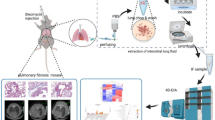
Abstract
Changes in lung and gut microbial communities have been associated with idiopathic pulmonary fibrosis (IPF). This study aimed to investigate correlations between microbial changes in the lung and gut and host physiological indices in an IPF model, exploring potential mechanisms of the lung-gut axis in IPF pathogenesis. IPF model rats were established via trans-tracheal injection of bleomycin, with assessments of hematological indices, serum cytokines, lung histopathology, and microbiome alterations. Significant differences in microbial structure and composition were observed in the IPF model compared to controls, with 14 lung and 7 gut microbial genera showing significant abundance changes. Further analysis revealed 20 significant correlations between pulmonary and gut genera. Notably, 11 pairs of correlated genera were linked to the same IPF-related physiological indices, such as hydroxyproline, mean corpuscular volume (MCV), and red cell distribution width-standard deviation (RDW-SD). We identified 24 instances where a lung and a gut genus were each associated with the same physiological index, forming "lung genus—index—gut genus" relationships. Mediation analysis showed that indices like hydroxyproline, MCV, and RDW-SD mediated correlations between 10 lung genera (e.g., Cetobacterium, Clostridium XVIII ) and the gut genus Allobaculum. This study first describes gut-lung microbial interactions in pulmonary fibrosis. Mediation analysis suggests pathways underlying "lung genus—host index—gut genus" and "gut genus—host index—lung genus" correlations, thus providing clues to further elucidate the mechanisms of the “gut-lung axis” in IPF pathogenesis.
Similar content being viewed by others
Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive interstitial lung disease of unknown etiology. It is characterized by proliferation of fibroblasts and accumulation of extracellular matrix. Histologically, IPF demonstrates usual interstitial pneumonia pattern with patchy fibrosis and honeycomb changes. Epidemiological studies have shown that the incidence of IPF has been increasing in recent years, but the prognosis has not significantly improved1. IPF presents with a clinical syndrome of insidious onset unexplained exertional dyspnea, chronic dry cough, and fatigue. The clinical course is variable but progressive in most patients. As the disease advances, restrictive pulmonary ventilatory defect develops along with impaired gas exchange. In severe cases, respiratory failure ensues. The nonspecific clinical presentation hampers early diagnosis, and the natural history is unpredictable. At present, no curative therapy for IPF exists2, underscoring the imperative for research to develop novel treatments.
Several animal models have been utilized to evaluate drug efficacy and investigate the disease mechanisms of IPF. Among them, the bleomycin-induced IPF model in rodents, particularly mice and rats, via intratracheal administration, recapitulates key pathological features of human IPF and closely mimics the progressive characteristics of the clinical disease3. Due to its ability to stably and reproducibly induce lung fibrosis phenotypes highly similar to human IPF, it has been widely used in related studies4.
IPF has traditionally been viewed as a sterile lung disease. However, with the revolutionary advances in microbiome research in recent years, the role of microbial communities in IPF pathogenesis has gained increasing attention. Multiple studies have shown that compared with healthy controls, IPF patients have significantly increased microbial burden and altered composition in the lungs, which correlate with disease progression and mortality5,6,7. Additionally, a prospective study on IPF patients and healthy controls demonstrated interactions between the lung microbiota and the host immune responses in IPF through integrated analysis of host transcriptomics and microbial features8. These findings suggest the hypothesis that alterations in the bacterial communities of the lower airways in IPF may not only act as persistent stimuli for repetitive alveolar injury but also lead to correlated changes in peripheral blood gene expression profiles, reflecting the host inflammatory responses.
