Abstract
The Developmental Origins of Health and Disease (DOHaD) concept explores the link between exposure to adverse conditions during fetal and early childhood development and the onset of chronic non-communicable diseases, such as prostate cancer (PCa). Changes in epigenetics that control gene expression have been identified as potential contributors to the developmental origin of PCa. Piwi-interacting RNAs (piRNAs), for example, control transposable elements (TEs) and maintain genome integrity in germ cells. However, stress-induced deregulation of TEs warrants investigating the role of piRNAs in the prostate gland from the DOHaD perspective, which remains underexplored. This study aimed to detect and characterize piRNA expression in the ventral prostate (VP) of Sprague Dawley rat offspring at 21 postnatal days (PND21) and PND540. The rats were subjected to maternal protein restriction during pregnancy and lactation to understand its impact on prostate development and aging. Histological analyses showed that the gestational and lactation low-protein diet (GLLP) group experienced a delay in prostate gland development, with increased stromal and epithelial compartments and decreased luminal compartments during early life. Aging in this group resulted in decreased luminal compartments and increased stromal areas. Epithelial atrophy was observed in both groups, with an increased incidence of carcinoma in situ in the GLLP group. Small RNA sequencing from control and restricted groups (at PND21 and PND540) identified piRNA clusters in both young and aged animals. We also detected the expression of PIWI genes (Riwi, Rili, Rili2) in the prostate. Our data highlight the key role of maternal malnutrition in modulating piRNA expression in the offspring’s VP, with the potential to influence prostate developmental biology and the risk of prostatic disorders with aging.
Similar content being viewed by others
Introduction
In the last decades, public health and preventive medicine improvements have contributed to more people reaching old age. Nevertheless, many elderly individuals face chronic illnesses that require specialized care. Identifying adjustable risk factors like unhealthy eating, lack of exercise, smoking, stress, and sleep deprivation can lead to a healthier lifestyle, providing an affordable public health strategy for a better quality of life in older age1,2. Intrauterine and early postnatal experiences can shape health throughout a person’s life. David Barker suggested that prenatal development could affect postnatal health in his “Fetal Origin of Adult Diseases” theory. This idea evolved into the “Developmental Origins of Health and Disease” (DOHaD) framework, which considers pre-conception and early postnatal life as critical periods of susceptibility3,4, which may interfere with health or disease during aging.
One model associated with this concept is Maternal Protein Restriction (MPR), which affects many systems and organs. Epidemiological studies elucidated that maternal malnutrition causes a decrease in birth weight, which is a potential factor in the development of chronic diseases with aging3,4. In this context, the prostate is an accessory gland of the male genital system that can be important for reproduction, however, has been recognized for the high incidence of diseases with aging, mainly benign prostatic hyperplasia and PCa5,6, especially the ventral prostate (VP), which is more responsive to hormones.
Several models with male rats elucidated that exposure to severe MPR leads to PCa oncogenic pathways in early life and aging, demonstrating the potential biomarkers of cancer already at the beginning of life7,8. Experimental studies showed that MPR decreases kidney function and increases blood pressure9, reduces insulin, testosterone, and albumin levels in early life7, and causes prostate cancer (PCa) with aging7,8. This condition highlights the severe adverse effects of MPR on hormone production, reproductive factors, and prostate disorders. Epigenetics is a significant factor related to this illness since it involves the effects of early-life exposure on the occurrence of chronic noncommunicable diseases in offspring and future generations10.
Among these mechanisms, the small non-coding RNAs (sRNAs) are key players in the posttranscriptional regulation processes by targeting transcripts. Specifically, the PIWI-Interecting RNAs (piRNAs) are 24–35 nucleotides long processed from precursor transcripts, the piRNA clusters11. These processed small molecules are loaded into PIWI proteins to form the RNA-induced silencing complex (RISC)12 piRNA-RISC captures and slices transposable element transcripts, which are “jumping-genes”, able to create copies of themselves and insert them elsewhere in the genomes. The action of piRNAs controls the TE mobilization, and that is why it is essential to germinative cells, guaranteeing genome integrity13. Although most studies describe the role of epigenetic mechanisms associated with embryonic development and cancer biology, the relationship between MPR, piRNAs, and PCa yet remains to be investigated.
