Abstract
The success of therapeutic monoclonal antibodies (mAbs) and their biosimilars, highlights the challenge to control their purity, identity, and stability. For this purpose, among several orthogonal methodologies, imaged capillary Iso Electric Focusing (icIEF) is one of the leading techniques. Despite the isoelectric point (pI) being a univocal parameter relying on the protein’s primary sequence and its post-translational modifications (PTMs), the current Charge Variants Profile Assessment (CVPA) carried out by cIEF relies on relative comparisons with corresponding reference standards, this is because the inconsistent outputs for the same sample across different instruments preclude the uniqueness of the measured parameters. We demonstrate that refining the current calibration approach in the iciEF method allows for obtaining more reliable and objective pIs, and a deeper understanding of the pH gradients along the capillaries. We are confident our advancements will enhance CVPA, by exploring the concept of univocal charge identity. This is crucial for constructing biobanks and developing algorithms to quickly identify divergences from the originators, thus ensuring drug quality, efficacy, and safety. Moreover, our method allows an experimental design optimized to minimize bias (“Unbiased” Experimental Design—UED) to study resolution as a multivariate function of different input variables. This endeavor aims to develop optimal methods tailored to specific pH ranges.
Similar content being viewed by others
Introduction
The success of mAbs and their biosimilars has highlighted the importance of product identity as a critical quality attribute1. Due to the higher complexity of mAbs, compared to chemical drugs, analytical characterization is complicated by numerous possible PTMs2 such as glycosylation and deamidation, which can occur during the manufacturing process3,4. These modifications may result in the formation of alternative product variants, potentially affecting drug safety and efficacy5,6,7. Consequently, careful monitoring of the manufacturing process and storage, particularly in the control and evaluation activities carried out by regulatory agencies, is essential for establishing their identity, purity, potency, and stability and is performed through comparative analysis across different batches with reference materials8.
Several complementary and orthogonal techniques are employed to monitor the Critical Quality Attributes (CQA) of BMPs, such as identity (i.e. by mass spectrometry, peptide mapping, surface plasmon resonance, and many others) and purity (i.e. cation exchange chromatography, capillary zone electrophoresis). Capillary isoelectric focusing (cIEF) and the more recently developed icIEF are emerging applications to characterize the charge heterogeneity of mAbs8,9,10 and their biosimilars. Both cIEF and icIEF are currently considered valuable options for studying identity and purity in terms of their charge variants profile. In the context of quality control activities, these methods are used to quickly assess the absence of unexpected peaks, the relative abundance of pI variants, and consequently the aggregation or degradation process in samples.
In cIEF, proteins are separated along a pH gradient, by exploiting their isoelectric points (pIs). A mixture of amphoteric species, known as “carrier ampholytes”, each possessing its own pI and specific buffering capacity, is loaded into the capillary alongside the sample. Upon applying an electric field, each charged ampholyte starts migrating towards the opposite pole, until it enters the buffering zone of a neighboring ampholyte species, resulting in a change in its net charge. In the end, this process shapes a pH gradient. Simultaneously, the sample isoforms start migrating (focusing phase) until they reach their pIs where their mobility is nulled11,12, due to a net charge equal to zero.
Modern devices have been developed to improve the separation and resolution capabilities by utilizing small-coated capillaries, reducing sample wasting, heating, and endosmotic flux13,14. In conventional cIEF, analytes follow two contiguous steps, (i) focusing—where they are separated along the pH gradient shaped by the carrier ampholytes, (ii) mobilization—where the separated analytes are moved towards a detection window by chemical mobilization or applying pressure and analyzed at 280 nm. In the more recent imaged-cIEF (icIEF), the sample is read by imaging the entire capillary, by a CCD detector, without any mobilization phase. Thus, in cIEF basic species are detected first and peak position is expressed as migration time (minutes) while, since in icIEF the electropherogram it’s a direct image of the analytes separation along the capillary, peaks are expressed in units of the capillary length (pixels)1,9. As a result, the electropherograms obtained from cIEF and icIEF are generally mirror images of each other. Due to the more complex procedures involved in conventional cIEF—specifically, the capillary preparation and mobilization phases—these steps can introduce potential sources of discrepancies between the two methods, affecting the consistency and accuracy of the results. Recently, a study carried out by the Official Medicines Control Laboratory (OMCL) network aimed to standardize analytical procedures of these techniques revealed significant inconsistencies in the electropherograms, pIs, and resolution between a conventional cIEF from Sciex PA800+™ and a BioTechne-Protein Simple Maurice™ icIEF1.
Recently, due to its simplicity and enhanced reproducibility15, the icIEF technique has emerged as the predominant choice for evaluating the charge heterogeneity of mAbs in quality assessment by numerous manufacturers and regulatory agencies.
The study of Goyon and colleagues, which examined the charge variants of a panel of 23 FDA- and EMA-approved mAbs using icIEF, found that the pIs of therapeutic mAbs typically fall within the pH range of 6.1–9.416. Therefore, this interval represents a critical range for optimizing flexible yet effective icIEF procedures tailored to mAb-based products.
Currently, there are inconsistencies in the pIs obtained for the same sample by different devices, and there are concerns that the isoelectric point may be perceived as an ambiguous and method-dependent parameter rather than reflecting an objective, intrinsic structural property of a product1. Thus, ensuring that observed pIs remain unaffected by measurement methodological inconsistencies is crucial.
As a result of the highly specific charge heterogeneity, each mAb has its specific fingerprint profile that can affect bioavailability, tissue distribution, and pharmacokinetics and hence safety and efficacy. Thus, it represents itself as a CQA1. Therefore, defining an “univocal identity”, in terms of pIs, would offer several new significant advantages and perspectives, such as a criterion for reproducibility between different laboratories, the adaptation of methods between different devices, as well as the implementation of biobanks and “Generative Pre-trained Transformer” algorithms able to rapidly identify possible alterations in the molecule structure and hence possible divergence from the originator, and in the control of counterfeited biological medicinal products (BMPs).
In this context, we share our experience with the ProteinSimple-Maurice™ apparatus and propose an innovative icIEF approach that overcomes the limitations typically encountered in analyzing therapeutic antibodies.
Our study started by observing biases in the obtained pIs of a mix of commercial and certified pI-markers when run as analytes. We found that the assumption of a linear calibration curve, currently enforced by both icIEF and cIEF ((i)cIEF) analysis software, introduces unpredictable errors in the expected pI-values, across the pH gradient, despite Pearson’s determination coefficient being very close to one. We assessed these biases and proposed a possible solution, employing non-linear regressions to enable recalibration of the data. Consequently, we obtained pI-values remarkably similar to those certified. The reliability of our approach is ultimately supported by the proximity of the main isoform pI value of Infliximab, to its theoretical values, derived from its primary amino acid sequence. Thus, we provide a new relevant improvement of the icIEF method toward achieving a univocal identity.