On the other hand, the gut microbiota, as an "endogenous microbial organ" closely related to various physiological functions of the human body, has also been considered to play an important role in the occurrence and development of pulmonary fibrosis in recent years9. A number of microbiota comparison studies have found that compared with healthy controls, pulmonary fibrosis patients/models exhibit significant alterations in gut microbiota composition10,11,12,13. Several studies suggest that changes in the gut microbiota may affect the progression of pulmonary fibrosis by regulating immune responses or through the ectopic effects of their metabolites9,14,15,16,17. For example, it was shown in IPF mice model that the changes in the gut microbiota can lead to significant alterations in the proportion of CD4 + IL-6 + and CD4 + IL-17A + T cells in the lungs and affect the progression of pulmonary fibrosis through activation of the IL-6/STAT3/IL-17A pathway15. In addition, intestinal microbial metabolites, including amino acids, short chain fatty acids, bile acids, and valproic acid have also been found to be involved in the process of pulmonary fibrosis through extracellular matrix accumulation, energy metabolism, epigenetics, immune regulation and other pathways18,19,20,21.
Regarding the impact of gut microbiota on the lungs, emerging epidemiological and experimental evidence supports the existence of the so-called gut-lung axis22. It has been proposed that the bidirectional communication between the gut and the lungs through the interaction and regulation of microorganisms, immune functions and metabolic products plays an important role in the pathogenesis and progression of various lung diseases including pulmonary fibrosis23. However, the specific bacterial species and mechanisms of interaction involved in the gut-lung axis during IPF pathogenesis remain unclear.
In the present study, a rat model of IPF was constructed via bleomycin administration. Compared to control animals, changes in peripheral blood and lung tissue physiological indices, as well as alterations in lung and gut microbiota composition, were analyzed in the constructed model. By examining the correlations between lung and gut microbiota profiles and physiological indices over the course of pulmonary fibrosis development, the potential mechanisms involving gut-lung microbiota interactions in the pathogenesis of IPF were explored.
Materials and methods
Animals and ethics statement
Sixteen male, 6-weeks-old, specific-pathogen-free (SPF) Sprague–Dawley (SD) rats weighing 250 ± 20 g were obtained from SPF Biotechnology Co., Ltd. (Beijing, China). Rats were housed in standard polypropylene shoebox cages (42 × 20.5 × 20 cm) on hardwood chip bedding in a designated room on alternate 12-h light/dark cycles at 24–26 °C and 50% humidity. They were allowed free access to water and fed a standard diet. All animal experiments were performed in accordance with Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. The study protocol was approved by the Ethics Committee of Minzu University of China (Beijing, China; No. ECMUC2023006AO). All experiments were performed in a Good Laboratory Practice (GLP)-accredited laboratory. All methods were carried out in accordance with relevant guidelines and regulations.
Bleomycin induced IPF model in rats
After adaptive feeding for 10 days, SD rats were randomly divided into 2 groups: control group (Ctrl) and model group (BLM), 8 rats in each group. The rats in model group were intracheally administrated with bleomycin (BLM) (5mg/kg) (Batch number: R25001, Beijing Saimo Fei’er Science & Technology Co., Ltd.) to induce IPF model. Rats in both groups were sacrificed for sample collection 49 days after modeling (supplemental Fig. 1).
Sample Collection
On day 49 after modeling, all rats were anesthetized with isoflurane and sacrificed by exsanguination after blood collection from the posterior vena cava. An aliquot of whole blood from each rat was used for blood routine examination, while the remaining blood samples were centrifuged at 3000 rpm for 10 min to obtain serum, which was then stored at -80 °C for subsequent cytokine detection. Immediately after opening the chest, the middle lobe of the right lung was dissected using sterile scissors and forceps. The dissected lung lobe was not de-hemoglobinized, but was thoroughly flushed with cold phosphate-buffered saline to remove residual blood before being placed in 5-ml sterile tubes containing 4 M guanidine thiocyanate solution as a preservative. The colon was transected 8–11 cm from the anus with a sterile instrument and the fecal samples were collected in sterile tubes containing 4 M guanidine thiocyanate solution. All samples of right middle lung lobe and feces in sterile tubes were stored in a -80 °C freezer for subsequent microbiota analysis. Finally, the left lung lobe was taken and fixed in 10% formaldehyde solution for subsequent histopathological examination.
