Abstract
Renal clear cell cancer (RCC) is a complex disease that is challenging to predict patient outcomes. Despite improvements with targeted therapy, personalized treatment planning is still needed. Artificial intelligence (AI) can help address this challenge by developing predictive models that accurately forecast patient survival periods. With AI-powered decision support, clinicians can provide patients with tailored treatment plans, enhancing treatment efficacy and quality of life. The study analyzed 267 patients with renal clear cell carcinoma, focusing on 26 who received targeted drug therapy. The data was refined by excluding 8 patients without enhanced CT scans. The research team categorized patients into two groups based on their expected lifespan: Group 1 (over 3 years) and Group 2 (under 3 years). The UPerNet algorithm was used to extract features from CT tumor markers, validating their effectiveness. These features were then used to develop an AI-based predictive model trained on the dataset. The developed AI model demonstrated remarkable accuracy, achieving a rate of 93.66% in Group 1 and 94.14% in Group 2. In conclusion, our study demonstrates the potential of AI technology in predicting the survival time of RCC patients undergoing targeted drug therapy. The established prediction model exhibits high predictive accuracy and stability, serving as a valuable tool for clinicians to facilitate the development of more personalized treatment plans for patients. This study highlights the importance of integrating AI technology in clinical decision-making, enabling patients to receive more effective and targeted treatment plans that enhance their overall quality of life.
Similar content being viewed by others
Introduction
Renal clear cell carcinoma (RCC) is a common and aggressive type of kidney cancer, with multiple treatment options available1,2. Renal cell carcinoma (RCC) is a type of kidney cancer that affects approximately 81,610 people in the United States annually, with a mortality rate of around 14,3903. Globally, RCC is the ninth most common cancer diagnosis and the 12th leading cause of cancer-related deaths4. In the United States, men are more likely to be affected by RCC than women, with a male-to-female ratio of approximately 2:13. Targeted therapy has shown promise in treating RCC, with studies suggesting that around 20–30% of patients respond well to treatment5. Among these, targeted drug therapy has emerged as a key approach due to its high specificity and minimal side effects1,2,6,7,8. Tyrosine kinase inhibitors (TKIs) have emerged as a targeted drug therapy in the treatment of various malignancies, revolutionizing the landscape of cancer therapy through targeted intervention. These agents specifically inhibit the activity of tyrosine kinases, which play a crucial role in the signaling pathways that regulate cell proliferation, differentiation, and survival. The advent of TKIs has led to significant improvements in clinical outcomes for patients with conditions such as chronic myeloid leukemia, non-small cell lung cancer, and gastrointestinal stromal tumors5. However, the response to targeted therapy varies significantly between patients, making it essential to develop personalized treatment plans that maximize therapeutic efficacy9,10,11.
In recent years, artificial intelligence (AI) has made significant strides in the medical field, leveraging its powerful data processing and deep learning capabilities to revolutionize healthcare12,13,14,15,16. AI has shown immense potential in RCC diagnosis and treatment, including assisting doctors in identifying abnormalities through image recognition technology, providing comprehensive diagnostic support through medical knowledge graphs, and tailoring treatment plans to individual patients’ characteristics and medical history17,18,19,20,21.
In RCC treatment, AI is also being increasingly applied. For instance, AI can analyze large patient datasets and genomic information to identify potential drug targets and predict the efficacy and side effects of drugs on patients17,19. Additionally, AI can optimize treatment plans and provide personalized treatment recommendations20,22,23.
This study aims to harness the power of AI to predict the survival time of patients with renal cancer undergoing targeted drug therapy. By leveraging data from the Cancer Imaging Archive (TCIA) database and analyzing clinical data, pathological information, and relevant data on targeted drug therapy for patients with renal cancer, we will develop a predictive model that accurately forecasts the survival time of patients receiving targeted therapy. This model will not only provide a scientific basis for doctors to develop personalized treatment plans but also offer more accurate treatment recommendations for patients, ultimately improving treatment efficacy and quality of life.
Materials and methods
Data collection
We collected data from the Cancer Imaging Archive (TCIA) public database24,25, which provides a large repository of medical images and related clinical data. The research involving human research participants was performed in accordance with the Declaration of Helsinki. The datasets consisted of 267 patients with renal clear cell carcinoma, of which 26 patients received targeted drug therapy. We further excluded 8 patients who did not have enhanced CT scans to ensure the integrity and accuracy of the data.
