations of MR assumptions. Nominally significant associations were noted for genetically predicted circulating concentrations of mannitol (odds ratio per standard deviation [ORSD] = 0.97; 95% confidence interval [CI]: 0.95–0.99, p = 0.006), methionine (ORSD= 0.97; 95%CI: 0.94-1.00, p = 0.031), stearic acid (ORSD= 0.93; 95%CI: 0.87–0.99, p = 0.027), carnitine = (ORSD=1.01; 95% CI: 1.00-1.03, p = 0.027), hippuric acid (ORSD= 1.02; 95%CI: 1.00-1.04, p = 0.038) and 3-methylhistidine (ORSD= 1.05; 95%CI: 1.01–1.10, p = 0.02). Two gut microbiome genera were associated with reduced PDAC risk; Clostridium sensu stricto 1 (OR: 0.88; 95%CI: 0.78–0.99, p = 0.027) and Romboutsia (OR: 0.87; 95%CI: 0.80–0.96, p = 0.004). These results, though based only on genetically predicted gut microbiome characteristics and circulating bacteria-related metabolite concentrations, provide evidence for causal associations with pancreatic carcinogenesis.
Similar content being viewed by others
Background
Pancreatic cancers rank 12th in global incidence and are the 7th leading cause of cancer death with over 450,000 deaths in 2022 according to GLOBOCAN estimates1. More than 90% of these tumours present as pancreatic ductal adenocarcinoma (PDAC)2. While most cancers have seen improvements in 5-year survival rates, those for pancreatic tumours remain low at 12%3. This can be attributed to the cancers remaining largely asymptomatic until they reach an advanced stage resulting in only 10-20% of patients presenting with non-advanced or resectable disease4. Environmental and life-style risk factors for PDAC include cigarette smoking, obesity, alcohol use, diabetes, pancreatitis, familial history of pancreatic cancer and stress5. In the last 15 years several susceptibility genetic loci have also been identified through genome-wide association studies (GWAS) or candidate region approaches6,7,8,9.
Emerging evidence suggests that differences in the gut, oral and intratumoural microbiomes play an important role in PDAC development and progression10,11,12. Intestinal microbiota disturbance has been linked with a myriad of conditions ranging from obesity to atherosclerosis as well as tumour development at several anatomical sites13,14,15.
It is challenging to establish a clear causal relationship between gut microbiome ‘dysbiosis’ (i.e., altered composition, diversity and metabolic activity of the gut microbiome with pathogenic relevance) and pancreatic diseases through observational studies due to confounding factors and the potential for reverse causality. One potential mechanism by which the gut microbiome may influence carcinogenesis is through the alteration of circulating metabolite concentrations. Increased circulating and tumoural levels of gut derived lipopolysaccharide (LPS), a pro inflammatory bacterial membrane component, have been reported in a murine PDAC model and are correlated with reduced gut barrier functioning16. Additionally, the concentration of many bacteria-related circulating metabolites has been associated with increased or decreased risk of gastrointestinal (GI) cancers including secondary bile acids17,18,19,20,21, amino acid derivatives22, tryptophan derivatives23 and short-chain fatty acids (SCFAs)24.
Mendelian randomisation (MR) is a genetic epidemiology technique that uses genetic variants as instruments to determine the effect of an exposure on an outcome. As alleles segregate randomly during meiosis, associations with environmental confounding variables are negated. Similarly, because alleles are present before the development of PDAC, there is no possibility of reverse causation25. Therefore, MR is the optimal strategy for assessing the causal relationship between the gut microbiome and related circulating metabolites on PDAC risk.
Materials and methods
Aim
To investigate the effect of the gut microbiome and circulating microbiome-related metabolites on the development of PDAC, we first performed a comprehensive search of the literature and existing databases to identify bacteria-related metabolites. This was followed by an exhaustive search to identify SNPs associated with circulating concentrations of these metabolites. We also leveraged summary statistics from the most comprehensive exploration of genetic influences on the human gut microbiome to date26. We then performed two-sample MR using summary statistics from the studies of the Pancreatic Cancer Cohort Consortium (PanScanI-III) and the Pancreatic Cancer Case-Control Consortium (PanC4)27,28,29,30.
Metabolites data
Data on circulating gut microbiome-related metabolites (amino acids, vitamins and cofactors, fatty acids, carbohydrates, organic acids, hormones, lipids, sterols, bile acids, nucleotides and derivatives of these classes) were obtained through an extensive search in literature through Pubmed (search terms in Supplementary note 1, Additional file 1), and two metabolite databases, namely Exposome Explorer and Human Metabolome Database31,32. Specifically, Exposome Explorer is a manually curated database containing information on biomarkers that may represent risk factors for human disease31, while the Human Metabolome Database is a large database containing molecular and clinical-related data on small molecules and metabolites32. Genetic associations with the selected metabolites were downloaded from GWAS Catalog and published GWAS studies33. Only data from populations of European ancestry were used, and whenever more than one GWAS was available for a given metabolite, the largest study was selected to rely on higher statistical power. The complete list of metabolites under analysis is reported in Supplementary Table 1, Additional file 1).
Microbiome data
Genetic associations between SNPs and microbial taxa were gathered from the MiBioGen Consortium34, which represents the most significant endeavour in investigating host genetics-microbiome associations on a population scale to date. The MiBioGen Consortium comprises data from 25 cohorts for a total of 18,340 participants26. The microbiome data mainly originated from Illumina sequencing of the V4 hyper-variable region of the 16 S rRNA gene. Technical variables related to stool processing and microbial DNA extraction were taken into account (e.g., extraction kit used, mechanical lysis, enzymatic lysis, sequencing technology). The Consortium identified several associations at different taxonomical levels. However, we only focused on summary statistics from genome-wide analyses at the genus level, which was the lowest level of taxonomical resolution and comprised 119 defined genera. The complete list of microbial genera analysed is reported in Supplementary Table 1, Additional file 1.