Histopathological examination
The lung tissues were fixed in formaldehyde solution for approximately 12 h, embedded in paraffin, cut into 5μm thick sections, and stained with hematoxylin and eosin (H&E), as well as with Masson’s stain. The slides were observed under an optical microscope by a pathologist blinded to the treatments.
Quantification of hydroxyproline content in lung tissue
The HYP content in lung tissue samples was quantitatively detected using a hydroxyproline quantification assay kit (catalog number: A030-2–1, Nanjing Jiancheng Bioengineering Institute, China) according to the manufacturer’s instructions. Briefly, the tissue was hydrolyzed and neutralized with sodium hydroxide. Chloramine-T and dimethylaminobenzaldehyde were then sequentially added. Finally, a spectrophotometer was utilized to determine the absorbance at 550 nm, allowing evaluation of the HYP levels in the lung tissue.
Quantification of TGF-β1 levels in serum
Serum TGF-β1 levels were quantified by enzyme-linked immunosorbent assay (ELISA) technique using a specific TGF-β1 kit (D751002, Saiguo Biotech Engineering (Shanghai) Co., Ltd. China). The immunoassay was performed according to the manufacturer’s instructions. Each sample was assayed in duplicate and the respective mean values were computed.
Whole blood and serum analyses
An automated hematology analyzer (Sysmex, Tokyo, Japan) was utilized to detect hemoglobin (HB), red blood cells (RBCs), white blood cells (WBCs), platelets (PLTs), and other routine indicators, with a total of 24 routine indicators assessed. The Bio-Plex MAGPIX System (Bio-Rad) suspension microsphere chip platform and Bio-Plex Pro Rat Cytokine Group I Panel 23-Plex Kit were employed to detect 23 cytokines in the serum samples.
Microbiome sequencing
Total bacterial DNA was extracted using the Power Soil DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA). After quality and quantity checks of the DNA samples at 260/280 nm and 260/230 nm ratios, the bacterial 16S ribosomal RNA (rRNA) gene V3-V4 hypervariable region was amplified using common primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) combined with adapter and barcode sequences. Purified PCR products were subjected to second generation sequencing on an Illumina HiSeq 2500 platform (Illumina, Inc., San Diego, California; 2 × 250 paired-end reads) at Biomarker Technologies Corporation (Beijing, China).
Bioinformatics analysis
The raw sequences were input into the UNOISE pipeline of Usearch v11.0.667linux64 program (www.drive5.com/usearch/) and high-quality sequences were classified into zero-radius operational taxonomic units (ZOTUs). Taxonomic information for each ZOTU sequence was annotated at 80% confidence threshold using RDP classifier. The ZOTU table was rarefied (42,030 reads, minimum number of reads in a sample) to obtain equal sequencing depth across samples. Chao1 and Shannon indices were calculated using QIIME 1.91 pipeline. Differences in bacterial communities were assessed based on Bray–Curtis distance matrices. Distance-based redundancy analysis (db-RDA) were performed using R “vegan” package. Linear discriminant analysis effect size (LEfSe) analysis was used to compare phylum to genus level changes between groups and analyze species with significant differences (LDA ≥ 3). Correlation coefficients between variables were computed along with significance testing using R “psych” package.
Statistical analysis
The statistical analyses were performed using R software (version 4.2.1; R Foundation for Statistical Computing, Vienna, Austria) and OriginPro, (Version 2025, OriginLab Corporation, Northampton, MA, USA, URL link: https://www.originlab.com/ ). LSD test was used to assess differences in continuous variables between groups and Bonferroni test for multiple comparisons between groups. Spearman’s ρ correlation analysis was used to analyze associations between different variables.
Mediation analysis
To further assess the contribution of alteration in IPF related physiological indices to the correlation between lung microbiota and gut microbiota abundances, mediation analyses were conducted using the “mediation” package in R. By treating IPF-related physiological indices as the mediating variable (M), mediation analyses were conducted for two hypotheses: lung microbiota as predictor(X) and gut microbiota as outcome(Y); and gut microbiota as predictor(X) and lung microbiota as outcome(Y). The total effect (TE) and average causal mediation effect (ACME) values were determined between predictors and outcomes via the mediators, and the percent of mediation effect was estimated using a two-tailed P-value cutoff of < 0.05 to assess statistical significance.