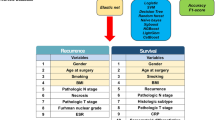
The collected data includes clinical information such as age, gender, and disease duration, as well as pathological information like pathological stage and histological type as showed in Table 1. Additionally, we obtained image data from enhanced CT scans. The flowchart depicting the clinical data collection and modeling process is illustrated in Fig. 1.
AI model
Features derived from the primary tumor were employed. 3D Slicer, Otsu’s thresholding method, and UPerNet were used to extract key features from the images26,27,28. Then, the features from radiomics and Otsu’s thresholding were applied to classify images using support vector machines. Finally, the UPerNet framework, a cutting-edge multi-task model developed by Tete Xiao and specifically designed to tackle complex scene understanding tasks, was utilized. UPerNet is a convolutional neural network (CNN) that captures multi-level information in images by combining different modules such as encoders, decoders, and pyramid pooling modules.The UperNet architecture utilized a unified perception analysis to construct a hierarchical network, enabling the simultaneous resolution of multiple levels of visual abstraction, the learning of distinct patterns in diverse image datasets, and the integration of these insights to facilitate joint reasoning and the discovery of complex visual relationships. By leveraging UPerNet capabilities, the goal is to develop a solution that helps healthcare professionals diagnose small intestinal angiodysplasias more accurately and quickly, ultimately improving patient outcomes and enhancing the diagnostic process29.
During training, UPerNet learns to extract information from heterogeneous annotations, including bounding boxes and semantic segmentation maps. With a large amount of labeled data, UPerNet can learn to recognize different objects, parts, and their textures and materials in images.
In the testing phase, UPerNet can receive a new image and output a semantic segmentation map containing category information for each region in the image.
Statistical methods
3dSlicer (version 5.6.1) was used to extract a comprehensive set of 1075 tumor-specific features. Subsequently, we refined the feature list by removing version-specific and non-essential information, resulting in a final count of 1037 relevant features.
The OpenCV library in Python was utilized to carry out Otsu threshold segmentation, followed by classification of the segmented images using a Support Vector Machine (SVM).
Two metrics was to evaluate the performance of our model: accuracy and Intersection over Union (IoU)30,31.
Accuracy measures the correctness of model predictions, calculated as: accuracy = (number of correctly predicted samples / total number of samples) × 100%.
IoU measures the degree of overlap between the predicted results and true labels, calculated as: IoU = (intersection area between the predicted result and the true label / union area between the predicted result and the true label) × 100%.
Results
In the comparative experiment, it was observed that the classification accuracy (ACC) of the model utilizing 3D Slicer for information extraction and applying it to the support vector machine (SVM) was only 0.33. In contrast, the data processed with the Otsu threshold segmentation method achieved a significantly higher classification accuracy of 0.59. In Fig. 2, the detailed process of establishing the AI model was outlined. The journey began with data collection, during which a comprehensive set of medical images containing tumors, along with their corresponding clinical information, was gathered. This data served as the foundation for training and testing the model.
Next, data preprocessing was conducted, a crucial step that involved the cleaning and enhancement of images to ensure their suitability for analysis. Data standardization was implemented as a comprehensive multi-step process that ensures the accuracy, consistency, and comparability of the data. Crucial steped such as data selection, format conversion, and data labeling were included to guarantee that the data was in a standardized and usable format for analysis and comparison.
After preprocessing, feature extraction was carried out. Advanced image processing techniques and deep learning algorithms were utilized to identify meaningful patterns and characteristics from the images. Features such as the shape, texture, and location of the tumors were formed as the basis for the model’s understanding of tumors.
Once the features were extracted, the model training phase was entered. The labeled data (images with known tumor characteristics and survival outcomes) was fed into the model, and training was conducted to recognize patterns and make predictions. This process involved optimizing the model’s parameters to minimize errors and maximize accuracy.
After training, the model’s performance was evaluated using independent test data. This assessment allowed for the evaluation of its generalization capabilities and the identification of any areas for improvement.
Finally, the model was iterated and refined based on the evaluation results. Adjustments to the network architecture were made, hyperparameters were changed, and additional data was incorporated to enhance the model’s performance. This iterative process continued until satisfactory results were achieved. By following this rigorous model-building process, as depicted in Fig. 2, a robust and accurate AI model was developed that can assist doctors in tumor diagnosis and treatment.
When describing the results in Table 2, specific analyses were conducted on the models, datasets, and corresponding accuracy and IoU ratios presented in the table. The table showed the performance of AI models on two different datasets, Group 1 and Group 2. By comparing the accuracy and IoU ratios of different models on the same dataset, a general understanding of the model performance was obtained.