Finally, we observed that the technique’s resolution power is uneven across the pH range, exhibiting unexpected fluctuations independent from the employed experimental conditions, and we demonstrated the possibility of investigating the resolution power across the entire capillary to identify the optimal focusing conditions and carrier ampholytes combination. This endeavor allows the comprehension of the pH gradient shapes that can be customized for specific purposes. Our approach allows for identifying protocols to achieve the maximal resolution power within a particular pH region relevant to the analyzed mAb.
Results
The suitability of calibration curves
In cIEF establishing an accurate calibration curve is essential. Typically, a mixture of pI-markers is run with the sample, under chosen experimental conditions. The positions of the markers in the electropherogram are identified and correlated with their certified pIs (Fig. 1A). The calibration curve is derived by fitting a model to the data allowing for extrapolation of the pI-values. The obtained pIs are highly reproducible, within and between the experiments (Table S1, Supplementary information).
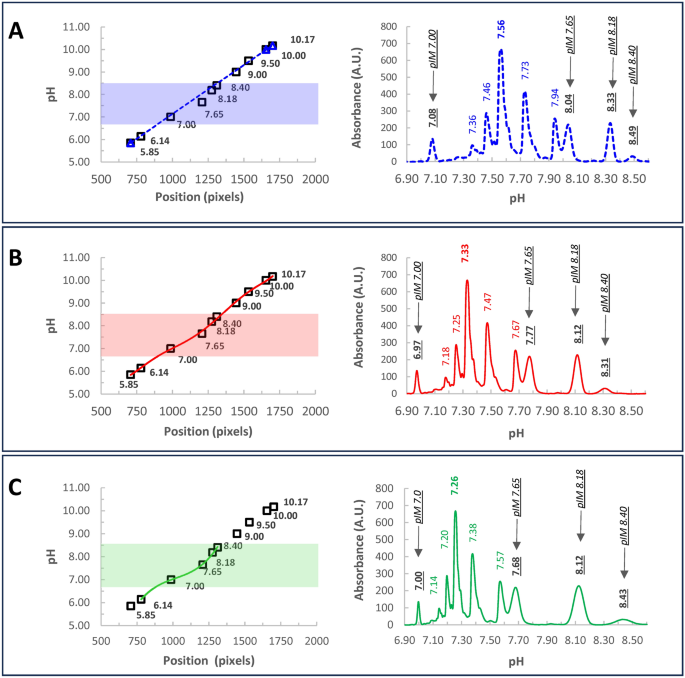
Setting of the calibration curve. (A) typical icIEF electropherogram obtained using ten certified pI-markers (5.85, 6.14, 7.05, 7.6, 8.18, 8.40, 9.0, 9.5, 9.99, 10.17). The sample mix containing pI-markers and ampholytes is loaded in the capillary and undergoes icIEF, sample mix has been run in twice. Subsequently, the whole capillary is imaged vs the absorbance (the reference system is set at the acidic end of the capillary, corresponding to 0). The resulting electropherogram plots the absorbance vs the position along the capillary (pixels). (B) A typical calibration curve, plots each pI-marker certified value vs its respective position, along the capillary. Linear regressions (blue dotted line: by using only 5.85, 9.99, 10.17 external pI-markers; black dotted line: by employing all the pI-markers) or non-linear regression (6th order polynomial regression, red line, all pI-markers) were applied to the scatterplot. Notice, that Pearson’s determination coefficient is always very near to one (R2Blue 0.9999, R2Black 0.9835, R2Red 0.9990), proving a good adaptation to each model. (C) Unfortunately, in linear regressions, significative errors are introduced in the measure, leading to unwanted biases in the estimation of the pI marker values. The average BIAS % has been quantified in the table inset and plotted as a barplot. Error bars are standard deviations. Maximizing the likelihood of the adaptation to the model allows for minimizing the errors and ensuring a good extrapolation of the pI values for each peak.**p value < 1%.
Nowadays, the software of (i)cIEF devices employs a linear model for the calibration curve, likely due to its simplicity, usually relying on two/three external pI-markers to avoid any superimposition17. To verify the suitability of linear fitting in icIEF, we decided to evaluate this adaptation by running ten pI-markers (certified as 5.85, 6.14, 7.05, 7.6, 8.18, 8.40, 9.0, 9.5, 9.99, 10.17) as samples, by using a 1:1—6% 3.0–10.0/8.0–10.5 Ampholytes (Fig. 1). The obtained values for these markers are reported in Fig. 1C (table inset).
The linear regression yielded a Pearson’s determination coefficient (R2 = 0.9999) very close to one, indicating a strong fit to the model. Nevertheless, the non-linearity in the data is evident (Fig. 1B). Despite this behavior is already well-known, for conventional cIEF18, the consequences of this data non-linearity haven’t been described yet. Although the linear regression assumption holds, it inevitably introduces biases in the measured values. To calculate this distortion, we compared expected and measured pI values of the markers through the regression equations (Fig. 1C). Subsequently, the percentage bias magnitude (BIAS%) for each peak was determined by subtracting the certified values from the measured values, and then dividing by the certified values. These results are summarized in Fig. 1C (table inset). The distortion observed is significant in the range of 6.0–8.5 pH (p-value ANOVA 8.30 E-03; linear 3 points vs non-linear 10 points post-hoc test p-value Bonferroni 9.31 E-03). An error of 8%, as in the worst case obtained, implies that if the true value of a mAb is 7.6, the measured value could vary between 6.99 and 8.21. Considering that pH is measured on a logarithmic scale, this corresponds to a 16-fold difference in absolute magnitude.
A regression that accurately represents the data ideally passes through all the points. As depicted in Fig. 1C polynomial regression can better accommodate the complexity of the data, proving to be an effective option (table inset). However, obtaining the most suitable non-linear regression often strikes a balance between fitting the data well and avoiding overfitting. Thus, choosing the most appropriate condition may involve a complex systematic approach, but in the end, mitigates the bias (Supplementary Information). Consequently, this approach significantly enhances the estimation of pI-values for samples.