Instrumental variant (IV) selection
To ensure adherence to the relevance, independence, and exclusion restriction core assumptions of the two-sample MR design, the model reported in Supplementary Fig. 1, Additional file 1 was strictly followed. Briefly, these three assumptions state that all genetic variants used as instrumental variables (IVs) should be associated with the exposure of interest (relevance), but not with risk factors and confounders (independence), and that the IVs should affect the outcome only through the exposure (exclusion restriction). For each exposure trait (metabolites or microbial taxa), SNPs with a p-value of association < 5 × 10−8 were selected. Whenever for this threshold there were no associations (i.e., all bacterial genera and 31 metabolites), a less stringent threshold of < 1 × 10−5 was adopted to gain sufficient IVs to perform sensitivity analyses. The SNPs were then pruned using a linkage disequilibrium (LD) threshold of 0.01 with Ldlink35. Palindromic SNPs (C/G or A/T) whose effect allele frequency ranged between 0.45 and 0.55 were removed. SNPs with a minor allele frequency (MAF) < 0.01 were also removed. To account for potential pleiotropy between the SNPs and the outcome, GWAS Catalog and Phenoscanner were screened for associations with known PDAC risk factors and potential confounders: alcohol drinking, allergy, tobacco consumption, diabetes, diet-related traits (e.g., meat, fruit, and vegetable consumption), pancreatitis, body mass index, and lipidic- and weight-related traits using a p-value threshold of 5 × 10−833,36. SNPs directly associated with PDAC risk were also removed. In addition, each SNP was removed whenever it was in LD with another SNP (r2 > 0.8) associated with one of the above-mentioned traits. The remaining SNPs were considered valid IVs.
Pancreatic cancer GWAS data
Genetic associations between SNPs and PDAC risk were obtained from four GWASs: three from the Pancreatic Cancer Cohort Consortium (PanScan I-III) and one GWAS from Pancreatic Cancer Case-Control Consortium (PanC4). All such studies are available in the Database of Genotypes and Phenotypes (dbGaP) with study accession nos. phs000206.v5.p3 and phs000648.v1.p1; project reference no. 1264427,28,29,30,37. Additional details about the original studies are reported in Supplementary note 2, Additional file 1.
After download, quality control procedures were applied using Plink 1.938 as reported previously39. Briefly, individuals with sex mismatches (genotyped sex Vs self-reported sex), heterozygosity rates higher or lower than three standard deviations from the mean, high levels of cryptic genetic relatedness (pi hat > 0.2), or call rates lower than 90% were eliminated. Genetic variants with a call rate lower than 95%, MAF < 0.5%, and not respecting Hardy-Weinberg equilibrium (p < 1 × 10−6) were removed. Imputation was carried out, separately for each dataset, using the Michigan Imputation Server, with the r1.1 Haplotype Reference Consortium HRC panel40. Imputed SNPs with an info score r2 < 0.7, a quality threshold for the selection of imputed genetic variants, and MAF < 0.5% were removed, and all four studies were merged. The final dataset comprised 7,543,430 SNPs and 15,824 individuals (among which 8,769 PDAC cases). Logistic regression adjusted for age, sex, and the top eight principal components was applied to calculate summary statistics.
Data harmonization
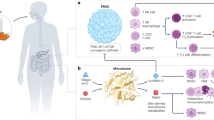
The datasets for each exposure and outcome were aligned and merged. For each IV that was absent from the outcome dataset, a proxy SNP was searched using the 1000 Genomes database (European population) with a r2 > 0.99 to replace the missing one. The resulting datasets were harmonized so that the beta for the association with the exposure was positive and the IVs had the same effect alleles for both the exposure and the outcome. Finally, all betas and standard errors were standardized to the standard deviation scale, to have comparable quantities for the exposure and outcome associations. The complete workflow is reported in Fig. 1.
Statistical analysis
The inverse-variance weighted (IVW) method with multiplicative random effects was used as the reference model for causal estimation. The potential heterogeneity in the estimate was calculated using Cochran’s Q and the I2 statistics41, and weak instrument bias was assessed using the approximated first-stage F statistic42 and applying a debiased IVW estimator that is advocated to be higher than 2043. The weighted median, MR-Egger, Lasso, RAPS, and contamination mixture MR methods were applied as additional sensitivity methods to verify the consistency of the IVW result under more flexible and varying assumptions44,45,46. To identify potential “outlier” SNPs driving the associations, forest plots for single estimates and leave-one-out method calculations were evaluated, together with data from funnel plots, and the PRESSO outlier test which tests for the presence of horizontal pleiotropy47. Finally, to account for both potential pleiotropy and outliers in a single easy-to-interpret model, the radial regression-based approach of the IVW and Egger methods was applied, and the relative radial plots were inspected48. Specifically, the radial methods identify outliers based on their contribution to the total heterogeneity as estimated by the Cochran’s Q and Rucker’s Q statistics for the radial IVW and Egger, respectively48. In this way, we were able to identify possible outliers based on multiple non-redundant tests. A summary is reported within the workflow in Fig. 1.
The estimates obtained by each MR method were converted to odds ratios (ORs). An estimate was considered statistically significant if the IVW method and at least two additional methods among the weighted median, contamination mixture, RAPS, and lasso were statistically significant, conditional to the absence of heterogeneity and pleiotropy.
An adjusted threshold for statistical significance was calculated based on Bonferroni correction, by dividing the nominal threshold of statistical significance by the number of traits analysed: 0.05/(109 + 119) = 2.19 × 10−4.
All statistical analyses were performed using the packages MendelianRandomization (version 0.9.0), MRPRESSO (version 1.0), mr.raps (version 0.2), and RadialMR (version 1.1) in RStudio version 4.3.
In this study, the exposure is the abundance of circulating metabolites and gut microbial taxa at the genus level of taxonomical classification. The outcome is PDAC risk. The workflow reported above shows the criteria for IV selection and the relative thresholds, and the methods applied to estimate the causal effect of each exposure on PDAC risk.