Group 1: An accuracy rate of 93.66% was achieved by the artificial intelligence model on Group 1, while the intersection-over-union ratio (IoU) reached 89.79%. This indicated that strong classification and localization capabilities were demonstrated by Model A on Group 1, allowing for accurate identification and localization of targets.
Group 2: On Group 2, the accuracy of Model A slightly decreased to 94.14%, while the IoU also decreased to 89.9%. This decrease may suggest that the target or background in Group 1 was more complex than that in Group 2, resulting in a decline in model performance.
The effect of our AI was evident. Tumor segmentation and survival prediction were depicted in Fig. 3. Firstly, the model was recognized for its exceptional ability to segment tumor regions accurately in medical images. Advanced image processing techniques and deep learning algorithms were employed, facilitating the precise differentiation between the boundaries of tumors and adjacent healthy tissues. As a result, essential information, including the tumor’s location, shape, and size, was captured with high accuracy. This step proved to be critical for doctors, as it enriched their understanding of tumor characteristics and provided vital support for subsequent treatment plans.
Secondly, in addition to tumor segmentation, the model was also capable of predicting whether a patient’s survival period would exceed three years. This prediction was based on a thorough analysis of various factors, including tumor characteristics such as size, location, and shape. Through meticulous training and optimization, a high level of prediction accuracy was attained by our model, supplying doctors with invaluable reference information.
The integration of these two functionalities positioned our AI model as a significant asset in the field of tumor diagnosis and treatment. It aided doctors in developing a deeper understanding of tumors and in crafting more targeted and personalized treatment strategies for patients, ultimately enhancing treatment efficacy and improving patient survival rates.
Discussion
The integration of clinical, genomic, and imaging data enables the development of AI-powered targeted drug therapy for RCC, which identifies patient-specific biomarkers and predicts treatment responses13,15,20. By leveraging machine learning algorithms and large datasets, AI models can uncover novel patterns and relationships that may not be apparent to human clinicians, thereby facilitating the development of more precise and effective treatment plans32.
Given the limitation of a relatively small sample size in this study, it was determined that the effects of feature extraction using 3Dslicer and Otsu were suboptimal, whereas the application of the artificial intelligence-based UPerNet model was found to demonstrate significantly better performance in feature extraction. A combination of traditional techniques, including 3D Slicer and Otsu thresholding, was utilized alongside the cutting-edge UPerNet AI model for image analysis. While the results from the traditional methods were less than satisfactory, the outcomes from the innovative UPerNet analysis were exceptionally promising.
Our research focuses on using limited 2D slices from patient CT scans for deep analysis. This approach aims to explore how to effectively utilize medical imaging technology to advance the boundaries of medical diagnosis and treatment evaluation in the context of data scarcity33,34. Our work is inspired by a series of cutting-edge research, such as the three major AI data challenges based on CT and ultrasound35, which not only promote the development of algorithms but also demonstrate the potential of AI in complex medical imaging data analysis. Similarly, we have drawn on research in the field of COVID-19 pneumonia, where newly developed AI algorithms predict the therapeutic effect of favipiravir through quantitative CT texture analysis33, revealing the great value of AI in predicting drug response. In addition, we have also been inspired by research on the use of AI tools to assess multiple myeloma bone marrow infiltration in [18 F]FDG PET/CT35, which demonstrates the broad application prospects of AI in precision medicine.
Our specific research will focus on AI-assisted CT segmentation technology, especially in the validation study of body composition analysis. Although existing studies have shown the high accuracy and reproducibility of AI in CT segmentation29,33,34,36, we hope to further explore how to achieve more accurate assessment of individual body composition through optimizing algorithms and data processing processes in the case of limited patient numbers. This research will not only help to improve the scientific nature of clinical decision-making but may also provide strong support for the development of personalized medical treatment plans.
Concurrently, our research also addresses the application of deep learning in medical image registration, with a particular interest in the potential of non-rigid image registration technology for high-dose rate fractionated cervical cancer brachytherapy36. While this research focus is distinct from our primary objectives in AI predictive modeling for renal cancer patients undergoing targeted therapy, it provides valuable perspectives on harnessing AI technology to tackle intricate challenges in medical image processing.