This conclusion is summarized by the following experiment where Infliximab was run in an icIEF experiment together with ten pI-markers spanning through the entire pH-gradient (Fig. 2). Using a linear calibration and three external pI-markers, ProteinSimple Maurice™ revealed a main peak pI of 7.56 (Fig. 2A, blue dash line), consistent with findings by Goyon et al. 201716. However, rescaling the data with a non-linear regression considering all pI-markers (Fig. 2B, red solid line), shifted the main peak near 7.33. Additionally, employing a narrower calibration based on 5 pIM (6.14, 7.0, 7.65, 8.18, 8.40) surrounding the sample (Fig. 2C, green solid line), the non-linear regression gets the Infliximab main species at 7.26. Contextually, we found the measured pI-markers peaks closer to their certified values. A main peak of 7.26 falls within the range computed by Goyon et al. using Vector NTI and MassLynx16. This value also matches the theoretical pI obtained by Prot pi software19, employing the most abundant PTMs described in the literature for Infliximab 2,20,21,22,23 (Supplementary Information).
Infliximab charge variants characterization through icIEF. Infliximab was run together with 10 pI-markers, spanning the gradient. The electropherogram was scaled considering: (A) Linear calibration (left panel, blue dash line) by using three external pI-markers (5.85, 10.0, 10.17), as usually done in our standard icIEF procedures. The correspondent electropherogram is reported in the right panel. (B) A non-linear calibration curve (red line) using ten pI-markers (5.85, 6.14, 7.0, 7.65, 8.18, 8.40, 9.0, 9.5, 10.00, 10.17). (C) A non-linear calibration curve optimized in a narrow window (green line) enclosing the sample and five pI-markers (6.14, 7.0, 7.65, 8.18, 8.40). Notice that, within the considered interval, the green line better approximates the data, as demonstrated by the proximity of measured pI-markers to their certified values (underlined Italic fonts, in the electropherograms). Conversely, under suboptimal conditions (blue and red lines), considerable deviations are evident. This underscores the critical importance of maximizing data adaptation, which not only enhances the accuracy of pI determination for markers but also ensures the Infliximab main peak exhibits a pI that closely aligns with theoretical values derived from the primary sequence (Supplementary Information). On the contrary, by using a poor calibration curve very biased values are observed. In the electropherograms, main peak-pIs are reported in bold. Filled areas in the calibration curves highlight the pH interval considered for the electropherograms.
To summarize, we have shown that the electropherogram scaled according to our analytical instrument software, which relies on linear regression, is affected by calibration errors; however, it is feasible to correct these errors through a non-linear rescaling algorithm, thereby obtaining a more accurate pI determination. From now on, we will use “unbiased” to refer to data where the calibration error has been minimized as much as possible (Supplementary Information).
Assessing the resolution of the method
In Europe, EDQM and the Ph. Eur. Commission, work to develop comprehensive and standardized recommendations for analytical testing strategies in response to stakeholder requests. These recommendations address the diverse needs for manufacturing processes and analytical testing of various classes of biotherapeutics. Such protocols deveoped to control a given class of biotherapeutics (i.e. monoclonal antibodies) are defined as “horizontal standards” and serve as a basis to develop and validate further product-specific standard operating procedures24. According to these “horizontal standards,” the efficacy of peak separation in CVPA carried out by cIEF is effectively measured using “chromatographic resolution,” which is the distance between two peaks of the electropherogram normalized for their average width. Unfortunately, since the resolution is dependent on the calibration curve, is itself biased by the same calibration errors described above. Moreover, this resolution is assumed to be constant within the intervals between the electropherogram peaks, without providing more detailed information along the whole pH range. Here, we propose a more suitable estimation of the resolution that is independent of the specific mAb under study, being based on a fine study of the pH gradient through an accurate internal calibration system consisting of a panel of several pI-markers that will be secondarily run along the sample in the injection. Thus, we named this resolution “a priori,” meaning that we could determine the resolving conditions of the chosen experimental setting independently of the analyte we plan to study and, therefore, choose the best option between different conditions.
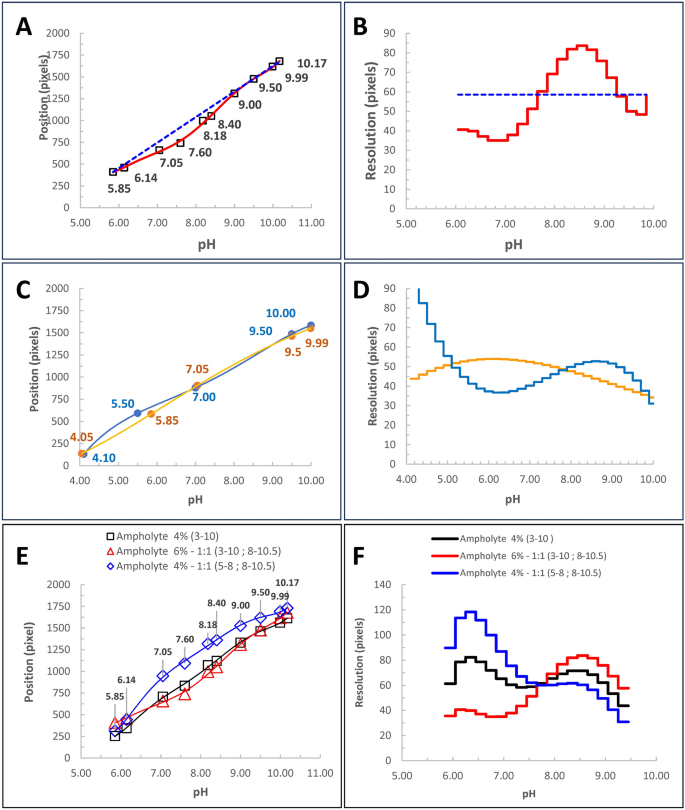
Knowing the optimal calibration curve, we can estimate the distance between consecutive points shifted by the same pH interval (ΔpH) along the gradient. These distances (pixels), represent a measure of the resolution, offering a predictive insight into the separation efficiency for a given molecule. Notably, assuming a linear calibration (Fig. 3A,B—blue dotted line) implies a constant a priori resolution over the pH gradient. On the contrary, the non-linear regression (Fig. 3A,B—red curves) reveals a non-homogeneous resolution along the gradient.