Results
Circulating metabolites
Among the 109 metabolites that were analysed, mannitol, methionine, and stearic acid were estimated as causally related with decreased PDAC risk, whereas carnitine, hippuric acid and 3-methylhistidine were causally associated with increased risk (Table 1). The results of the IVW method for mannitol and methionine (odds ratio per standard deviation [ORSD] = 0.97; 95% CI: 0.95–0.99, p = 0.006) and (ORSD=0.97; 95% CI: 0.94-1.00 p = 0.031 respectively) were supported by all other MR- and non-MR-based sensitivity analyses (Supplementary Figs. 2–3, Additional file 1), and no pleiotropy, outliers and heterogeneity were detected using Egger, radial Egger methods, PRESSO, Cochran’s Q (p > 0.05), and I2. One outlier was identified according to the radial IVW method for mannitol, but after its removal, the results were not significantly different (Supplementary Fig. 4, Additional file 1). Moreover, the potential issue of weak instrumental bias was excluded based on the approximated F statistic, which was 21.2 and 21.3 for mannitol and methionine, respectively, and based on the debiased IVW estimator (66.9 and 93.2, respectively).
Since only two IVs were used as proxy for stearic acid levels, only the debiased and radial IVW methods could be applied as sensitivity analyses (Supplementary Fig. 5, Additional file 1). Both alternative IVW methods supported the result of the standard IVW model (ORSD=0.93; 95%CI: 0.87–0.99, p = 0.027), and no evidence of weak instrument bias (approximated F statistic = 55.7, debiased IVW estimator = 77.4) was observed.
The causal effects of carnitine, hippuric acid and 3-methylhistidine on PDAC risk (ORSD=1.01; 95% CI: 1.00-1.03, p = 0.027, ORSD=1.02; 95% CI: 1.00-1.04, p = 0.038 and ORSD=1.05; 95% CI: 1.01–1.10, p = 0.02 respectively) were supported by most MR methods, and no specific issue related to heterogeneity in the estimates, weak instruments, pleiotropy, or the presence of outliers affecting the results was detected (Supplementary Figs. 6–9, Additional file 1). When considering the Bonferroni threshold, however, none of the reported causal associations between metabolites and PDAC risk remained statistically significant.
Summary statistics for mannitol, methionine, stearic acid, carnitine, hippuric acid and 3-methylhistidine are reported in Supplementary Table 3, Additional file 1.
Microbial taxa
Among the 119 genera analysed, a causal relationship was observed between increased abundance of the Romboutsia genus and decreased risk of PDAC, with an OR of 0.87 (95% CI 0.80–0.96) and p = 0.004 (IVW method). Notably, the approximated first-stage F statistic was 21.2, excluding potential bias due to weak instruments. Moreover, no heterogeneity was detected using Cochran’s Q (p = 0.565) and I2 (0%). The weighted median and contamination mixture methods supported the result, with ORs of 0.85 (95% CI: 0.74–0.97) and 0.80 (95% CI: 0.69–0.89), and p-values of 0.019 and 0.002, respectively. Additionally, both the radial IVW and the debiased IVW confirmed the direction and effect size of the IVW method (Table 2). Moreover, the reliability of the asymptotic approximation of the IVW estimate was checked, yielding a value of 75.65. The radial IVW and leave-one-out forest plots and PRESSO method did not show the presence of influential or outlier IVs (Supplementary Fig. 10, Additional file 1), and neither the Egger nor the radial Egger methods detected pleiotropy (pintercept > 0.05).
Supportive evidence of a causal effect between Clostridium sensu stricto 1 genus and lower PDAC risk was also observed, with an OR of 0.88 (95% CI: 0.78–0.99) and p = 0.027. No heterogeneity was observed based on Cochran’s Q (p = 0.625) and I2 (0%), and no weak instrument bias was suggested (approximated F statistic of 20.3). Lasso and RAPS methods supported the result with similar effect sizes: ORs of 0.88 (95% CI: 0.78–0.99) and 0.87 (95% CI: 0.79–0.97), respectively. The weighted median and contamination mixture approaches did not support the result, however, there were no outliers or influential IVs detected by either forest and funnel plots or the PRESSO method. The standard and radial Egger methods suggested an inverse direction of the effect, even though not statistically significant. The results of scatter, forest, and funnel plots are reported in Supplementary Fig. 11, Additional file 1. However, none of these two causal associations between microbial taxa and PDAC risk remained statistically significant after Bonferroni correction.
Summary statistics for Romboutsia and Clostridium sensu stricto 1 are reported in Supplementary Table 3, Additional file 1.
Discussion
To our knowledge, this MR study is the largest to date to investigate causal associations between gut microbiota abundances using genetic associations reported by the MiBioGen consortium, having leveraged summary statistics from the PanScanI-III and PanC4 studies, including a total of 8,769 PDAC cases and 7,055 controls. Previous studies assessing the impact of gut microbiota abundances on PDAC risk have been conducted using the more modestly sized United Kingdom Biobank (case n = 587) and FinnGen (case n = 1416) GWAS summary statistics49,50. Additionally, we conducted thorough searches of the literature and existing databases to identify bacteria associated metabolites and related SNPs.
Metabolites
Higher circulating mannitol was found to be associated with reduced PDAC risk. Mannitol is a sugar alcohol produced by lactic acid bacteria and used as a sweetener as it does not induce hyperglycaemia due to its slow absorption by the body51,52. Dietary mannitol has been suggested as a prebiotic and has been seen to significantly increase concentrations of gut SCFAs butyrate and propionate in animal models53. There is a paucity of reported associations of circulating mannitol with PDAC risk, however metabolomic analysis of fecal samples from CRC cases and controls revealed mannitol was present exclusively in samples from controls54. A similar analysis performed on CRC tumour and adjacent tissue found mannitol was found exclusively in adjacent mucosa55. Additionally, dietary supplementation of mannitol has been demonstrated to promote lipid metabolism and may prevent obesity in mouse models56. Mechanistically, mannitol may exert indirect effects through stimulation of SCFA production and obesity prevention or through direct anticancer effects such as free radical scavenging and antiproliferative activities57,58.