Results
Model evaluation
H&E-stained sections revealed regular lung tissue structure and neatly arranged lung cells without obvious inflammatory cell infiltration in the control group. In the model group, lung tissue cells were misshapen with adhesions, nuclear disappearance, and abundant inflammatory cell infiltration in necrotic foci. In Masson-stained sections, collagen fibers were stained blue, whereas cytoplasm, muscle, cellulose and neuroglia were stained red, and nuclei stained dark blue. Masson staining displayed normal lung tissue cell structure with scarce collagen fibers in the control group. In contrast, severely damaged cell structure and extensive collagen fiber deposition were observed in the model group, indicating increased pulmonary fibrosis (Fig. 1 A).
Evaluation of the pulmonary fibrosis model. (A) Representative H&E staining and Masson’s trichrome staining lung sections (× 200) showing morphological changes in lung tissue. (B) Comparison of serum TGF-β1 levels among different rat groups. (C) Comparison of lung tissue HYP contents among different rat groups.
The results of HYP content in lung tissues and TGF-β1 levels in serum showed that the HYP content in lung tissues (Fig. 1B) and TGF-β1 levels in serum (Fig. 1C) were significantly higher in the model group compared to the control group (p < 0.05). This further confirms that the rats in the model group have developed pulmonary fibrosis.
Whole blood and serum analyses
Routine whole blood examination showed that 6 hematological indices changed significantly after BLM stimulation compared to the control group (Fig. 2). The model group showed significantly increased mean platelet volume (MPV) (Fig. 2A) and neutrophil percentage (NEU%) (Fig. 2B), while red blood cell distribution width standard deviation (RDW-SD) (Fig. 2C), mean corpuscular volume (MCV) (Fig. 2D), lymphocyte percentage (LYM%) (Fig. 2F) and plateletcrit (PCT) (Fig. 2G) were significantly decreased compared to the control group.
Changes in routine blood indices and serum cytokines in a bleomycin-induced rat model of pulmonary fibrosis. Of the 24 hematological indices examined, six showed significant changes in the model group compared to control: (A) mean platelet volume (MPV), (B) neutrophil percentage (NEU%), (C) red blood cell distribution width standard deviation (RDW-SD), (D) mean corpuscular volume (MCV), (E) lymphocyte percentage (LYM%), and (F) plateletcrit (PCT). Serum levels of four cytokines were also significantly altered in the model group: (G) IL-4, (H) IL-5, (I) IL-10, and (J) IFN-γ.
For serum cytokine detection, the levels of 4 cytokines changed significantly. Compared to the control group, the model group had significantly decreased IL-4 (Fig. 2G), IL-5 (Fig. 2H), IL-10 (Fig. 2I), and IFN-γ levels (Fig. 2J).
Overall assessment of lung microbiota
A total of 18,730 zero-radius operational taxonomic units (ZOTUs) were generated from 1,058,479 high-quality reads. Using the Ribosomal Database Project (RDP) classifier, 17,290 ZOTUs were successfully annotated at different taxonomic levels, including 22 phyla, 49 classes, 87 orders, 155 families and 289 genera.
Comparison of diversity of microbial communities among model and control group
For the α-diversity analysis, the Chao1 and Shannon indices were calculated to reflect species richness and evenness of each microbiota sample. The indices were compared between the control and model groups across different sampling sites (lung and gut). Overall, α-diversity indices of the lung microbiota were markedly lower than those of the gut microbiota, for both the control and model groups. In the comparison within the same sampling site, the results showed that compared to the control group, both diversity indices of the lung microbiota in the model group were significantly increased (P < 0.01). In contrast, the diversity of the gut microbiota showed no significant changes between the control and model groups (Fig. 3A, B).