Assessing the Resolution of the Method. (A) Calibration curves of the same icIEF experiment. Certified values of ten pI markers (5.85, 6.14, 7.05, 7.60, 8.18, 8.40, 9.00, 9.50, 10.00, 10.17) are plotted against their positions along the capillary. Linear regression (blue dotted line: using only 5.85, 9.99, 10.17 external pI markers) and non-linear regression (6th-order polynomial, red line, using all pI markers) were applied. (B) Method resolution along the pH gradient, corresponding to A. This graph shows the distance (pixels) between different points of the pH gradient, spanning a ΔpH of 0.2 units. Resolution values were derived from the regressions in A. Notably, linear calibration assumes constant resolution, while non-linear regression allows resolution to vary along the gradient. (C) Calibration curves for two different pI marker sets: ProteinSimple (4.05, 5.85, 7.05, 9.50, 9.99—orange) and SCIEX (4.10, 5.50, 7.00, 9.50, 10.00—blue) diluted in the same parent solution. (D) Resolution corresponding to the marker sets in C. Using the same parent solution highlights that differences in resolution curves are due to solvent effects of the two pI marker formulations, significantly influencing the pH gradient. (E) Ten pI markers (5.85, 6.14, 7.00, 7.60, 8.18, 8.40, 9.00, 9.50, 10.00, 10.17) run in icIEF with different ampholyte stratifications (4% 3–10—black line; 6% 1:1 3–10/8–10.5—red line; 4% 1:1 5–8/8–10.5—blue line). (F) As shown by the calibration curves in E, ampholyte composition strongly impacts resolution along the pH gradient. However, these variations are difficult to predict without systematic experimental design.
Our resolution is useful, for instance, when we need to consider the solvent effects wherein key components such as pI-markers are diluted, which can significantly impact the shape of the pH gradient and the resolution of the method. This scenario is summarized by the experiment depicted in Fig. 3C,D where two different pI-markers’ sets, one from Protein Simple and the other from SCIEX, have been dissolved in two vials of the same parent solution. The choice of different pI markers is sufficient to affect the two calibration curves and thus the resolution.
As mentioned above, in cIEF, the configuration of this pH gradient is the most critical factor for successfully identifying all isoforms as distinct peaks. While certain variables influencing the gradient shape are beyond the operator’s control, such as the capillary embedded in a closed cartridge, other parameters can be adjusted for specific purposes, allowing the device to operate under customized conditions. Above all, formulation, concentration, and stratification of carrier ampholytes are the most significant (Fig. 3C,E). They are usually supplied as ready-to-use mixtures, and unfortunately, their composition is not disclosed. The only information typically provided is the range of the pH gradient generated. According to this information, ampholytes can be classified as wide- or narrow-range. In Fig. 3F, by using the same mixture of ten pI-markers we evaluated the resolution of the pH gradient against different ampholyte conditions. Generally, we didn’t find a uniform resolution throughout the whole pH range but rather there are intervals where it is better than others. We always observed the poorer resolutions near pH 7 and above 9. The knowledge of these effects raises the idea that whatever method should be accurately designed considering all the variables affecting the resolution of the system.
The development of icIEF mAbs-tailored methods through an unbiased experimental design
Recently, a new trend has emerged aimed at adapting and transferring previously validated methods to different technological tools and developing platform methods for controlling mAb-based drugs to harmonize different analytical procedures. This is reflected in recent texts published by international standardization bodies, such as European Pharmacopoeia17, to be used by both manufacturers and OMCLs. In this context, developing a method able to reveal the identity of biotherapeutics where no reference standard (RS) is available, is key in the fight against the counterfeiting of biological medicinal products (BMPs). In all these cases, OMCLs that currently carry out analytical controls should be able to perform an easy and independent evaluation of the BMPs’ quality and efficacy. To pursue such an approach, the only possibility is to carry out an UED, which is applicable to any specific drug under study, independently of the device to analyze them, or of the operators.
Following this strategic approach, it becomes fundamental to examine the variables that significantly influence the outcome. We considered that key input variables impacting the method’s resolution include: the total percentage of ampholytes, ampholyte ratio, pH, voltage, and separation time. We posit that an optimal method should adhere to an ideal design capable of discerning conditions for achieving the highest resolution possible across the specified pH range, regardless of the sample. This condition can only be fulfilled through a multiparametric analysis of the resolution, treated as a scalar function of multiple real input variables, changing simultaneously. Within each pH interval, this function should exhibit relative maxima whose coordinates represent the optimal compromise for achieving the desired resolution.
Consequently, we conducted a multivariate experiment utilizing a sample consisting of ten certified pI-markers (ranging from 5.85 to 10.17) carrying out icIEF, under various combinations (N = 45) of the input variables: (i) Total ampholyte percentage (4%, 6%, 8%); (ii) narrow-range ampholyte ratio (pH 5–8/pH 8–10.5 = 0.33, 0.66, 1.00, 1.66, 3.00); (iii) separation times (9.0’, 10.5’, 12.0’); (iv) pH (each pI marker peak in the electropherograms flags the pH value that was present at that point of the gradient at the end of the experiment).
The only output variable is the resolution. Therefore, the “resolution function” is the scalar relationship defined in a set of the input real variables (domain) returning its values in a set of real numbers (codomain). To study this function, by a multiparametric analysis, our first concern was to achieve a good sampling of the domain, so we designed all the combinations of the input variables reported above to achieve this purpose.
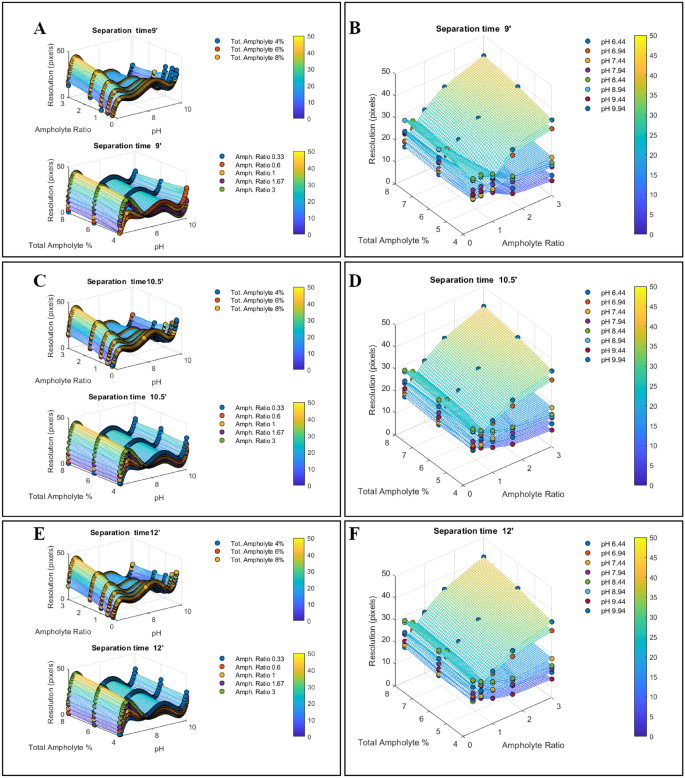
To better show the results we plotted the resolution function against the input variables, two at a time (Fig. 4). We plotted the simultaneous dependence of the resolution on pH and the specified ratio of pH 5–8/pH 8–10 narrow range ampholytes or total ampholyte percentage in Fig. 4-panel A, C, E; while, the effects of the simultaneous variation of ampholyte ratio and total ampholyte percentage in different set of pH, is displayed in Fig. 4-panel B, D, F. To be noticed, the focusing time effects on the resolution can be analyzed by a direct comparison of the results summarized in Fig. 4 panels A, C, E, or panels B, D, F.