Many cancer cells including tumour initiating cells are dependent on the essential amino acid methionine for progression in a process known as the Hoffman effect59,60. Further, a methionine restriction diet has been seen to inhibit cancer growth and enhance the efficacy of existing therapies in cell culture and animal models61,62. In contrast, we found that increased circulating methionine was protective against PDAC risk. Methionine is essential for T-cell proliferation and function indicating that methionine may be a double-edged sword63,64. Ming and colleagues found low methionine intake reduces T cell abundance, exacerbates intestinal tumour growth and impairs tumour response to immunotherapy in mice. Low methionine intake was seen to reduce the conversion of methionine to hydrogen sulfide by the gut microbiome, a mechanism critical for immune cell activation and survival65. In a sub-study (n = 31,626), they also found that UK Biobank participants with low protein/methionine intake had significantly higher overall cancer risk compared to participants in the high-intake group65. In vivo, circulating methionine has previously been noted to be higher in healthy volunteers relative to pancreatic cancer patients in two Japanese case-control studies66,67. In experimental studies it was observed that proliferation was reduced and apoptosis increased in pancreatic cancer cells following methionine treatment in vitro68. Methionine intake was also significantly inversely associated with pancreatic cancer risk in a large prospective cohort from Sweden (n = 81,922) and nested case-control samples within prospective cohorts from China (n = 387) and Singapore (n = 162)69,70. Therefore, our result expands the current knowledge of an existing association between circulating methionine levels and PDAC risk to a plausible causal association.
The long-chain saturated fatty acid stearic acid was associated with reduced PDAC risk. This finding should be interpreted with caution as only two IVs were available for our analysis; therefore, sensitivity MR analyses could not be run. Dietary stearic acid has been reported to promote the relative abundance of Akkermansia and Lactobacillus in the gut and bolster gut barrier integrity in mouse models71. Circulating stearic acid has been seen to be lower in PDAC patients relative to controls72,73. PDAC tissue was seen to have lower stearic acid concentration relative to healthy adjacent tissue in two separate cohorts74. Stearic acid has also been seen to significantly induce expression of the TNF-related apoptosis-inducing ligand (TRAIL), trigger apoptosis, and inhibit proliferation in pancreatic cancer cells in vitro74.
Higher carnitine was also found to be associated with an increased PDAC risk. Circulating carnitine has previously been associated with a reduced risk of multiple GI cancers and has established anti-inflammatory and antioxidant activities75,76,77. However, high circulating levels of carnitine have also been reported in patients with colon cancer78. Carnitine plays a significant role in shuttling fatty acids into the mitochondria for fatty acid oxidation, one of the most common ATP-generating processes of PDAC cells79,80. Expression of multiple carnitine transporters is altered in many cancer types, conferring a survival advantage by supplying carnitine essential for fatty acid oxidation81. In the case of PDAC, expression of the carnitine transporter SLC22A5 is associated with tumour progression82. Finally, knockdown of carnitine palmitoyltransferase 1 C (CPT1C), an enzyme that catalyses fatty acid carnitinylation, inhibited the tumourigenesis of PANC-1 cells in vivo and suppressed xenograft tumour growth in situ, illustrating the role of the carnitine system in PDAC83. Taken together, these studies suggest increased circulating carnitine may facilitate increased fatty acid oxidation and consequent cancer cell survival and progression.
Higher concentrations of hippuric acid, a glycine conjugate of benzoic acid, were found to be associated with increased risk of PDAC. Hippuric acid is a microbial–host co-metabolite produced in the liver, following the metabolism of phenylalanine to benzoic acid by the gut microbiome. Levels of circulating hippuric acid rise with the consumption of phenolic compounds (such as fruits and whole grains) and are associated with increased gut bacterial Shannon diversity and improved metabolic health84. However, circulating levels have been seen to be higher in colorectal cancer (CRC), pancreatitis and PDAC relative to controls85,86. Additionally, β-cell proliferation is increased by infusions of hippuric acid in mouse models87. A tissue metabolomic study also revealed an increase in hippuric acid between pancreatic tumour tissues and adjacent pancreatic tissues as well as its utility as prognostic marker in PDAC88. In mouse CRC models, circulatory hippuric acid levels positively associated with tumour weight and the expression of oncogenes, including ROBO3, JAK3 and BEST489. Additionally, at high concentrations, hippuric acid can disrupt the redox balance by inducing mitochondrial ROS production in vitro90,91. Therefore, our result is supported by the current knowledge and adds further evidence to a link between circulating hippuric acid and PDAC risk.
Increase in circulating concentrations of the post-translationally modified amino acid 3-methylhistidine was found to be associated with increased PDAC risk. A potential relationship between 3-methylhistidine and carcinogenesis has not been well characterised. Circulating levels have been observed to be increased in both case-control and prospective studies of prostate cancer but few other associations have been reported92,93.
Overall, considering the relatively small effect sizes, alterations in the individual assessed metabolite concentrations may not be clinically relevant in stratifying the population for PDAC risk. However, future studies may assess their utility as components of multifactorial risk assessments and may lead focused studies to explore the correlation between the metabolome and PDAC risk.
Gut microbiome
Clostridium sensu stricto 1 of the Firmicutes phylum, a genus that includes beneficial and pathogenic strains, was also found to be protective. Previously, Clostridium sensu stricto 1 has been shown to have lower abundance in fecal samples of patients with pancreatic cancer and pre-cancerous lesions relative to healthy and non-alcoholic fatty liver disease controls94. Higher gut abundances of Clostridium sensu stricto 1 have also been associated with a reduced risk of hepatocellular cancer in a study on 142 individuals95.
The butyrate producing Romboutsia genus of the phylum Firmicutes was found to be associated with reduced PDAC risk. Low abundances of Romboutsia have previously been associated with other GI tract cancers. Markedly lower Romboutsia levels were measured in colorectal polyp and tumour tissue relative to healthy colonic tissue96. Fecal abundances of Romboutsia have also been shown to be significantly lower in patients with hepatocellular carcinoma when compared to their healthy first-degree relatives97. Higher abundances of Romboutsia in stool samples have been associated with a decreased risk of esophageal cancer in a previous MR analysis98. Fecal abundances of Romboutsia have also been inversely correlated with serum markers of inflammation TNF-α and IL-1β, and markers of reduced gut barrier integrity D-lactic and diamine oxidase in patients with severe pancreatitis, an established PDAC precursor disease99,100. Members of the Romboustia genus, namely Romboutsia timonensis and Romboutsia ilealis were, respectively, observed to be depleted in the faeces of Spanish and Chinese patients with PDAC relative to controls through shotgun and 16 S sequencing approaches101,102.