In addition to germinative cells, piRNA expression has also been observed in somatic tissues, such as the prostate, and in certain types of cancer14,15. In this study, we employed small RNA sequencing (sRNAseq) to investigate the presence of piRNA pathways during early life and aging in prostate rats subjected to MPR. Our objective was to characterize piRNAs in the VP of male Sprague Dawley rats exposed to MPR during pregnancy, lactation, early life, and aging. We confirmed the expression of piRNA sequences in the rat prostate and linked this mechanism to dysregulation associated with aging. These findings provided a foundation for pioneering research into the role of piRNA clusters in the prostate of rats exposed to MPR.
Materials and methods
Animals and experimental design
The detailed experimental design is described by Santos et al.7. Briefly, after the determination of pregnancy on gestational day 1 (GD1), pregnant rats were distributed into two experimental groups (n = 6/group): Control (CTR): dams fed a normal protein diet (17% protein) and gestational and lactational low protein (GLLP): dams fed a low protein diet (LPD) during gestational and lactational periods. Normal and LPD diets were provided by PragSoluções (PragSoluções, SP, Brazil). All diets were isocaloric and normosodic7. The male offspring were euthanized on a postnatal day (PND) 21 (weaning) (n = 12/group) and PND 540 (n = 12/group). The offspring, which were euthanized on PND 540, had free access to a normal protein diet after weaning until the end of the experiment.
The animals were euthanized by an overdose of anesthesia (ketamine/xylazine) followed by decapitation and weighing, and the blood and ventral prostate (VP) were collected and processed by a different analysis as described below. The body weight, VP weight, and hormonal levels were analyzed using a Student t-test, and statistical differences were considered when p < 0.05. Throughout the experiment, housing and use of animals were performed accordingly with the appropriate guidelines and regulations. Efforts were made to minimize suffering and to reduce the animal numbers used in the experiments. The acquisition and description of data followed the recommendations set out in the ARRIVE guidelines.
Morphological analysis
VP samples were collected from the CTR and GLLP groups (n = 06 per group) at PND21 and PND540 and fixed in Methacarn solution (70% methanol + 20% chloroform + 10% acetic acid) for 4 h. They then underwent dehydration in ethanol, clearing in xylene, and inclusion in Paraplast (Sigma). For general morphological analysis, 5 μm histological sections were obtained and stained with Hematoxylin/Eosin. Maternal and offspring weight and size assessments were carried out individually, as already described in other works by the group16,17,18 Histological slides were evaluated for the animals’ prostate cells, stained with hematoxylin–eosin (HE) for an overview of glandular morphology.
Total RNA extraction and sequencing
RNA extraction was performed with Trizol (Ambion, USA), following the manufacturer’s instructions. Four samples from the CTR group and 4 samples from the GLLP group were used for RNA sequencing, at both ages. The samples were quantified using the Nanodrop and had their integrity inferred by the Bioanalyzer, keeping only samples with RIN (RNA Integrity Number) values greater than 8.
An aliquot of unfractionated total RNA was submitted for library construction and sequencing. Ribo-Zero was used during library preparation for rRNA depletion. Purification of small RNAs was carried out using the TruSeq Standard mRNA Sample Preparation Kit (Illumina) and TruSeq Small RNA Library Preparation Kit (Illumina), respectively, and construction of the libraries followed the manufacturer’s specifications. Sequencing was performed with the HiSeq Sequencing System. Macrogen (Seoul-South Korea) processed the entire library preparation and sequencing process, and data was downloaded via an HTTP link. The sequencing data is deposited in the Gene Expression Omnibus (GEO DataSets) under accession code GSE180674 (small non-coding RNAs).
Identification and organization of piRNA clusters in the Rattus norvegicus genome
The reading quality of the reads was accessed using the FastQC software (FastQC: A quality control tool for high throughput sequence data. Version 0.11.9, available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc)19. Then, the sequences were filtered to remove adapters using the software Cutadapt (Version 3.5.0, available online at: https://cutadapt.readthedocs.io/en/stable/guide.html)20, resulting in the selection of reads with sizes between 24 and 35 nucleotides (Supplementary Table S1). Reads shorter than 24 or longer than 35 nucleotides were removed from the libraries. The prediction of piRNA clusters expressed in the sRNA libraries was performed using the program proTRAC (proTRAC: A software for probabilistic piRNA cluster detection, visualization and analysis)21 based on the rat reference genome (Gold standard version: Rattus_norvegicus mRatBN7.2), as well as the annotations of transposable elements obtained by RepeatMasker (Available at: https://www.repeatmasker.org/RepeatMasker/). HTML and FASTA files were generated for each predicted piRNA cluster and are available in Supplementary Data 1. Detailed information for each cluster, including coordinates, location, normalized hit counts, read numbers, and the sequences of individual piRNAs within the clusters, is provided in Supplementary Table S3. Testis samples from 90-day-old rats (Rattus norvegicus) available at GEO (SRP309332) were used as a comparative model for data found in the prostate (Supplementary Table S4). Then, the genomic coordinates of each piRNA cluster predicted in the prostate and testis were compared using the bedtools intersect package22.