The development of a transversal unbiased method for mAbs icIEF by studying the resolution function. We performed a multivariate icIEF experiment where 4 input variables are changing together, while we measure the resolution of the methods along the pH gradient. In each injection a sample consisting of a mix of ten pI-markers (5.85, 6.14, 6.61, 7.05, 7.40, 7.90, 8.40, 9.00, 9.50, 9.99, 10.17) is run under an array of different input variables: (i) total ampholytes percentage (4%, 6%, 8%); (ii) narrow range pH 5–8/pH 8–10.5 ampholytes ratio (0.33, 0.66, 1.00, 1.66, 3.00); (iii) separation times (9.0’, 10.5’, 12.0’). Due to the presence of the pI-markers in each injection, we performed an internal optimized calibration allowing us to define the shape of the pH that was present in the capillary at the end of the measure. This pH is independent of the measure since the pI-markers are only flags of the pH gradient that affected the experiments. (A,C,E) Based on the calibration curves, we computed the resolutions every 0.05 ΔpH units from pH 6 to pH 10, for each array of variables, these data are presented as scatter plots of the different arrays of variables presented in two at a time. The remaining input variables are reported as different color datasets superimposed on the same graph. The effects of the migration times can be observed by a direct comparison of panels (A,C,E). (B,D,F) These data show the average resolution in simultaneous variations of total ampholyte percentage and narrow range pH 5–8/pH 8–10.5 ampholytes ratio, in defined slots of pH. These scatterplots demonstrate that pH and ampholyte ratio strongly affect the resolution of the methods. The effects of the migration times can be observed by a direct comparison of panels B, D, F. All the scatter plots were interpolated by non-linear polynomial surfaces to obtain each corresponding-colored mesh, these are the 3D representations of the resolution functions. The Colormaps highlight the resolution values by color-coded scales.
Overall, our results strongly demonstrate that the resolution function is not a linear relationship and that both pH and the ratio of pH 5–8/pH 8–10.5 narrow-range ampholytes mainly affect the resolution by directly shaping the gradient. On the contrary, both the total percentage and the focusing time only slightly affect the overall resolution. However, at the tails of the gradients sometimes this scenario could change. We also noticed that in all the cases the lower resolution occurs near pH 7.5, probably due to the narrow range ampholyte-specific composition (unknown to us). Since a similar effect has been observed even by using only 3–10 wide-range ampholytes Fig. 3 (black trace), this effect cannot arise from a gap between the narrow-range ampholytes but is rather dependent on carrier molecule enrichment.
Discussion
Since the early 1980s, there has been a significant acceleration in the development of new patented mAbs-BMPs and their biosimilars, leading to a market estimated to be worth over a hundred billion dollars today25,26,27. This rapid growth prompted regulatory agencies to swiftly define policies aimed at controlling all aspects of the manufacturing process, including all aspects of the manufacturing and distribution process, to ensure both the efficacy and purity of drugs and, hence the safety of patients. Within this framework, the purity and identity of a product play a pivotal role.
Capillary Isoelectric Focusing is one of the most commonly used techniques, among several complementary and orthogonal methods, for testing the identity and purity of mAbs. It provides a mAb fingerprint based on charge profile and heterogeneity arising from the presence of different pI variants. However, significant discrepancies may arise in evaluating charge variant isoforms among different laboratories and devices, rendering the concept of “product identity” not objective but only relative to a reference standard.
In the attempt to study the reliability of our experimental methods, we noticed that our calibration curves, despite the good adaptation to the linear trend, displayed local inconsistencies; a feature yet described elsewhere28,29,30,31. We asked how the assumption of a linear calibration curve would affect the coherence of our data. We concluded that effectively this assumption introduces a strong bias in the extrapolated pIs. To counteract this bias, an optimized regression model has been applied to the data, which effectively minimized the calibration error.
To demonstrate the efficacy of our approach, we run an Infliximab sample along with several pI-markers in the same injection, to achieve an optimal calibration of the electropherogram. By comparing the pIs determined through the optimized method as compared to those obtained by a standard procedure, the main peak for the infliximab reported a pI of 7.26 instead of 7.57 as determined with the standard procedure. The latter is very similar to that reported by Goyon using a similar device16. On the contrary, the value of 7.26 is very consistent with the theoretical pI we calculated with Prot pi19, starting from the Infliximab’s primary sequence (Table S2—Supplementary Information). At the same time, this value is coherent to those found by Goyon by using two different pI calculators—Vector NTI (7.10) and MassLynx (7.40)16. The evidence that a more accurate calibration strategy brings the estimated Infliximab pI closer to its theoretical pI is significant. This approach must be confirmed and verified on a larger panel of therapeutic antibodies to extend its general applicability, which we plan to address in future work; it is also important to cross-validate the obtained pI-values with other experimental techniques, not simply based on a sequence of tabulated pKa values, since they can vary according to the specific algorithm to compute them (Supplementary Information).
Noteworthy, at present different cIEF devices often produce inconsistent results for the same analyte, despite the comparable procedure1. We believe that an optimized calibration approach on each device could mitigate these discrepancies in measured pIs and improve reproducibility across laboratories, particularly when different icIEF devices are employed. These inconsistencies may arise from different calibration errors between devices or suboptimal experimental designs. Therefore, we suggest manufacturers incorporate such corrective routines into their software, ensuring more accurate and consistent results.