To examine bacterial community structural differences between the model and control groups, Bray–Curtis distance matrices were generated from subsampled ZOTU abundance data separately for lung and fecal samples. Principal coordinate analysis (PCoA) based on the distance matrices revealed distinct clustering trends between the model and control groups for both lung and fecal samples. Additional distance-based redundancy analysis (db-RDA) confirmed the significant inter-group differences in microbiota structure, suggesting that the composition of genera in both lung and gut microbiota was substantially altered after pulmonary fibrosis development (Fig. 4).
Differences in microbiota composition among model and control group
In the comparison of lung microbiota between model and control groups, LEfSe analysis identified 45 differentially abundant taxa belonging to 14 genera. At the genus level, Prevotellamassilia, Bacteroides, Limosilactobacillus, Prevotella, Muribacter, Rodentibacter, Helicobacter, Clostridium_sensu_stricto, Mailhella, Parabacteroides, Perlucidibaca and Clostridium_XVIII were enriched in the model group, while Cetobacterium and Sutterella were depleted compared to the control (Fig. 5A).
In the comparison of gut microbiota between model and control groups, LEfSe analysis identified 22 discriminative taxa affiliated to 7 genera. Specifically at the genus level, the model group was significantly enriched with Elusimicrobium, Paramuribaculum, Allobaculum and Romboutsia, but depleted of Parasutterella, Muribaculum and Lactobacillus compared to the control (Fig. 5B).
Correlation analysis between lung and gut microbiota composition
To explore the potential correlations between lung and gut microbiota composition, Spearman correlation analysis was performed between lung and gut microbial genera that showed significant changes during pulmonary fibrosis modeling. The analysis identified 20 significant correlations (P < 0.05; |r|> 0.6) between 12 lung genera and 4 gut genera (Fig. 6A).
Correlation analysis between microbial genera and physiological indices significantly associated with IPF. (A) Heatmap showing the correlations between microbial genera from lung (X-axis) and gut (Y-axis) samples. (B) Heatmap showing the correlations between lung microbial genera and IPF-associated indices. (C) Heatmap showing the correlations between gut microbial genera and IPF-associated indices.
Among them, the lung genus Muribacter exhibited the most correlations with gut genera (4 correlations). In contrast, the gut genus Allobaculum showed the highest number of correlations with lung genera (11 correlations).
Correlation analysis between microbiota composition and IPF-related physiological indices
Further correlation analyses were performed between the above mentioned 16 genera (12 in lung and 4 in gut) and indices showing significant changes during IPF modeling, including TGF, HYP levels, blood routine indices and cytokine levels. The results found that 10 of the 12 differential lung genera exhibited 29 significant correlations with 6 indices related to pulmonary fibrosis development, including lung content of HYP, serum levels of TGF-β1 and IL-10, and hemogram indices of MCV, RWV-SD and MPV (Fig. 6B).
Meanwhile, among the 4 differential gut genera, 2 genera namely Allobaculum and Romboutsia showed 4 significant correlations with 4 indices related to IPF, including lung content of HYP, serum levels of TGF-β1, and hemogram indices of MCV and RWV-SD (Fig. 6C).
Mediation analysis of IPF-related indices on correlation between lung and gut microbiota composition
By summarizing the results of the associations between lung genera and gut genera, lung genera and IPF-related indices, and gut genera and IPF-related indices, we found that among the 20 identified pairs of associated lung and gut genera, 11 groups demonstrated concurrent significant associations with one or more IPF-related indices. These interrelated lung and gut genera and their commonly associated indices were summarized in Fig. 7A. As shown in the figure, a total of 24 triangle correlations consisting of "lung genus—index—gut genus" were identified.
Mediation analysis of the IPF-related indicators on association of lung-gut or gut-lung microbiota interactions. A) Interrelated lung (X-axis) and gut (Y-axis) genera and their commonly associated indices. B) The significant mediation effect of IPF-related indices (including mean corpuscular volume, MCV; and HYP) on the correlations between specific lung genera (including Mailhella, Prevotella, Muribacter, Perlucidibaca, Prevotellamassilia, Clostridium_XVIII, Rodentibacter, Parabacteroides, Bacteroides, Cetobacterium) and the gut genus Allobaculum. C) The significant mediation effect of IPF-related indices (including MCV and red cell distribution width standard deviation, RDW-SD) on the correlation between the gut genus Allobaculum and the lung genus Clostridium_XVIII. ADE (Average Direct Effect), ACME (Average Causal Mediation Effect), * indicates p < 0.05, ** indicates p < 0.01.