There is little in the literature on interactions between Clostridium Sensu Stricto 1, Romboutsia and the reported metabolites. Circulating concentrations of methionine have previously been positively associated with abundances of Romboutsia in rodent models and abundances of Romboutsia and Clostridium were both negatively correlated with urine concentrations of hippuric acid in individuals treated with metformin103,104. However, these associations are tenuous and require functional studies for verification.
To our knowledge, only two studies assessing the contribution of genetically predicted variance in the microbiome or metabolites to pancreatic cancer risk using the PanScan and PanC4 genetic data from dbGaP have been published, both by Zhong and colleagues105,106. They identified 5 metabolites and 1 bacterium, as well as 44 metabolites, to be causally associated with PDAC risk, respectively; however, none of these were replicated in the present study. These discrepancies can likely be attributed to the lower info score of 0.3 used in the previous studies, whereas we included only those with an info score > 0.7, increasing reliability. Additionally, in the first study by Zhong and colleagues, they used the TWAS/FUSION framework based on four methods, specifically, best linear unbiased predictor (BLUP), least absolute shrinkage and selection operator (LASSO), Elastic Net (enet) and top SNPs to develop prediction models to identify associated metabolites and applied these models to the PanScan and PanC4 genetic data. The microbiome data used in their MR analysis was obtained from the Finnish FINRISK study, which comprised 5,959 individuals, making comparisons with the current study challenging105.
This study has several advantages. Firstly, we leveraged the most comprehensive summary statistics available for both gut microbiome populations and pancreatic cancer from the MiBioGen Consortium (18,340 individuals) and PanScan-PC4 studies (8,769 cases and 7055 controls), respectively. Moreover, being an MR-based study, any potential issue of reverse causation and confounding was eliminated. Finally, we have applied the most current MR methods and sensitivity analysis including stringent tests for invalid instruments as well as adhering to STROBE-MR guidelines107.
Our study is not without limitations. As SNPs at the more stringent genome-wide significant threshold 5 × 10−8 were not available for all exposures analysed, we used SNPs at the more relaxed threshold of 1 × 10−5. Our findings rely on genetic data exclusively from individuals of European ancestry. As such, they cannot be generalised to other populations. Additionally, for the gut microbiome taxa analysis, no species-level data are available. We therefore assessed features at a higher rank which may not be as biologically relevant due to the broad range of functions by members within these genera. Finally, many of the assessed circulating microbiome-associated metabolites have sources such as diet and endogenous metabolism. As such it is difficult to assess relative contributions.
In conclusion, we used two-sample MR to assess the impact of gut microbiome characteristics and circulating concentrations of gut microbiota-associated metabolites on PDAC risk. The genera Romboutsia and Clostridium sensu stricto 1 and metabolites mannitol, methionine and stearic acid were estimated to be causally related to decreased PDAC risk, whereas carnitine, hippuric acid, and 3-methylhistidine were implicated as causally related to increased PDAC risk.
Data availability
The datasets supporting the conclusions of this article are available in the PanScan data from the dbGaP repository, (phs000206.v5.p3), https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi? study_id=phs000206.v6.p3, the PanC4 data from the dbGaP repository, (phs000648.v1.p1) https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi? study_id=phs000648.v1.p1 and within the MiBioGen consortium data: https://mibiogen.gcc.rug.nl/menu/main/home/. Data sources for SNPs associated with circulating metabolite concentrations are found in Additional file 1, supplementary Table 1.
Change history
19 December 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41598-024-83618-7
Abbreviations
- CI:
-
Confidence Interval
- CPT1C:
-
Carnitine Palmitoyltransferase 1 C
- CRC:
-
Colorectal Cancer
- dbGaP:
-
Database of Genotypes and Phenotypes
- GI:
-
Gastrointestinal
- GLOBOCAN:
-
Global Cancer Observatory
- GWAS:
-
Genome-Wide Association Study
- HMDB:
-
Human Metabolome Database
- HRC:
-
Haplotype Reference Consortium
- IV:
-
Instrumental Variable
- IVW:
-
Inverse-Variance Weighted
- Lasso:
-
Least Absolute Shrinkage and Selection Operator
- LD:
-
Linkage Disequilibrium
- Ldlink:
-
Linkage Disequilibrium Link
- LPS:
-
Lipopolysaccharide
- MAF:
-
Minor Allele Frequency
- MR:
-
Mendelian Randomization
- OR:
-
Odds Ratio
- ORSD:
-
Odds Ratio per Standard Deviation
- PanC4:
-
Pancreatic Cancer Case-Control Consortium
- PanScan:
-
Pancreatic Cancer Cohort Consortium
- PDAC:
-
Pancreatic Ductal Adenocarcinoma
- PRESSO:
-
Pleiotropy RESidual Sum and Outlier
- RAPS:
-
Robust Adjusted Profile Score
- ROS:
-
Reactive Oxygen Species
- rRNA:
-
Ribosomal RNA
- SCFA:
-
Short-Chain Fatty Acid
- SD:
-
Standard Deviation
- SNP:
-
Single Nucleotide Polymorphism
- STROBE-MR:
-
Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization
- TNF:
-
Tumour Necrosis Factor
- TRAIL:
-
TNF-related Apoptosis-Inducing Ligand
References
Bray, F. et al. Global cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clin. ; n/a.
Hidalgo, M. et al. Addressing the challenges of pancreatic cancer: future directions for improving outcomes. Pancreatology. 15, 8–18 (2015).
American Cancer Society Facts and Fig. (2023). https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2023/2023-cancer-facts-and-figures.pdf
Gillen, S., Schuster, T., Meyer Zum Büschenfelde, C., Friess, H. & Kleeff, J. Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages. PLoS Med.7, e1000267 (2010).