Since our recalibration approach needs to run several internal pI-markers together with the sample, naturally, one potential concern is that samples with broad charge variants could interfere with the signals of pI-markers, impairing the estimate of the peak identity and consistency in the electropherograms. However, taking advantage of modern icIEF devices’ capability to measure signals in both absorbance and fluorescence modes by using non-fluorescent pI-markers, it would be feasible to identify pI-markers’ positions on the absorbance channel and the sample electropherogram on the fluorescence one, avoiding their superimposition at least on one channel. At the same time, we strongly encourage the manufacturer to design a new generation of pI-markers/sample probes visible only in the absorbance or fluorescence mode, thus allowing to obtain a sort of independent “dual-channel electropherogram” without any interference. Thus, for each injection, an optimal internal calibration curve and a clean molecule electropherogram can be achieved at once (for more details, see Supplementary Information).
An optimal calibration curve allows to study, a priori, the resolution function, along the pH gradient, regardless of the sample. This might be relevant for quality control labs of manufacturers or regulatory agencies to develop or improve more effective and independent methods to analyze BMPs. In addition, pursuing the concept of a univocal identity could provide several advantages, such as the construction of a biobank able to rapidly identify possible alterations in the molecule structure and, hence, possible divergence from the originator, such as in the control of counterfeited BMPs.
Concerning the pH gradient, a remarkable observation is its significant instability against bulk effects. However, in the present work, we demonstrate that it is possible to perform a UED that can support a therapeutic choice of the most reliable and effective protocol to study mAbs within a specific pH interval by studying the scalar resolution function.
In conclusion, we have shown that the current approach, which relies on linear fitting assumptions, results in systematic calibration errors, making it difficult to achieve accurate and consistent pI estimations across different devices, reagents lots, and laboratories. On the contrary, our approach enables more effective pI measurements and facilitates an UED to identify resolution variations across the pH gradient. This allows the development of robust protocols tailored for specific mAbs and others BMPs. Thus, our results open new perspectives to achieve innovative solutions in the field of CVPA of mAb-based drugs by exploring the concept of univocal charge identity to be extended potentially to all BMPs such as biosimilars, fusion proteins32 (Fig. 5), but also for Lipid nanoparticles33, where the icIEF has been successfully used to characterize the surface charge of mRNA lipid nanoparticle vaccines. Thus, employing our method could lead to results that have a strong impact on social healthcare.
Enhanced calibration for an accurate and robust assessment of charge variant profile through iciEF. (Red panel). The current approach, based on the linear calibration assumption implies biased pI measurements and a constant resolution along the pH gradient. The former could explain the pIs inconsistencies reported in the literature for the same mAb, measured by different instrumentations. (Green panel). On the contrary, under the proposed approach a more accurate calibration curve is obtained through a non-linear regression, thus minimizing the bias and resulting in more effective measured pIs. This allows a direct comparison of the results carried out by different instrumentations. (Yellow panel). Additionally, the effective resolution pattern of each run injection can be analyzed along the pH gradient, allowing an UED to study resolution as a multivariate function of different key input variables. (Blue panel). Our approach allows us to find methods customized in a specific pH range, with a robust and accurate pI estimation and resolution leading to a univocal charge profile assessment.
Materials and methods
Equipment
IcIEF analysis was performed using a system based on the iCE platform (Maurice™) from Biotechne/ProteinSimple equipped with a whole capillary imaging detection device. The system encompasses an autosampler, a Charge Coupled Device (CCD) camera to capture the UV absorption image from the UV detector operating at 280 nm. For data evaluation, the instrument control software (ProteinSimple—Compass for iCE -2.0.10) was used.
Reagents and sample
TRIS (Tris(hydroxymethyl) aminomethane) 99.9 + % ultrapure grade (trometamol R), Urea R, L-Arginin and Iminodiacetic acid R were obtained from Merck -Sigma-Aldrich (Italy). Pharmalyte 3–10, 5–8, and 8–10.5 were obtained from GE Healthcare (Italy). pI markers 4.05, 5.85, 6.14, 7.05, 8.18, 8.40, 9.5, 9.99, 10.17 were purchased by Biotechne/ProteinSimple. A Peptide Marker Kit, consisting in pI markers pI 4.1, pI 5.5, pI 7.0, pI 9.5, pI 10.0, was purchased by Sciex (Framingham, MA USA). pI markers 7.65 and 9.0 were purchased by Sigma-Aldrich. Pre-assembled icIEF cartridges together with all batch reagents including ICE electrolyte kit, 0.5% methyl cellulose solution (MC), and 1% MC, were purchased from Biotechne/ProteinSimple (San Jose, CA, USA).
Infliximab CRS was purchased from EDQM.
Samples treatment
The formulation buffer of Infliximab was replaced with TRIS buffer (20 mM, pH 8.0) using a Microcon -10 Centrifugal Filter Device (Millipore) containing an Ultracel©PL-10 membrane (Regenerated cellulose 10,000NMWL). Briefly, 100 µL of the sample was mixed with 400 µL TRIS buffer in a 0.5 mL centrifugal filter device and then centrifuged at 14 500 rpm for 15 min. After discarding the flow-through, 400 µL TRIS buffer was again loaded onto the filter, and the centrifugation at 14 500 rpm was repeated for 10 min. Subsequently, the filter was inverted and placed into a clean tube, and the eluate was recovered by centrifuging at 100 rpm for 2 min. The obtained volume was adjusted to 100 µL with TRIS buffer, to restore the original sample concentration (10 mg/mL). For icIEF analysis, the final concentration of Infliximab was 0.4 mg/mL.
Shared reagents and procedures
Batch reagents
For an icIEF experiment, batch reagents have been prepared according to the “ProteinSimple Maurice cIEF Method Development Guide”. Analytes (Infliximab and/or pIMs spiked as ‘samples’) were added to a mixture (parent solution) containing a list of common reagents: the MC1% (Separation matrix, 0,35ultra%), TRIS buffer (0,8 M, pH 8), L-Arginine (as cathodic stabilizer, 10 mM), Iminodiacetic Acid (as anodic stabilizer, 4 mM), Urea (2,4 M) and tri-distilled Milli-Q ultrapure water. For all the experiments, 2 µL of each pI-marker (pIM) was used.
The separation was carried out with a pre-focusing at 1500 V for 1 min, followed by a focusing at 3000 V for 8 min, except the ED experiment (as described below).
The specific conditions in terms of ampholytes and pI markers—eventually alongside sample(s)—for each experiment session are detailed beneath, referring to their relative Figures.
Offline analysis of the experiments
As yet mentioned, ProteinSimple Maurice™ is equipped with a control software (Compass for iCE™ -2.0.10), allowing to analyze the experiments and produce reports as “.pdf”, as well to export raw data as “.txt” files. The calibration curves have been constructed by using offline algorithms. Beginning with the raw data, the position (pixels) of each pI marker from the injection was determined and correlated with its certified pI value, resulting in a scatterplot. Regression equation parameters for both linear and non-linear models were obtained by minimizing the sum of the squares of the residuals34,35.