To evaluate whether the IPF-related indices play a mediating role in these triangle paths representing lung-gut microbiota interactions, mediation analyses were conducted treating the indices as mediators and switching the microbiota variables between predictor and outcome positions. Specifically, the IPF-related physiological indices were treated as the mediating variable, the lung microbiota was treated as the predictor and the gut microbiota as the outcome in one analysis, and vice versa in the other analysis.
In the result, out of the 48 potential mediating effects (24 lung-gut and 24 gut-lung), 12 significant mediating effects of the IPF-related indices were found. Among them, 10 involved the IPF-related indices mediating the association between lung genera and gut genera (Fig. 7B), and 2 involved the IPF-related indices mediating the association between gut genera and lung genera (Fig. 7C).
Specifically, in the setting of lung microbiota as the predictor, and gut microbiota as the outcome, the mediation effect of MCP on the correlation between the abundances of various lung genera (Mailhella, Prevotella, Muribacter, Perlucidibaca, Prevotellamassilia, Clostridium_XVIII, Rodentibacter, Parabacteroides and Bacteroides) and the gut genus Allobaculum was statistically significant. The mediation effect of HYP on the correlation between the abundance of the lung genus Cetobacterium and the gut genus Allobaculum was also statistically significant. And in the setting of gut microbiota as the predictor, and lung microbiota as the outcome, the mediation effect of MPV and RDW-SD on the correlation between the abundance of the gut genus Allobaculum and the lung genus Clostridium_XVIII was statistically significant. This suggests that these physiological markers may play a mediating role in the above lung-gut or gut-lung microbial abundance associations.
Discussion
The main findings of this study reveal that in the context of pulmonary fibrosis, significant changes occurred in the microbial community structures and compositions in both the lung and gut. Correlations were observed among multiple lung and gut genera as well as IPF-related physiological indices. These results suggest that the composition and interaction of lung and gut microbiota may play a role in pulmonary fibrosis pathogenesis. Furthermore, mediation analysis demonstrated that several IPF-related indices played substantial mediating roles in the associations between lung and gut genera, lending further support to the gut-lung axis concept, shedding light on elucidating the specific mechanisms underlying lung-gut interactions in pulmonary fibrosis.
After 49 days of bleomycin stimulation in rat lungs, significant fibrotic changes were observed, as evidenced by increased hydroxyproline content, extensive infiltration of inflammatory cells, and collagen deposition. These changes confirm the model’s effectiveness in inducing lung fibrosis. Additionally, a series of hematological and immunological indices, including MPV, MCV, RDW-SD, NEUT%, LYMPH%, PCT, IL-4, IL-5, IL-10, IFN-γ, and TGF-β1, showed significant alterations. These systemic changes indicate that the model recapitulates the multi-systemic pathophysiological characteristics of human IPF.
Changes in MPV, RDW-SD, and MCV are particularly relevant. MPV alterations have been associated with lung function decline, and can indicate IPF severity and prognosis24. Abnormal RDW levels correlate positively with IPF severity, while MCV changes reflect red blood cell distribution status25,26. The observed increase in NEUT% and decrease in LYMPH% suggest a persistent inflammatory response, which may exacerbate fibrosis through the release of pro-inflammatory factors and neutrophil elastase27. Elevated levels of IL-4, IL-5, IL-10, and the pro-fibrotic cytokine TGF-β1 further reflect the inflammatory and fibrotic processes. The decrease in IFN-γ levels aligns with previous reports in IPF patients28, highlighting the cytokine’s role in disease progression.