Klein, A. P. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat. Reviews Gastroenterol. Hepatol. 18, 493–502 (2021).
Klein, A. P. et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat. Commun. 9, 556 (2018).
López de Maturana, E. et al. A multilayered post-GWAS assessment on genetic susceptibility to pancreatic cancer. Genome Med. 13, 15 (2021).
Lin, Y. et al. Genome-wide association meta-analysis identifies GP2 gene risk variants for pancreatic cancer. Nat. Commun. 11, 3175 (2020).
Campa, D. et al. The PANcreatic Disease ReseArch (PANDoRA) consortium: ten years’ experience of association studies to understand the genetic architecture of pancreatic cancer. Crit. Rev. Oncol. Hematol. 186, 104020 (2023).
Akshintala, V. S., Talukdar, R., Singh, V. K. & Goggins, M. The gut Microbiome in Pancreatic Disease. Clin. Gastroenterol. Hepatol. 17, 290–295 (2019).
Herremans, K. M. et al. The oral microbiome, pancreatic cancer and human diversity in the age of precision medicine. Microbiome. 10, 93 (2022).
McAllister, F., Khan, M. A. W., Helmink, B. & Wargo, J. A. The Tumor Microbiome in Pancreatic Cancer: Bacteria and Beyond. Cancer Cell. 36, 577–579 (2019).
Aoun, A., Darwish, F. & Hamod, N. The influence of the gut microbiome on obesity in adults and the role of Probiotics, Prebiotics, and Synbiotics for Weight loss. Prev. Nutr. Food Sci. 25, 113–123 (2020).
Duttaroy, A. K. Role of gut microbiota and their metabolites on atherosclerosis, hypertension and human blood platelet function: a review. Nutrients 13 (2021).
Cullin, N., Azevedo Antunes, C., Straussman, R., Stein-Thoeringer, C. K. & Elinav, E. Microbiome and cancer. Cancer Cell. 39, 1317–1341 (2021).
Yin, H. et al. Gut-derived lipopolysaccharide remodels tumoral microenvironment and synergizes with PD-L1 checkpoint blockade via TLR4/MyD88/AKT/NF-κB pathway in pancreatic cancer. Cell Death Dis. 12, 1033 (2021).
Jusakul, A. et al. Identification of biliary bile acids in patients with benign biliary diseases, hepatocellular carcinoma and cholangiocarcinoma. Asian Pac. J. Cancer Prev. 13 Suppl, 77–82 (2012).
Shukla, V. K., Tiwari, S. C. & Roy, S. K. Biliary bile acids in cholelithiasis and carcinoma of the gall bladder. Eur. J. Cancer Prev. 2, 155–160 (1993).
Dai, J. et al. Impact of bile acids on the growth of human cholangiocarcinoma via FXR. J. Hematol. Oncol. 4, 41 (2011).
Wang, W., Yin, X., Li, G., Yi, J. & Wang, J. Expressions of farnesoid X receptor and myeloid cell leukemia sequence 1 protein are associated with poor prognosis in patients with gallbladder cancer. Chin. Med. J. (Engl). 127, 2637–2642 (2014).
Su, H. et al. Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G1245–1253 (2012).
Knudsen, C., Neyrinck, A. M., Lanthier, N. & Delzenne, N. M. Microbiota and nonalcoholic fatty liver disease: promising prospects for clinical interventions? Curr. Opin. Clin. Nutr. Metab. Care. 22, 393–400 (2019).
Badawy, A. A. Tryptophan metabolism and disposition in cancer biology and immunotherapy. Biosci. Rep. 42 (2022).
Alvandi, E., Wong, W. K. M., Joglekar, M. V., Spring, K. J. & Hardikar, A. A. Short-chain fatty acid concentrations in the incidence and risk-stratification of colorectal cancer: a systematic review and meta-analysis. BMC Med. 20, 323 (2022).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Kurilshikov, A. et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165 (2021).
Amundadottir, L. et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 41, 986–990 (2009).
Childs, E. J. et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat. Genet. 47, 911–916 (2015).
Petersen, G. M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 42, 224–228 (2010).
Wolpin, B. M. et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat. Genet. 46, 994–1000 (2014).
Neveu, V., Nicolas, G., Salek, R. M., Wishart, D. S. & Scalbert, A. Exposome-explorer 2.0: an update incorporating candidate dietary biomarkers and dietary associations with cancer risk. Nucleic Acids Res. 48, D908–d912 (2020).
Wishart, D. S. et al. HMDB 5.0: the human metabolome database for 2022. Nucleic Acids Res. 50, D622–d631 (2022).
Sollis, E. et al. The NHGRI-EBI GWAS catalog: knowledgebase and deposition resource. Nucleic Acids Res. 51, D977–d985 (2023).
Wang, J. et al. Meta-analysis of human genome-microbiome association studies: the MiBioGen consortium initiative. Microbiome. 6, 101 (2018).
Machiela, M. J. & Chanock, S. J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 31, 3555–3557 (2015).
Kamat, M. A. et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 35, 4851–4853 (2019).
Sharma, S., Tapper, W. J., Collins, A. & Hamady, Z. Z. R. Predicting Pancreatic Cancer in the UK Biobank Cohort using polygenic risk scores and diabetes Mellitus. Gastroenterology. 162, 1665–1674e1662 (2022).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Gentiluomo, M. et al. Physical activity, sedentary behavior, and pancreatic cancer risk: a mendelian randomization study. J. Endocr. Soc. 8 (2024).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Burgess, S. et al. Guidelines for performing mendelian randomization investigations: update for summer 2023. Wellcome Open. Res. 4, 186 (2019).
Patel, A. et al. MendelianRandomization v0.9.0: updates to an R package for performing mendelian randomization analyses using summarized data. Wellcome Open. Res. 8, 449 (2023).
Ye, T., Shao, J. & Kang, H. Debiased inverse-variance weighted estimator in two-sample summary-data mendelian randomization. Annals Stat. 49, 2079–2100 (2021).