Expression of Piwi genes
The primers were designed using the PrimerExpress 3.0.1® software (Life Technologies, USA) based on sequences from NCBI for Piwil1, Piwil2, and Piwil4 (Supplementary Table S5). The appropriate sample and primer concentration tests were carried out. Total RNA from rats was used, extracted from VP samples (n = 6/group) from both experimental groups at PND 21 and 540, using Trizol reagent (Thermo Fisher), according to the manufacturer’s instructions. The samples were treated with the DNase I Amplification Grade kit (Invitrogen Life Technologies, USA) to eliminate any contaminants from the genomic DNA. According to the manufacturer’s instructions, cDNA conversion was performed with the High-Capacity RNA-to-cDNA Kit (Life Technologies) with 2 μg aliquots in 10 μl reaction. The relative quantification of each gene was performed using the 2-∆∆CT method23. Finally, RT-qPCR reactions were performed in duplicates for each target gene on the QuantStudio 12 K flex Real-Time PCR System (Applied Biosystems, USA) in 96 wells per plate and normalized by the ratio between the reference genes (Gapdh and Gusb). After collecting the results, the appropriate statistical analyses were carried out.
Blast of known piRNA sequences against the clusters expressed in the prostate
We used gold-standard reference sequences from the piRBase database (http://bigdata.ibp.ac.cn/piRBase/index.php, last accessed in February 2024—Supplementary Data 2) to compare with the sequences of piRNAs clusters from our VP samples, using the BLAST (Basic Local Alignment Search Tool) bioinformatics tool24 in BLASTN 2.12.0+ 25 (BLAST Nucleotide) mode to identify similarities between nucleotide sequences (Supplementary Table S1).
Statistical analysis
Statistical analyses were performed in the RStudio® development environment (RStudio for Ubuntu version 9.3-2024) and using the ggplot2 package26. The results were subjected to the normalization test (Shapiro–Wilk), the Student’s t-test was applied, and the others were subjected to the Mann–Whitney test. Data were expressed as mean ± Standard Deviation (SD) and differences were considered statistically significant when p < 0.05.
Ethical approval
This study was performed in line with the principles of ethics. Approval was granted by the Biosciences Institute/UNESP Ethics Committee for Animal Experimentation (Protocol #1178).
Results
Development of the prostate of rats subjected to MRP
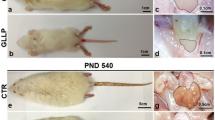
There was a delay in glandular development in the early life (postnatal day PND21), with a stromal and epithelial increase and lumen decrease (Fig. 1A,B) in the gestational and lactational low protein (GLLP) group. At PND540 (Fig. 1C,D), there was a luminal decrease and stromal increase in the GLLP group, and an increased incidence of carcinoma in situ, corroborating our previous data7. The morphological results (lumen size, stromal, and epithelial changes) have already been described in detail in other works by the group7,8.
Representative histological sections of the VP lobes from the CTR and GLLP groups on PND 21 and 540, stained with hematoxylin–eosin (HE). (A,C) Glandular growth in the GLLP group on PND 21 was impaired compared to the CTR. (B,D) At PND 540, the carcinoma in situ was highlighted by the dashed circle. S Stroma, L Lumen, E Epithelium, Scale bar: 500 μm.