In addition to the aforementioned pathophysiological changes, we also observed significant alterations in lung and gut microbiota. Analysis revealed an elevated lung microbiota α-diversity in the pulmonary fibrosis model compared to the control group, consistent with previous reports of increased lung microbiota diversity in IPF patients that correlates with disease status29. The possible explanation for the increased microbiota diversity in fibrotic lung tissues is that obstruction of native clearance mechanisms against exogenous microbes, due to lung tissue damage, may result in aggregation of environmental microorganisms in the lungs, which could, in turn, lead to an increase in the microbiota α-diversity. With regard to gut microbiota, previous studies have reported no significant difference in the alpha diversity of gut microbiota in silica and bleomycin-induced pulmonary fibrosis models. However, significant differences were observed in the β-diversity between lung and gut microbiota genera between bleomycin and control groups17. Our results are consistent with these findings. In our study, no noticeable changes were detected in the α-diversity of gut microbiota after modeling, yet the β-diversity of gut and lung microbiota structures exhibited significant variations. This indicates that substantial modifications had occurred in the composition and abundance of both lung and gut microbiota.
To further investigate the microbial compositional basis for the changes in lung tissue and gut microbiota structure after modeling, LEfSe analysis was utilized to identify differential taxa between the model group and control group. The results showed that the abundance of 14 lung genera and 7 gut genera were found to be significantly different between the model group and control group. Current studies on lung microbiota have shown that Bacteroides and Prevotella can promote pulmonary fibrotic pathogenesis via IL-17R signaling30. Helicobacter can induce production of IL-8 and IL-6 in lung cells, participating in fibrosis development and decline of lung function in IPF patients31. Clostridium sensu stricto significantly correlates with the inflammatory marker CRP32, indicating its potential role in the inflammatory stage of pulmonary fibrosis. Parabacteroides possesses glycose-fermenting abilities and positively correlates with IL-6 and TNF-α, significantly elevated in radiation-induced pulmonary fibrosis models33. Additionally, studies have found Sutterella significantly decreases in IPF patients34. Our results align with these findings. Regarding differential gut genera, studies have reported that Elusimicrobium positively correlates with proinflammatory factors and is highly expressed in COVID-19 rat models35. Allobaculum is significantly elevated in the gut of cystic fibrosis patients36. Romboutsia is a commensal bacterium with anti-inflammatory properties, and is markedly increased in the gut of bleomycin-induced pulmonary fibrosis mice models37. Additionally, Muribaculum and Lactobacillus were found to decrease in the gut of radiation-induced pulmonary fibrosis models33. The above results are also consistent with our findings, indicating that the compositional changes of gut and lung microbiota in our IPF model align with characteristics of IPF, and suggest these multiple genera in lung and gut tissues may participate in IPF progression through different aspects.
Furthermore, this study’s results indicate that during the development of pulmonary fibrosis, the abundance of several differential lung and gut genera was significantly correlated with IPF-related physiological indicators. Among these microbiota-associated indicators, HYP content is one of the important criteria for judging pulmonary fibrosis developmen38. We found that lung abundance of Cetobacterium was negatively correlated with HYP content. Previous research have shown that gut Cetobacterium can catalyze anaerobic degradation of HYP via HypD39. Therefore, we speculate lung Cetobacterium may play a role in degrading HYP, and reducing HYP accumulation by regulating lung Cetobacterium levels could have potential therapeutic value for IPF. In addition, we found that lung Cetobacterium was also significantly correlated with serum TGF-β1 levels, MCV and RDW-SD. TGF-β1 is a key promoter of pulmonary fibrogenesis. MCV and RDW-SD are important hematological indicators reflecting red blood cell distribution40. The significant correlations between lung Cetobacterium and these critical factors involved in pulmonary fibrosis pathogenesis further highlight the potentially profound connection between Cetobacterium and disease progression. Therefore, comprehensively exploring the intrinsic link between lung Cetobacterium and pulmonary fibrosis development is of great significance.