Burgess, S., Small, D. S. & Thompson, S. G. A review of instrumental variable estimators for mendelian randomization. Stat. Methods Med. Res. 26, 2333–2355 (2017).
Slob, E. A. & Burgess, S. A comparison of robust mendelian randomization methods using summary data. Genet. Epidemiol. 44, 313–329 (2020).
Zheng, J. et al. Recent developments in mendelian randomization studies. Curr. Epidemiol. Rep. 4, 330–345 (2017).
Verbanck, M., Chen, C-Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Bowden, J. et al. Improving the visualization, interpretation and analysis of two-sample summary data mendelian randomization via the Radial plot and radial regression. Int. J. Epidemiol. 47, 1264–1278 (2018).
Su, Q. et al. Association between gut microbiota and gastrointestinal cancer: a two-sample bi-directional mendelian randomization study. Front. Microbiol. 14, 1181328 (2023).
Jiang, Z. et al. Causal effect between gut microbiota and pancreatic cancer: a two-sample mendelian randomization study. BMC Cancer. 23, 1091 (2023).
Msomi, N. Z., Erukainure, O. L. & Islam, M. S. Suitability of sugar alcohols as antidiabetic supplements: a review. J. Food Drug Anal. 29, 1–14 (2021).
Wisselink, H. W., Weusthuis, R. A., Eggink, G., Hugenholtz, J. & Grobben, G. J. Mannitol production by lactic acid bacteria: a review. Int. Dairy J. 12, 151–161 (2002).
Maekawa, M. et al. Butyrate and propionate production from D-mannitol in the large intestine of pig and rat. Microb. Ecol. Health Disease. 17, 169–176 (2005).
Yang, Y. et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 9, 4101–4114 (2019).
Brown, D. G. et al. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metabolism. 4, 11 (2016).
Jeon, H-J. et al. D-Mannitol induces a Brown Fat-like phenotype via a β3-Adrenergic receptor-dependent mechanism. Cells. 10, 768 (2021).
Feige, K. et al. Cardioprotective properties of Mannitol—involvement of mitochondrial potassium channels. Int. J. Mol. Sci. 22, 2395 (2021).
Ujlaki, G. et al. Identification of bacterial metabolites modulating breast Cancer cell proliferation and epithelial-mesenchymal transition. Molecules. 28, 5898 (2023).
Kaiser, P. Methionine dependence of cancer. Biomolecules 10 (2020).
Wang, Z. et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 25, 825–837 (2019).
Wanders, D., Hobson, K. & Ji, X. Methionine restriction cancer biology. Nutrients 12 (2020).
Kubota, Y. et al. Synergy of combining methionine restriction and chemotherapy: the disruptive next generation of Cancer Treatment. Cancer Diagn. Progn. 3, 272–281 (2023).
Sinclair, L. V. et al. Antigen receptor control of methionine metabolism in T cells. Elife 8 (2019).
Roy, D. G. et al. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell. Metab. 31, 250–266e259 (2020).
Ji, M. et al. Methionine restriction-induced sulfur deficiency impairs antitumour immunity partially through gut microbiota. Nat. Metab. 5, 1526–1543 (2023).
Kobayashi, T. et al. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol. Biomark. Prev. 22, 571–579 (2013).
Fukutake, N. et al. A novel Multivariate Index for Pancreatic Cancer Detection based on the plasma free amino Acid Profile. PLoS One. 10, e0132223 (2015).
Benavides, M. A. et al. L-Methionine inhibits growth of human pancreatic cancer cells. Anticancer Drugs. 25, 200–203 (2014).
Larsson, S. C., Giovannucci, E. & Wolk, A. Methionine and vitamin B6 intake and risk of pancreatic cancer: a prospective study of Swedish women and men. Gastroenterology. 132, 113–118 (2007).
Huang, J. Y. et al. A prospective evaluation of serum methionine-related metabolites in relation to pancreatic cancer risk in two prospective cohort studies. Int. J. Cancer. 147, 1917–1927 (2020).
Nie, W. et al. Stearic acid prevent alcohol-induced liver damage by regulating the gut microbiota. Food Res. Int. 155, 111095 (2022).
Nishiumi, S. et al. Serum metabolomics as a novel diagnostic approach for pancreatic cancer. Metabolomics. 6, 518–528 (2010).
Tao, L. et al. Metabolomics identifies serum and exosomes metabolite markers of pancreatic cancer. Metabolomics. 15, 86 (2019).
Zhang, G. et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 19, 4983–4993 (2013).
Han, Y., Yoo, H. J., Jee, S. H. & Lee, J. H. High serum levels of L-carnitine and citric acid negatively correlated with alkaline phosphatase are detectable in koreans before gastric cancer onset. Metabolomics. 18, 62 (2022).
Liu, T. et al. The association of serum L-carnitine concentrations with the risk of cancer in Chinese adults with hypertension. Nutrients 14 (2022).
Farahzadi, R. et al. Clinical significance of Carnitine in the treatment of Cancer: from traffic to the regulation. Oxid. Med. Cell. Longev. 2023, 9328344 (2023).
Ni, Y., Xie, G. & Jia, W. Metabonomics of human colorectal cancer: new approaches for early diagnosis and biomarker discovery. J. Proteome Res. 13, 3857–3870 (2014).
Lee, J. S. et al. ATP production relies on fatty acid oxidation rather than glycolysis in pancreatic ductal adenocarcinoma. Cancers (Basel) 12 (2020).
Console, L. et al. Carnitine traffic in cells. Link with Cancer. Front. Cell. Dev. Biol. 8, 583850 (2020).
Juraszek, B. & Nałęcz, K. A. SLC22A5 (OCTN2) carnitine transporter-indispensable for cell metabolism, a Jekyll and Hyde of Human Cancer. Molecules 25 (2019).
Bharadwaj, R., Jaiswal, S., Velarde de la Cruz, E. E. & Thakare, R. P. Targeting solute carrier transporters (SLCs) as a therapeutic target in different cancers. Diseases 12 (2024).