Characterization of piRNAs clusters in prostate and comparison to testis data
piRNA clusters are transcriptional active genomic regions that generate the piRNA precursors to be processed in mature piRNA sequences11. Based on the sRNAseq libraries, we predicted the piRNA clusters potentially expressed in prostate. In total, 123 clusters were predicted in prostate, 23 of which were expressed in the four groups. In young animals, 66 piRNA clusters were identified in the CTR group and 61 in the GLLP group. At PND540, 57 clusters were identified in the control group (CTR540), while 53 clusters were identified in the restricted group (GLLP540). Since piRNAs are well-documented in germ cells, we also compared the predicted clusters found in the prostate with those expressed in the testis to confirm that the characteristics of the predicted sRNA sequences align with piRNAs. A total of 20 clusters from testis are shared with the prostate samples (Fig. 2A). Initially, 7 clusters were detected in the 21-day groups, suggesting a direct impact on the animals’ development and growth. In contrast, 8 clusters were common to the 540-day groups, indicating that aging itself influences cluster expression. Additionally, 10 clusters were found exclusively in the GLLP540 group, linking the combined effects of maternal diet and aging. Another 14 clusters were uniquely identified in the four prostate groups, while 9 clusters were shared between both groups and tissues, highlighting essential clusters for prostate function and other key clusters present across all tissues.
Similarly, we compared the distribution size of the piRNAs expressed in the prostate, miRNAs characterized from the same prostate samples27, and piRNAs generated from the testis28 to verify how the predicted sequences overlap with other sRNAs. miRNAs have a greater abundance of sequences of length between 21 and 23 nucleotides, while prostate and testis piRNAs have a greater abundance of sequences of 24 and 27 nucleotides, respectively (Fig. 2B). In short, this comparison showed that there was no overlap between miRNA from the same RNAseq, meaning that the sequences predicted in the cluster have a profile more similar to piRNA. With all this evidence, we can conclude that VP expresses piRNAs.
Impact of MPR and aging on prostate piRNAs
To identify similarities with previously described sequences, the predicted clusters in the prostate were blasted against known piRNAs from the testis, as deposited in piRBase (Fig. 3). A total of 54 piRNAs found in the prostate showed similarities to those in the database. Specifically, 3 known piRNAs (piR-2047424, piR-2122556, and piR-2131909) have similarity with clusters expressed in CTR21, suggesting an essential role during critical developmental periods. Additionally, 21 known piRNAs showed similarity with sequences in both the CTR21 and GLLP21 groups, indicating a role in early life and growth. A unique sequence in the GLLP21 group was similar to piR-140328, which may be related to the impact of diet. The CTR540 group shared one piRNA (piR-2163700) with the GLLP540 group, suggesting a potential link between aging and diet in regulating this specific piRNA. Lastly, 12 known piRNAs presented similarity with sequences expressed across both groups and age ranges, indicating their activity regardless of age or treatment. Additionally, distinct patterns of piRNA expression were observed across different groups. The Venn Diagram (Fig. 3) displays the distribution of piRNAs across the evaluated groups, highlighting both the piRNAs unique to each group and those shared between them. The cluster and chromosome location of all the known piRNAs can be found in Supplementary Table S1.
Differences in piRNA cluster expression between samples
Based on the normalized reads expressed by cluster, we detailed the variation of the expression across the treatments (Fig. 4). For example, some clusters (IDs 59, 35 and 31) are constantly expressed across the conditions, representing the clusters found in the prostate despite the age or treatments. More specifically, cluster 35 is expressed during the early stage without restriction. Additionally, there are clusters specifically expressed in some groups. For instance, several clusters are only expressed in GLLP21 group, representing the early restriction effect, while other clusters are exclusively expressed during the late stages. Lastly, we detected some clusters that are more expressed in GLLP540, depicting the late effect of maternal restriction in the prostate. Overall, the variation in piRNA cluster expression across the samples suggests distinct regulation of piRNAs, potentially reflecting differences in the biological characteristics of the groups. The impact of differential piRNA expression in the prostate is further explored in the following section which provides an overview of putative piRNA targets (all intersections of piRNA clusters in the samples are available in Supplementary Table S6).
Heatmap of cluster expression across aging and under different experimental conditions. We showed only the sequences presenting at least 10 normalized reads of expression per cluster in at least one sample. The expression is shown in log scale, and 0 represents no expression. Each row depicts a piRNA cluster assigned with an ID that can be cross-checked in Table S6. The chromosome where a certain piRNA is located is also represented by the side column.