Experimental scheme and calculation
The application of calibration curves
To assess the biases introduced by a not-optimal calibration curves, we run in twice 10 pI-markers (5.85, 6.14, 7.05, 7.60, 8.18, 8.40, 9.0, 9.50, 9.99, 10.17) in a parent solution containing 1:1—6% Pharmalyte 3.0–10.0 /8.0–10.5 according to the icIEF method for mAbs recently published in European Pharmacopeia (Fig. 1). After the run the same electropherogram was scaled trough two different linear regressions (by using only 5.85, 9.99, 10.17 external pI-markers or all the pI-markers) or by a 6th order non-linear polynomial regression (based on all ten pI-markers). Pearson’s determination coefficients were obtained to test the adaptation to each model. For each pI-marker peak of the electropherogram, the percentage biases introduced by the models were estimated according to the Eq. (1). Their average values (mean + SD) were finally plotted as a bar plot.
To highlight the effects of these biases, we run the Infliximab in duplicate together with 10 pI-markers (5.85, 6.14, 7.65, 8.18, 8.40, 9.00, 9.50, 9.99, 10.17) chosen to minimize sample overlapping in the electropherogram (Fig. 2). In this case, we used only Pharmalyte 3–10 (4%) to unmask any potential data distortion that might occur even in a more homogeneous gradient condition, which is commonly expected to best fit a linear model. A ready-to-use ProteinSimple System Suitability was added in the first injection. As a blank, we used the mixture of pI-markers alone (data not shown). As a control for the identity of the 7.65 pI-marker, expected to be very close to the last basic peak of Infliximab, we prepared an analogous sample without this marker. The overlay of these two last electropherograms has been reported in Fig. S2.
Assessing the resolution of the method
Starting from the optimized calibration curve, we assessed the resolution as follows. For each considered i-th pH along the gradient, by knowing the calibration curve (pH vs position), we assessed the “a priori” resolution along the capillary, as:
That is the distance between two points shifted by a constant pH interval (ΔpH), along the whole gradient. These distances (pixels) measure the resolution of the method. A linear calibration implies a constant resolution over the pH gradient, because according to the equation, for every ΔpH we have the same Δposition (pixels), along the gradient.
We illustrated this concept in Fig. 3A,B where 10 pI-markers and 1:1—6% Pharmalyte 3.0–10.0 /8.0–10.5 were used.
To better investigate the bulk effects of the pI-markers buffers on the resolution, the same parent solution was aliquoted into two different vials, and two different sets of pI-markers were added as samples (pI-markers: 4.1, 5.5, 7.0, 9.5, 10.0 from ABSCIEX and 4.05, 5.85, 7.05, 9.5, 9.99 from ProteinSimple—Fig. 3C,D). To study how wide and narrow range ampholyte stratification affected the calibration curve shape (Fig. 3E,F), we prepared several parent solutions, differing for their ampholyte stratification percentages and ratio: 4% 3–10 only, 6% 3–10/ 8–10.5 (1:1), 4% 5–8/8–10.5 (1:1). As usual, a System Suitability, designed to check that all was working well, was run in the first injection. Each sample was run in duplicate.
The development of icIEF mAbs-tailored methods through an unbiased experimental design
To better investigate how the resolution functions are affected by the input variables (ampholyte total percentage, ampholyte ratio, pH, voltage, and separation time) we performed an icIEF experiment using different parent solutions where all the combinations of the input variable described in the Table S3 are used (N = 45). Here, we employed only the Pharmalytes 5–8 and Pharmalytes 8–10.5, to avoid, unpredictable complexities due to the superimpositions of the carriers. To deeply investigate the entire pH-gradient we run as a sample a mixture of ten pI-markers: 6.14, 6.61, 7.05, 7.40, 7.90, 8.40, 9.00, 9.50, 9.99, 10.17.
A ready-to-use System Suitability from ProteinSimple was run in the first injection. Here a different protocol consisting of a prefocusing step at 1500V for 1 min followed by focusing at 3000V for different separation times (9.0’, 10.5’, 12.0’) was utilized. At the end of the experiment, the raw data obtained by the instrument control software were exported and analyzed by custom routines in Matlab (R2023b-MathWorks™). For each injection (corresponding to every array of variables), the peak positions (pixels) corresponding to the specific pI-markers of the injection were obtained and related to their certificated pI values finding the calibration curves Eqs. (6th order polynomial regressions), and the correspondent resolution functions every 0.05 ΔpH units, from pH 6 to pH 10. These data were presented as colour-coded scatter plots where the input variables were presented two at a time. The remaining input variables were reported as different colour datasets superimposed on the same graph. All scatter plots depict experimental data. These graphs were interpolated by non-linear polynomial surfaces to generate corresponding-colored meshes, effectively representing the 3D resolution functions. Colormaps highlight the resolution values on each 3D function.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ascione, A. et al. Charge heterogeneity of therapeutic monoclonal antibodies by different cIEF systems: Views on the current situation. MAbs 16 (2024).
Walsh, G. & Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24, 1241–1252 (2006).
Liu, H., Caza-Bulseco, G., Faldu, D., Chumsae, C. & Sun, J. Heterogeneity of monoclonal antibodies. J. Pharm. Sci. 97, 2426–2447 (2008).
Harris, R. J. et al. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J. Chromatogr. B Biomed. Sci. Appl. 752, 233–245 (2001).
Singh, S. K., Kumar, D., Nagpal, S., Dubey, S. K. & Rathore, A. S. A charge variant of bevacizumab offers enhanced FcRn-dependent pharmacokinetic half-life and efficacy. Pharm. Res. 39, 851–865 (2022).
Higel, F., Seidl, A., Sörgel, F. & Friess, W. N-glycosylation heterogeneity and the influence on structure, function and pharmacokinetics of monoclonal antibodies and Fc fusion proteins. Eur. J. Pharm. Biopharm. 100, 94–100 (2016).
Beck, A. et al. Risk-based control strategies of recombinant monoclonal antibody charge variants. Antibodies 11, 73 (2022).
Ambrogelly, A. et al. Analytical comparability study of recombinant monoclonal antibody therapeutics. MAbs 10, 513–538 (2018).
Gahoual, R., Beck, A., Leize-Wagner, E. & François, Y. N. Cutting-edge capillary electrophoresis characterization of monoclonal antibodies and related products. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1032, 61–78 (2016).