Wang, Y. et al. Carnitine palmitoyltransferase 1 C regulates cancer cell senescence through mitochondria-associated metabolic reprograming. Cell. Death Differ. 25, 735–748 (2018).
Pallister, T. et al. Hippurate as a metabolomic marker of gut microbiome diversity: modulation by diet and relationship to metabolic syndrome. Sci. Rep. 7, 13670 (2017).
Adam, M. G. et al. Identification and validation of a multivariable prediction model based on blood plasma and serum metabolomics for the distinction of chronic pancreatitis subjects from non-pancreas disease control subjects. Gut. 70, 2150–2158 (2021).
Zhu, J. et al. Colorectal Cancer detection using targeted serum metabolic profiling. J. Proteome Res. 13, 4120–4130 (2014).
Brial, F. et al. Human and preclinical studies of the host–gut microbiome co-metabolite hippurate as a marker and mediator of metabolic health. Gut. 70, 2105–2114 (2021).
Liu, C. et al. Tissue metabolomics identified new biomarkers for the diagnosis and prognosis prediction of pancreatic cancer. Front. Oncol. 12, 991051 (2022).
Lo, E. K. K. et al. Mechanistic insights into zearalenone-accelerated colorectal cancer in mice using integrative multi-omics approaches. Comput. Struct. Biotechnol. J. 21, 1785–1796 (2023).
Sun, B. et al. Hippuric Acid promotes renal fibrosis by disrupting redox homeostasis via facilitation of NRF2-KEAP1-CUL3 interactions in chronic kidney disease. Antioxidants (Basel) 9 (2020).
Huang, M. et al. The uremic toxin hippurate promotes endothelial dysfunction via the activation of Drp1-mediated mitochondrial fission. Redox Biol. 16, 303–313 (2018).
Dereziński, P., Klupczynska, A., Sawicki, W., Pałka, J. A. & Kokot, Z. J. Amino acid profiles of serum and urine in search for prostate Cancer biomarkers: a pilot study. Int. J. Med. Sci. 14, 1–12 (2017).
Lécuyer, L. et al. NMR metabolomic profiles associated with long-term risk of prostate cancer. Metabolomics. 17, 32 (2021).
Half, E. et al. Fecal microbiome signatures of pancreatic cancer patients. Sci. Rep. 9, 16801 (2019).
Albhaisi, S. et al. Gut Microbial signature of Hepatocellular Cancer in Men with cirrhosis. Liver Transpl. 27, 629–640 (2021).
Mangifesta, M. et al. Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer. Sci. Rep. 8, 13974 (2018).
Feng, J. et al. Gut microbial signatures of patients with primary hepatocellular carcinoma and their healthy first-degree relatives. J. Appl. Microbiol. 134. (2023).
Gao, X., Wang, Z., Liu, B. & Cheng, Y. Causal association of gut microbiota and esophageal cancer: a mendelian randomization study. Front. Microbiol. 14 (2023).
Jiao, J. et al. Gut microbiota-derived Diaminopimelic Acid promotes the NOD1/RIP2 signaling pathway and plays a key role in the progression of severe Acute Pancreatitis. Front. Cell. Infect. Microbiol. 12, 838340 (2022).
Gandhi, S., de la Fuente, J., Murad, M. H. & Majumder, S. Chronic pancreatitis is a risk factor for pancreatic Cancer, and incidence increases with duration of Disease: a systematic review and Meta-analysis. Clin. Transl Gastroenterol. 13, e00463 (2022).
Kartal, E. et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut. 71, 1359–1372 (2022).
Wang, X. et al. Gut Microbial Profile in patients with pancreatic Cancer. Jundishapur J. Microbiol. 15, e122386 (2022).
Miao, J. et al. Er-Chen Decoction alleviates high-Fat Diet-Induced nonalcoholic fatty liver disease in rats through remodeling gut microbiota and regulating the serum metabolism. Evidence-Based Complement. Altern. Med. 2022, 6221340 (2022).
Lee, Y. et al. Changes in the gut microbiome influence the hypoglycemic effect of metformin through the altered metabolism of branched-chain and nonessential amino acids. Diabetes Res. Clin. Pract. 178, 108985 (2021).
Zhong, H., Liu, S., Zhu, J. & Wu, L. Associations between genetically predicted levels of blood metabolites and pancreatic cancer risk. Int. J. Cancer. 153, 103–110 (2023).
Zhong, H. et al. Elucidating the role of blood metabolites on pancreatic cancer risk using two-sample mendelian randomization analysis. Int. J. Cancer. 154, 852–862 (2024).
Skrivankova, V. W. et al. Strengthening the reporting of Observational studies in Epidemiology using mendelian randomization: the STROBE-MR Statement. Jama. 326, 1614–1621 (2021).
Acknowledgements
We acknowledge the participants and investigators of PanScan, PanC4, MiBioGen studies and consortia. This article is based upon work from COST Action TRANSPAN, CA21116, supported by COST (European Cooperation in Science and Technology).
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.
Funding
This work was partially supported by Fondazione ARPA (www.fondazionearpa.it) and by Fondazione Tizzi (www.fondazionetizzi.it). ND was funded by a UCD College of Science Ad Astra PhD scholarship (R20443; supervisor, DH).
Author information
Authors and Affiliations
Contributions
DH, ND and DC conceived the study. DH, DC, CR and AT supervised the work of ND, RF and FB, and reviewed and approved the submitted version. CC, ALM, PR and MJ identified bacteria associated metabolites. ND drafted the manuscript and collated the instruments. RF and FB performed the analysis and wrote the statistics section of the manuscript. All authors revised and approved the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
All data included in this study were obtained from publicly available studies that are available in controlled access repositories. Each of these studies received approval from the appropriate institutional review board or committee.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error, where the disclaimer was omitted from the text. As a result, it now reads: “Disclaimer: Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.”
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Daniel, N., Farinella, R., Chatziioannou, A.C. et al. Genetically predicted gut bacteria, circulating bacteria-associated metabolites and pancreatic ductal adenocarcinoma: a Mendelian randomisation study. Sci Rep 14, 25144 (2024). https://doi.org/10.1038/s41598-024-77431-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-77431-5