Composition of piRNA clusters expressed in prostate
Usually, piRNA clusters originate from regions enriched by degenerated TEs. Contradictory, the composition of TEs can indicate the potential target of the piRNA cluster due to the sequence similarity29. Thus, we detailed the composition of piRNA clusters predicted in prostate to investigate their potential targets and TE regulation in this tissue. The rat genome is mainly composed of retrotransposons, such as SINE (45.93%), LTR (26.82%), and LINE (21.27%), while DNA transposons (5.53%) are the least abundant22. Specifically in the prostate, TEs of the order SINE confer half the composition of expressed piRNA clusters, followed by LTR (27.53%) and LINE (13.94%). The least abundant TEs in prostate-expressed clusters were DNA transposons (7.49%), similar to what is found in the entire genome. The piRNA clusters expressed in the testis have the highest composition of LTR (42.37%), followed by SINE (36.16%), LINE (17.51%), and the least abundant, DNA transposons (3.67%). In summary, the general composition of piRNA clusters expressed in the prostate follows the same pattern found in the testis. The differences reflect the number of clusters expressed in each group, and consequently, the abundance of specific TEs within these clusters. This indicates that retrotransposons are potential targets and may be more regulated (Fig. 5). Detailed information on transposable elements is in Supplementary Table S2.
Distribution of TEs present in the piRNA clusters expressed in testis and prostate. The diameter of the circles indicates the abundance of each TE in the piRNA clusters expressed in the samples. The squares highlight a lesser abundance of piRNA derived from retrotransposons L1, Alu, and ERVK expressed in older (PND 540) animals.
Expression of PIWI genes in prostate samples
We also evaluated the expression of the main genes involved in the piRNA biogenesis. For instance, Riwi, Rili, and Rili2 are orthologues of PIWIL1, PIWIL2, and PIWIL4 in humans. Therefore, the expression of PIWI protein genes was evaluated in the prostate from both groups and ages. The expression of these genes was detected in both experimental groups, initially in young animals there was no difference in expression for any of them (Fig. 6A–C). In aged animals, there was an increase in Riwi expression in the restricted group. However, Rili expression decreased in the restricted group. The expression of Rili2 showed no differences in any of the groups. Therefore, these findings confirm the activity of these genes (essentially characterized in germinal tissues) in somatic tissues, in addition to highlighting the impact of aging and diet on the expression of these proteins.
Gene expression of PIWI proteins. (A) Riwi (PIWI1) expression at ages 21 and 540 days. (B) Rili (PIWI2) expression at PND 21 and 540. (C) Rili2 (PIWI4) expression at PND 21 and 540. Fold change is indicated by the lateral vertical bars. The asterisk indicates the statistical difference between samples and was applied to Student’s t-test (p < 0.05). The central bars indicate the standard deviation (SD).
The Fig. 7 summarizes the workflow depicting the effects of maternal exposure to a low-protein diet on the modulation of piRNA cluster expression in the offspring VP. We further demonstrated differences in the VP piRNAs expression profile in young and old male offspring rats. Overall, these data highlighted the involvement of piRNAs in altering prostate developmental biology with long-lasting consequences for prostatic disorders in old male offspring rats.
We analyzed RNAseq data of the ventral prostate to provide new insights into the association of maternal malnutrition with the deregulation of piRNAs in male offspring rats. Moreover, we identified the differential expression profile of the VP piRNAs between young and old offspring. These analyses revealed that both aging and maternal malnutrition altered epigenetic markers involved in prostate developmental biology and aging, contributing to the increased incidence of prostate disorders in old male offspring rats.
Discussion
There is an extensive discussion about how exposure to adverse conditions during the first stages of life can increase the propensity for the development of chronic diseases in offspring in adulthood. Our results demonstrated an increased incidence of carcinoma in situ in the GLLP group, demonstrating the potential of gene expression and epigenetic modulating mechanisms involved. Compiling epigenomic analyses of VP in early life and aging, we found a profile of unique and differentially found piRNA clusters in the prostate of young and old rats exposed to MPR, characterizing piRNAs as potential modulators of prostate carcinogenesis. These results show the epigenetic potential of maternal exposure to nutritional adversities, in addition to providing the characterization of piRNA in prostate tissue, being the first study to highlight the correlation of this pathway between prostate and MPR.