This study demonstrates the clinical value of AI predictive modeling in personalizing treatment decisions for renal cancer patients undergoing targeted therapy. By analyzing patient data, the AI model was able to identify specific patient subgroups that were more likely to respond well to particular treatments, allowing clinicians to make more informed decisions about therapy selection. This has significant implications for improving patient outcomes, as patients who are most likely to benefit from a particular treatment can receive it earlier and avoid unnecessary exposure to ineffective or toxic therapies. Moreover, the model’s ability to identify patients at high risk of poor outcomes enables early intervention and adjustment of treatment plans, which can lead to better survival rates and improved quality of life. The use of AI predictive modeling in this study highlights its potential to transform the way we approach personalized medicine in oncology, enabling clinicians to deliver more effective and efficient care for patients with renal cancer.
Our study has led to the development of a novel survival prediction model for targeted drug therapy in patients with RCC, leveraging AI to analyze tumor characteristics from CT imaging data. The model integrates a small-scale clinical dataset, CT imaging data, and targeted therapy information to predict patient survival outcomes. Our findings demonstrate exceptional prediction accuracy on the validation set, with accurate forecasting of patient survival outcomes. This finding has significant implications for personalized treatment strategies in RCC patient management, ultimately enhancing patient outcomes and quality of life.
Limitations of the Study.
While this study has yielded promising results, it is not without limitations. Notably, the sample size is relatively small, which may not fully capture the diversity of patients with renal cell carcinoma (RCC). To address this, future research should prioritize expanding the sample size and enhancing the model’s generalizability. Furthermore, this study’s focus on predicting survival outcomes is a crucial first step, but it is equally important to explore the potential of AI technology in optimizing and personalizing treatment plans. By doing so, we can provide more accurate and effective treatment plans tailored to individual patients’ needs. Future studies should aim to integrate AI-driven decision-making into treatment planning, ultimately improving patient outcomes.
The model was trained on a small dataset, which increased the risk of overfitting; it learned the noise and random fluctuations in the data, leading to overly optimistic results that may not generalize to new data. The validation process revealed a drop in predictive accuracy and an increase in error rates on a separate validation set, indicating that the model’s performance might not be as robust in a broader patient population. This will raise concerns about the generalizability of the findings and the applicability of the model to other datasets or patient groups. To further investigate, a larger dataset would be employed, which confirmed the presence of overfitting, as the model’s performance varied across different data subsets.
AI has been leveraged to reconstruct three-dimensional (3D) models from computed tomography (CT) images to personalize surgical treatment of renal cell carcinoma (RCC). Researchers have employed deep learning algorithms to create 3D models from CT images, achieving improved surgical planning and outcome prediction. For instance, deep learning algorithms was used to create 3D models for surgical planning and outcome prediction37. Similarly, convolutional neural networks was used to segment and reconstruct CT images for RCC surgery planning, achieving comparable results38. AI-assisted 3D reconstruction significantly improved surgical accuracy and reduced complications in RCC surgery39. These studies exemplify the potential of AI in enhancing surgical planning and outcome prediction for RCC patients.
This study’s findings and limitations serve as a foundation for future research directions, which can be guided by the following key areas:
-
1.
Expanding the Horizon: Scalability and Generalizability.
Increasing the sample size will enable the model to generalize more accurately to diverse patient populations, thereby enhancing its predictive capabilities and broadening its applicability.
-
2.
Tailoring Treatment: AI-Driven Personalization.
Investigating the potential of AI technology in developing and optimizing treatment plans will allow for the creation of personalized, data-driven treatment strategies that improve patient outcomes and enhance patient-centered care.
-
3.
Navigating Ethical Landscapes: Responsible Adoption and Protection.
Strengthening research in this area will ensure that AI technology applications in the medical field adhere to established ethical norms, laws, and regulations, thereby safeguarding patient confidentiality and trust, and promoting responsible innovation.
Conclusion
In summary, our study successfully developed a survival prediction model for targeted drug therapy in patients with RCC by AI. The model demonstrated its immense potential in the field of medical prediction, offering a promising approach to improving patient outcomes. As we continue to explore the applications of AI in the medical field, future research should focus on developing more accurate and personalized treatment plans for patients. Furthermore, it is essential that we also prioritize ethical and privacy considerations to ensure that AI technology is used in a responsible and compliant manner, aligned with relevant laws, regulations, and ethical norms.
The flowchart shows the whole process from data collection to data preprocessing, model construction, model training and evaluation in detail.
It outlines the AI model development process: data collection, preprocessing to standardize and enhance images, feature extraction using image processing and deep learning, model training with labeled data, performance evaluation using test data, and iterative refinement to optimize accuracy.