Suba, D., Urbányi, Z. & Salgó, A. Capillary isoelectric focusing method development and validation for investigation of recombinant therapeutic monoclonal antibody. J. Pharm. Biomed. Anal. 114, 53–61 (2015).
Isoelectric Focusing - 1st Edition. https://www.elsevier.com/books/isoelectric-focusing/catsimpoolas/978-0-12-163950-1.
Hjertén, S. & de Zhu, M. Adaptation of the equipment for high-performance electrophoresis to isoelectric focusing. J. Chromatogr. A 346, 265–270 (1985).
Crosnier de Bellaistre, M., Randon, J. & Rocca, J. L. Hydrodynamic flow and electroosmotic flow in zirconia-packed capillaries. Electrophoresis 27, 736–741 (2006).
Slater, G. W., Tessier, F. & Kopecka, K. The electroosmotic flow (EOF). Methods Mol. Biol. 583, 121–134 (2010).
Kinoshita, M., Nakatsuji, Y., Suzuki, S., Hayakawa, T. & Kakehi, K. Quality assurance of monoclonal antibody pharmaceuticals based on their charge variants using microchip isoelectric focusing method. J. Chromatogr. A 1309, 76–83 (2013).
Goyon, A. et al. Determination of isoelectric points and relative charge variants of 23 therapeutic monoclonal antibodies. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1065–1066, 119–128 (2017).
European Directorate for the Quality of Medicines & HealthCare. Drafts Published in Pharmeuropa 35.4 (End of Commenting Period: 31 December 2023) 2.5.44. Capillary Isoelectric Focusing for Recombinant Therapeutic Monoclonal Antibodies. European Directorate for the Quality of Medicines & HealthCare https://pharmeuropa.edqm.eu/app/phpa/content/issue35-4/20544E.htm (2023).
Mack, S., Cruzado-Park, I., Chapman, J., Ratnayake, C. & Vigh, G. A systematic study in CIEF: Defining and optimizing experimental parameters critical to method reproducibility and robustness. Electrophoresis 30, 4049–4058 (2009).
Reinert, T., Houzé, P., Mignet, N., Francois, Y. N. & Gahoual, R. Post-translational modifications comparative identification and kinetic study of infliximab innovator and biosimilars in serum using capillary electrophoresis-tandem mass spectrometry. J. Pharm. Biomed. Anal. 234, 115541 (2023).
Martín, S. M., Zaborowska, I., Jakes, C., Carillo, S. & Bones, J. Comparability Study for The Determination of Post-Translational Modifications of Infliximab Innovator and Biosimilars by Automated High-Throughput Peptide Mapping Analysis. https://assets.thermofisher.com/TFS-Assets/CMD/Application-Notes/an-21850-lc-ms-comparability-biosimilars-an21850-en.pdf.
Kim, H. et al. Advanced assessment through intact glycopeptide analysis of Infliximab’s biologics and biosimilar. Front. Mol. Biosci. 9 (2022).
Hong, J. et al. Physicochemical and biological characterization of SB2, a biosimilar of Remicade® (infliximab). MAbs 9, 364–382 (2017).
Capillary Isoelectric Focusing for Recombinant Therapeutic Monoclonal Antibodies.
Ecker, D. M., Jones, S. D. & Levine, H. L. The therapeutic monoclonal antibody market. MAbs 7, 9–14 (2015).
Grilo, A. L. & Mantalaris, A. The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. 37, 9–16 (2019).
Doortje Reijman, M., Meeike Kusters, D. & Wiegman, A. Expert opinion on biological therapy ISSN: (Print) (Online) Journal homepage: www.tandfonline.com/journals/iebt20 Current and emerging monoclonal antibodies for treating familial hypercholesterolemia in children Current and emerging monoclonal antibodies for treating familial hypercholesterolemia in children. https://doi.org/10.1080/14712598.2024.2330948 (2024).
Wu, J. & Huang, T. Peak identification in capillary isoelectric focusing using the concept of relative peak position as determined by two isoelectric point markers. Electrophoresis 27, 3584–3590 (2006).
Štastná, M. & Šlais, K. Colored pI standards and gel isoelectric focusing in strongly acidic pH. Anal. Bioanal. Chem. 382, 65–72 (2005).
Mack, S., Cruzado-Park, I. D. & Ratnayake, C. K. High-resolution CIEF of therapeutic monoclonal antibodies a platform method covering PH 4–10.
Fast preparation of monolithic immobilized pH gradient column by photopolymerization and photografting techniques for isoelectric focusing separation of proteins. https://doi.org/10.1002/elps.201100195.
Wu, G. et al. A platform method for charge heterogeneity characterization of fusion proteins by icIEF. Anal. Biochem. 638 (2022).
Loughney, J. W., Minsker, K., Ha, S. & Rustandi, R. R. Development of an imaged capillary isoelectric focusing method for characterizing the surface charge of mRNA lipid nanoparticle vaccines. Electrophoresis 40, 2602–2609 (2019).
Motulsky, H. & Christopoulos, A. Fitting models to biological data using linear and nonlinear regression: A practical guide to curve fitting. In Fitting Models to Biological Data Using Linear and Nonlinear Regression. https://doi.org/10.1093/OSO/9780195171792.001.0001 (2004).
Riazoshams, H., Midi, H. & Ghilagaber, G. Robust nonlinear regression in R. In Robust Nonlinear Regression 207–214. https://doi.org/10.1002/9781119010463.CH9 (2018).
Acknowledgements
We thank Dr. Niccolò Candelise, Dr. Barbara Ridolfi, and Dr. Francesca Carlini, for their helpful suggestions in the revision of this manuscript.
Funding
This work has been supported by research funds of the National Centre for Preclinical and Clinical Evaluation and Research of Drugs (FARVA), Istituto Superiore di Sanità. Viale, Regina Elena 299, 00161 ROME, ITALY and the Centre for the Control and Evaluation of Medicines (CNCF), Istituto Superiore di Sanità. Viale, Regina Elena 299, 00161 ROME, ITALY.
Author information
Authors and Affiliations
Contributions
M.B. & A.A. conceived the work, performed most of the experiments, and wrote the manuscript. M.B. performed the data analysis, V.G. performed the experiments and revised the manuscript, SD performed the experiments, F.L. revised the manuscript, and coordinated and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Belfiore, M., Ascione, A., Ghizzani, V. et al. A new paradigm for optimized experimental design in cIEF platforms aimed at an accurate robust and reliable mAbs charge-variant assessment. Sci Rep 14, 28087 (2024). https://doi.org/10.1038/s41598-024-79108-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-79108-5