In this context, a global understanding of aspects relating to the DOHaD can provide essential biological and molecular aspects for the discovery of new prognostic and therapeutic targets7,30. These studies corroborate our findings, from biometric data, such as low offspring weight and smaller size visible on the first postnatal day8.
piRNAs are well characterized in the testis, where they regulate transposable elements to preserve genomic integrity. Deactivating piRNA clusters has been shown to cause problems in sperm maturation, even with subtle transcriptome changes14. It is crucial to understand the role of piRNAs in other tissues, including their potential link to diseases like prostate cancer31,32. Similar piRNA clusters found in both testis and prostate confirm that piRNAs are not exclusive to germinal tissue but are also present in other tissues in smaller amounts15. Recent studies have reported piRNAs in various cell types, including the mouse brain, somatic tissues, and human plasma, suggesting that piRNAs have broader functions than previously thought33,34. In a recent study conducted by Ben et al., it was found that the overexpression of a human prostate piRNA called PROPER interacts with the m6A reader protein YTHDF2. This interaction promotes RNA circularization and leads to the degradation of the dual-specificity phosphatase DUSP1. The resulting suppression of DUSP1 translation is associated with enhanced tumor metastasis through the p38 MAPK signaling pathway. These findings underscore the critical role of piRNAs as a key regulator in cancer progression34.
Therefore, our findings highlight the emerging role of piRNAs in somatic tissues, especially in the prostate. The nine clusters commonly found between the prostate and testis indicate that these are specifically essential for regulatory cell functions, as they were not affected independently by diet or aging in the prostate and are present in germinal tissues (testis). This finding correlates to the following, where sequence sizes were compared. Both prostate and testis overlapped in the analysis, which in addition to differing in miRNAs also confirmed that they were piRNAs being expressed. Finally, about reads in the association of a prostate CTR 21 cluster in common with the testis, the number of reads was statistically higher in the prostate, again emphasizing that the role of piRNAs is essentially characterized in gonads but is differentially regulated in somatic cells.
From the perspective of the role of piRNAs in aging, the impacts on the body are demonstrated, in addition to being one of the biggest risk factors for the development of diseases such as PCa35,36. From the perspective of the DOHaD concept, it was demonstrated that there is a decrease in the quantities of piRNA clusters in 540-day-old animals37. These results highlight the impact of aging when correlating the expressed piRNA clusters, where the young control group presents 66 clusters and 58 clusters in aging. Furthermore, when comparing the CTR 21 sample with the GLLP 540 we have an even greater variation of 66 to 53. Therefore, not only aging itself, but the restriction environment was able to modulate this expression. Regarding transposable elements, the correlation with piRNAs favors a deeper understanding of the processes that control development, cell differentiation, and the maintenance of genomic integrity. In the context of diseases, especially cancer, where genomic instability is a common feature, understanding the regulation of transposable elements and piRNAs can lead to possible therapeutic approaches36.
When we analyzed piRNA clusters in the testis, LTR elements, especially LTR/ERVK, were the most abundant, playing a crucial role in cell differentiation and sperm formation. We found more SINE elements in the prostate clusters, mainly SINE/Alu, known for regulating gene expression and genome stability37,38. In the GLLP 540 group, fewer SINE/Alu-derived piRNA clusters are expressed, suggesting that aging and environment impact these piRNAs and potentially their targets (TEs), possibly contributing to tumorigenesis, as carcinoma in situ was observed in these animals. On the contrary, young animals’ prostates have more piRNA clusters derived from SINE/Alu and LINE/L1 elements than the GLLP 540 group, with overall fewer piRNA clusters in the latter, reflecting their role in maintaining TEs. Decreased piRNA in the testis leads to retrotransposon hypomethylation, mainly LINE-1, affecting the expression or regulation of other genes in the genome, resulting in germ cell tumors due to effects on genomic stability. Not only in piRNAs expressed in the testis but also in the genome as a whole, SINE-Alu is the most abundant TEs, which suggests an impact of this element in situations of reactivation38.
The results obtained from RT-qPCR are associated with similar studies in the literature, demonstrating an interface between Prostate-piRNA-DOHaD. In recent studies, Zhang et al. (2020) investigated the role of piRNA sequences in human prostate cancer, revealing that these RNAs reduce the expression of the tumor suppressor protein PCDH9. This suppression leads to increased activity in the AKT signaling pathway, which promotes tumor growth and metastasis15. The research further demonstrated that inhibiting these piRNAs significantly curbs cancer cell growth in both laboratory experiments and animal models39. Furthermore, our results demonstrated that maternal protein restriction combined with aging led to the identification of 10 unique clusters in the GLLP 540 group. These findings highlight the potential of piRNAs in understanding the developmental origins of diseases, including cancer. In other organisms, such as arthropods, using mutated silkworm (Bombyx mori) with loss of function of Siwi (homolog to PIWI), there was a delay in larval growth, and they were unable to become adult moths40. They also exhibited defects in wing development and sexual differentiation. Transcriptome analysis revealed that loss of somatic piRNA biogenesis pathways results in abnormal expression of not only transposons but also host genes, presumably causing severe growth defects39. In particular, the maternal exposure to a toxin called MC-LR (microcystin-leucine arginine) during pregnancy in mice, observed a decrease in the expression of Rili and an increase in Riwi genes, which promoted the activation of the signaling pathway (PI3K/AKT) inducing prostate hyperplasia in the offspring41. In our results, there was also a decrease in Rili expression and an increase in Riwi in aged animals in the restricted group, suggesting not only an effect of aging but one enhanced by maternal protein restriction. This supports the idea that early-life factors affect aging (DOHaD). These findings show that the piRNA pathway works in the prostate and suggest that piRNAs might help detect cancer early and offer new treatment options, improving patient outcomes42. The PI3K/AKT pathway, which controls cell growth and survival, links piRNAs to cancer development41.