Legend: This figure illustrates the dual capabilities of our AI model: tumor segmentation and survival prediction. The AI model precisely segments tumors from CT images, capturing crucial information, and forecasts patient survival beyond three years with high accuracy, positioning it as a promising tool for tumor diagnosis and treatment, enabling more targeted and personalized strategies.
Data availability
The original contributions of this study are included in the article and supplementary material. Further inquiries can be directed to the corresponding authors.We provide the Python source code of AI model training, which is freely available at https://figshare.com/s/7bfbba953a552656e884.
References
Gulati, S., Labaki, C., Karachaliou, G. S., Choueiri, T. K. & Zhang, T. First-line treatments for metastatic clear cell renal cell carcinoma: An ever-enlarging Landscape. Oncologist. 27 (2), 125–134 (2022).
Benamran, D. et al. Treatment options for de novo metastatic clear-cell renal cell carcinoma: current recommendations and future insights. Eur. Urol. Oncol. 5 (1), 125–133 (2022).
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74 (1), 12–49 (2024).
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74 (3), 229–263 (2024).
Motzer, R. J. et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl. J. Med. 373 (19), 1803–1813 (2015).
Boyle, J. J., Pfail, J. L., Lichtbroun, B. J. & Singer, E. A. Adjuvant therapy for renal cell carcinoma: End points, outcomes, and risk assessments. JCO Precis Oncol. 7, e2200407 (2023).
Jin, J. et al. Sunitinib resistance in renal cell carcinoma: From molecular mechanisms to predictive biomarkers. Drug Resist. Updat. 67, 100929 (2023).
Bakouny, Z. et al. Upfront cytoreductive nephrectomy for metastatic renal cell carcinoma treated with immune checkpoint inhibitors or targeted therapy: An observational study from the International Metastatic Renal Cell Carcinoma Database Consortium. Eur. Urol. 83 (2), 145–151 (2023).
Dong, Y., Xu, J., Sun, B., Wang, J. & Wang, Z. MET-targeted therapies and clinical outcomes: A systematic literature review. Mol. Diagn. Ther. 26 (2), 203–227 (2022).
Rotte, A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J. Exp. Clin. Cancer Res. 38 (1), 255 (2019).
Winer, A. G., Motzer, R. J. & Hakimi, A. A. Prognostic biomarkers for response to vascular endothelial growth factor-targeted therapy for renal cell carcinoma. Urol. Clin. North. Am. 43 (1), 95–104 (2016).
Guan, Z. et al. Artificial intelligence in diabetes management: Advancements, opportunities, and challenges. Cell. Rep. Med. 4 (10), 101213 (2023).
Huang, X. et al. Artificial intelligence promotes the diagnosis and screening of diabetic retinopathy. Front. Endocrinol. (Lausanne). 13, 946915 (2022).
Kelly, B. S. et al. Radiology artificial intelligence: A systematic review and evaluation of methods (RAISE). Eur. Radiol. 32 (11), 7998–8007 (2022).
Mann, M., Kumar, C., Zeng, W. F. & Strauss, M. T. Artificial intelligence for proteomics and biomarker discovery. Cell. Syst. 12 (8), 759–770 (2021).
Zhong, F. et al. Artificial intelligence in drug design. Sci. China Life Sci. 61 (10), 1191–1204 (2018).
Barkan, E. et al. Artificial intelligence-based prediction of overall survival in metastatic renal cell carcinoma. Front. Oncol. 13, 1021684 (2023).
Chen, S. et al. Machine learning-based pathomics signature could act as a novel prognostic marker for patients with clear cell renal cell carcinoma. Br. J. Cancer. 126 (5), 771–777 (2022).
Knudsen, J. E., Rich, J. M. & Ma, R. Artificial intelligence in pathomics and genomics of renal cell carcinoma. Urol. Clin. North. Am. 51 (1), 47–62 (2024).
Prelaj, A. et al. Artificial intelligence for predictive biomarker discovery in immuno-oncology: A systematic review. Ann. Oncol. 35 (1), 29–65 (2024).
Raman, A. G., Fisher, D., Yap, F., Oberai, A. & Duddalwar, V. A. Radiomics and artificial intelligence: Renal cell carcinoma. Urol. Clin. North. Am. 51 (1), 35–45 (2024).
Nie, P. et al. A CT-based deep learning radiomics nomogram outperforms the existing prognostic models for outcome prediction in clear cell renal cell carcinoma: A multicenter study. Eur. Radiol. 33 (12), 8858–8868 (2023).