In conclusion, for the first time, we presented the piRNA expression profile in rat VP within the context of the DOHaD concept. The young rats had more piRNA clusters than old ones, and diet influenced these cluster expressions. Moreover, the number of transposable elements in piRNA clusters varied with age and the dam’s diet. Overall, these data highlighted the key role of maternal malnutrition in altering epigenetic mechanisms involved in prostate developmental biology early in life, with long-lasting consequences for the incidence of prostate disorders in old offspring rats.
Data availability
The data that support the findings of this study are openly available in The Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/gds) on accession number(s) GSE180674 and SRP309332.
References
Chatterji, S., Byles, J., Cutler, D., Seeman, T. & Verdes, E. Health, functioning, and disability in older adults—Present status and future implications. Lancet 385, 9967 (2015).
Newman, C. B. et al. Statin safety and associated adverse events: A scientific statement from the American Heart Association. Arterioscler. Thromb. Vasc. Biol. 39, e38-81 (2019).
Barker, D. J., Osmond, C., Golding, J., Kuh, D. & Wadsworth, M. E. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ 298, 564–567 (1989).
Wadhwa, P. D., Buss, C., Entringer, S. & Swanson, J. M. Developmental origins of health and disease: Brief history of the approach and current focus on epigenetic mechanisms. Semin. Reprod. Med. 27(5), 358–368 (2009).
Verze, P., Cai, T. & Lorenzetti, S. The role of the prostate in male fertility, health and disease. Nat. Rev. Urol. 13(7), 379–386 (2016).
Sfanos, K. S., Yegnasubramanian, S., Nelson, W. G. & De Marzo, A. M. The inflammatory microenvironment and microbiome in prostate cancer development. Nat. Rev. Urol. 15(1), 11–24 (2018).
Santos, S. A. A. et al. Maternal low-protein diet impairs prostate growth in young rat offspring and induces prostate carcinogenesis with aging. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 74, 751–759 (2019).
Colombelli, K. T. et al. Impairment of microvascular angiogenesis is associated with delay in prostatic development in rat offspring of maternal protein malnutrition. Gen. Compar. Endocrinol. 246, 258–269 (2017).
Rizzi, V., Sene, L., Fernandez, C., Gontijo, J. & Boer, P. Impact of long-term high-fat diet intake on kidney morphology and function in gestational protein-restricted offspring. J. Dev. Origins Health Dis. 8, 89–100 (2017).
Burton, G., Fowden, A. & Thornburg, K. Placental origins of chronic disease. Physiol. Rev. 96, 1509–1565 (2016).
Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 20(2), 89–108 (2019).
Wu, X. et al. The biogenesis and functions of piRNAs in human diseases. Mol. Ther. Nucleic Acids 21, 108–120 (2020).
Iwasaki, Y. W., Siomi, M. C. & Siomi, H. PIWI-interacting RNA: Its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433 (2015).
Liu, Y. et al. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 18, 1–15 (2019).
Zhang, L., Zheng, X., Zhang, L., Zhang, Z. & Wang, Z. piR-001773 and piR-017184 promote prostate cancer progression by interacting with PCDH9. Cell. Signal. 76, 109780 (2020).
Camargo, A. C. L. L. et al. Streptozotocin-induced maternal hyperglycemia increases the expression of antioxidant enzymes and mast cell number in offspring rat ventral prostate. Anat. Rec. 300, 291–299 (2017).
Santos, S. A. A. et al. Identification of potential molecular pathways involved in prostate carcinogenesis in offspring exposed to maternal malnutrition. Aging 12, 19954–19978 (2020).