Rallis, K. S. et al. Radiomics for renal cell carcinoma: Predicting outcomes from immunotherapy and targeted therapies-a narrative review. Eur. Urol. Focus. 7 (4), 717–721 (2021).
Akin, O. et al., J. The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma Collection (TCGA-KIRC) Version 3 ed 2016.
Clark, K. et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging. 26 (6), 1045–1057 (2013).
Fedorov, A. et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging. 30 (9), 1323–1341 (2012).
Kurt, B., Nabiyev, V. V. & Turhan, K. A novel automatic suspicious mass regions identification using Havrda & Charvat entropy and Otsu’s N thresholding. Comput. Methods Programs Biomed. 114 (3), 349–360 (2014).
Xiao, T., Liu, Y., Zhou, B., Jiang, Y. & Sun, J. Unified perceptual parsing for scene understanding. ArXiv ; (2018). abs/1807.10221.
Chu, Y. et al. Convolutional neural network-based segmentation network applied to image recognition of angiodysplasias lesion under capsule endoscopy. World J. Gastroenterol. 29 (5), 879–889 (2023).
Madani, A. et al. Artificial intelligence for intraoperative guidance: using semantic segmentation to identify surgical anatomy during laparoscopic cholecystectomy. Ann. Surg. 276 (2), 363–369 (2022).
Vinayahalingam, S. et al. Intra-oral scan segmentation using deep learning. BMC Oral Health. 23 (1), 643 (2023).
Lang, O. et al. Using generative AI to investigate medical imagery models and datasets. EBioMedicine. 102, 105075 (2024).
Ohno, Y. et al. Newly developed artificial intelligence algorithm for COVID-19 pneumonia: utility of quantitative CT texture analysis for prediction of favipiravir treatment effect. Jpn J. Radiol. 40 (8), 800–813 (2022).
Lassau, N. et al. Three artificial intelligence data challenges based on CT and ultrasound. Diagn. Interv Imaging. 102 (11), 669–674 (2021).
Sachpekidis, C. et al. Application of an artificial intelligence-based tool in [(18)F]FDG PET/CT for the assessment of bone marrow involvement in multiple myeloma. Eur. J. Nucl. Med. Mol. Imaging. 50 (12), 3697–3708 (2023).
Salehi, M. et al. Deep learning-based non-rigid image registration for high-dose rate brachytherapy in inter-fraction cervical cancer. J. Digit. Imaging. 36 (2), 574–587 (2023).
Grosso, A. A. et al. 3D virtual model for robot-assisted partial nephrectomy in highly-complex cases (PADUA ⩾ 10). Urologia :3915603241252905. (2024).
Grosso, A. A. et al. Three-dimensional virtual model for robot-assisted partial nephrectomy: A propensity-score matching analysis with a contemporary control group. World J. Urol. 42 (1), 338 (2024).
Grosso, A. A. et al. Robot-assisted partial nephrectomy with 3D preoperative surgical planning: Video presentation of the florentine experience. Int. Braz J. Urol. 47 (6), 1272–1273 (2021).
Acknowledgements
We thanks to TCGA Renal Phenotype Research Group for the open datasets.
Funding
This study was supported by the scientific research of the Heilongjiang. Provincial Health Commission [20210404050337], [20220404050660], and [20220404050752].
Author information
Authors and Affiliations
Contributions
YY and JN drafted the manuscript. YY and JN analyzed the data using AI models. SX and YY drew the pictures. Yaoqi Yu, Jirui Niu and Silong Xia shared the co-first authorship. SS reviewed and revised the manuscript. All the authors have read and approved the final version of the manuscript. All the authors contributed to the manuscript and approved the submitted version.
Corresponding author
Ethics declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Ethics approval and participant consent
The study protocol was reviewed and approved by the ethics board of the Heilongjiang Provincial Hospital. Due to the open datasets, the requirement for signed informed consent from the participants or their guardians was acquired by the TCGA Renal Phenotype Research Group. The requirement of informed consent for studying the open datasets is waived by the ethics board of the Heilongjiang Provincial Hospital.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yu, Y., Niu, J., Yu, Y. et al. AI predictive modeling of survival outcomes for renal cancer patients undergoing targeted therapy. Sci Rep 14, 26156 (2024). https://doi.org/10.1038/s41598-024-77638-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-77638-6
Keywords
This article is cited by
-
Radiology-based artificial intelligence for predicting targeted therapy response in pan-cancer: a comprehensive review
Journal of Translational Medicine (2025)