Portela, L. et al. Increased oxidative stress and cancer biomarkers in the ventral prostate of older rats submitted to maternal malnutrition. Mol. Cell. Endocrinol. 523, 1 (2021).
Wingett, S. & Andrews, S. FastQ screen: A tool for multi-genome mapping and quality control. F1000Research 7, 1338 (2018).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Rosenkranz, D. & Zischler, H. Protract—A software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinform. 13, 1–10 (2012).
Quinlan, A. R. & Hall, I. M. M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Zhang, Z., Schwartz, S., Wagner, L. & Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214 (2000).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Portela, L. M. et al. Early-life origin of prostate cancer through deregulation of miR-206 networks in maternally malnourished offspring rats. Sci. Rep. 13, 18685 (2023).
Hu, L. et al. MicroRNA regulation of the proliferation and apoptosis of Leydig cells in diabetes. Mol. Med. 27, 1–14 (2021).
Fueyo, R., Judd, J., Feschotte, C. & Wysocka, J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 23, 481–497 (2022).
Langley-Evans, S., Phillips, G. & Jackson, A. In utero exposure to maternal low protein diets induces hypertension in weanling rats, independently of maternal blood pressure changes. Clin. Nutr. 13, 319–324 (1994).
Markert, L. et al. Small RNAs as biomarkers to differentiate benign and malign prostate diseases: An alternative for transrectal punch biopsy of the prostate? PLoS ONE 16, 1–12 (2021).
Qian, L. et al. Piwi-interacting RNAs: A new class of regulator in human breast cancer. Front. Oncol. 11, 2525 (2021).
Chirn, G.-W. et al. Conserved piRNA expression from a distinct set of piRNA cluster loci in eutherian mammals. PLoS Genet. 11, e1005652 (2015).
Ben, S., Cai, Z., Li, Z., Chen, L. & Li, D. piRNA PROPER suppresses DUSP1 translation by targeting N6-methyladenosine-mediated RNA circularization to promote oncogenesis of prostate cancer. Adv. Sci. 11, 2402954 (2024).
Peng, J. C. & Lin, H. Beyond transposons: The epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr. Opin. Cell Biol. 25, 190–194 (2013).
Yu, Y., Xiao, J. & Hann, S. S. The emerging roles of PIWI-interacting RNA in human cancers. Cancer Manag. Res. 1, 5895–5909 (2019).
Noshay, J. M., Anderson, S. N., Zhou, P., Ji, L. & Springer, N. M. Monitoring the interplay between transposable element families and DNA methylation in maize. PLoS Genet. 15, e1008291 (2019).
Grundy, E. E., Diab, N. & Chiappinelli, K. B. Transposable element regulation and expression in cancer. FEBS J. 289, 1160–1179 (2022).
Aquino, A. M. et al. Integrated transcriptome and proteome analysis indicates potential biomarkers of prostate cancer in offspring of pregnant rats exposed to a phthalate mixture during gestation and lactation. Chemosphere 341, 140020 (2023).
Kiuchi, T. et al. Non-gonadal somatic piRNA pathways ensure sexual differentiation, larval growth, and wing development in silkworms. PLoS Genet. 19, e1010912 (2023).
Han, R. et al. piRNA-DQ722010 contributes to prostate hyperplasia of the male offspring mice after the maternal exposed to microcystin-leucine arginine. The Prostate 79, 798–812 (2019).
Hanusek, K. et al. piRNAs and PIWI proteins as diagnostic and prognostic markers of genitourinary cancers. Biomolecules 12, 186 (2022).
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brazil (CAPES)—Finance Code 001; Sao Paulo State Research Foundation FAPESP (2022/04339-1 and 2022/03990-0); Conselho Nacional de Desenvolvimento Científico e Tecnológico (311481/2021-3).
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. B.H.S, P.L.M.F, O.J.I.N, and J.L.A designed the experiment and wrote the manuscript. B.H.S and P.L.M.F performed the data analysis. F.M.N, M.R, R.I.T, L.A.B.L, sample collecting and processing. J.L.A and O.J.I.N supervised all the processes.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Baptista, H.S., Portela, L.M.F., Fioretto, M.N. et al. Influence of aging and maternal protein restriction on PIWI-interacting RNA expression in the offspring rat ventral prostate. Sci Rep 14, 30372 (2024). https://doi.org/10.1038/s41598-024-77901-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-77901-w