Among the physiological indices associated with microbiota, MCV and RDW-SD MCV and RDW-SD exhibited the strongest correlations with a diverse range of genera, spanning both gut and lung microbiota. This suggests a pivotal role for MCV and RDW-SD in shaping the intricate interplay between gut and lung microbiota. The “gut-lung axis” theory proposes that the gut and lung can exert long-distance bidirectional interactions on immune functions through commensal microorganisms and their metabolites, forming complex interconnections between respiratory microbiota, gut microbiota, and respiratory and digestive diseases41. However, the precise microbial taxa and metabolites responsible for mediating the "gut-lung axis," as well as the intricate mechanisms governing their interactions, remain largely elusive, especially the role of the “gut-lung axis” in the pathogenesis of IPF which still holds many unknowns.
By summarizing the correlations between gut and lung microbiota, lung microbiota and physiological indices, as well as gut microbiota and physiological indices, this study identified 24 triangle correlations consisting of “lung genus—index—gut genus”. Furthermore, mediation analysis revealed 12 significant mediation effects of physiological indices in the gut-lung interaction, offering insights into potential mechanisms of gut-lung microbiota interaction. For instance, the results showed that lung HYP levels played significant mediating role in the correlation between lung Cetobacterium and gut Allobaculum abundance. Given that Cetobacterium has HYP degradation activity39, its reduction may facilitate HYP deposition in the lung. In pulmonary fibrosis animal models, elevated lung HYP levels are usually accompanied by increased blood HYP levels42,43, suggesting the possibility that elevated blood HYP levels can affect the distal gut to promote the enrichment of gut Allobaculum. Indeed, current studies have shown that HYP can inhibit IL-6 expression through NF-κB signal transduction and alleviate DSS-induced colonic injury in mice, suggesting it may have the ability to influence gut microbiota44. The mediation analysis also showed that MCV exhibited significant mediating effects in the correlations between 9 lung genera and gut Allobaculum abundance. At the same time, MCV and RDW-SD together played significant mediating roles in the correlation between gut Allobaculum and lung Clostridium XVIII abundance. Decreased MCV is associated with microcytic anemia, while decreased RDW-SD may indicate more uniform red blood cell size and shape, both of which can affect oxygen transport45. Considering Allobaculum and Clostridium XVIII are strict anaerobes, their significant enrichment in the model group may be related to the hypoxic state. This notion is supported by evidence showing that causing local hypoxia by enhancing the oxygen metabolism of intestinal epithelial cells, thereby stimulating the growth of anaerobic bacteria, is a key mechanism mediating the interactions between intestinal microbial communities46,47. Although the mediating effects described above are inferences based on statistical data, they still provide valuable hints for further elucidating the specific mechanisms of gut-lung microbiota interaction. However, it’s crucial to emphasize that these specific causal mechanisms will necessitate further experimental validation.
In summary, this study discovered several differential lung and gut genera associated with the occurrence of pulmonary fibrosis. By analyzing the correlations between these lung and gut genera as well as their associations with host physiological indices, we first described the phenomenon of gut-lung interaction at the microbial level during pulmonary fibrosis, and explored some of the potential pathogenesis mechanisms from the perspective of interactions between microbes and the host. Furthermore, mediation analysis in this study proposed several possible pathways underlying the correlations of lung genus—host index—gut genus and gut genus—host index—lung genus, providing clues to further elucidate the mechanisms of the “gut-lung axis”. Although these findings are intriguing, it is important to acknowledge that the small sample size may have affected the statistical power of the study. To substantiate these findings, future research with more extensive sample sizes is necessary, allowing for a clearer understanding of the gut-lung axis in the context of pulmonary fibrosis.
Data availability
The datasets generated and/or analyzed during the current study are available in the SRA database repository, Accession No: PRJNA1028659.
References
Sergio, H. et al. Epidemiology of idiopathic pulmonary fibrosis: a population-based study in primary care. Intern. Emerg. Med. 15(3), 437–445 (2020).
Biondini, D. et al. Acute exacerbations of idiopathic pulmonary fibrosis (AE-IPF): an overview of current and future therapeutic strategies. Expert Rev Respir Med 14(4), 405–414 (2020).
Carrington, R. et al. Use of animal models in IPF research. Pulm Pharmacol Ther 51, 73–78 (2018).
Liu, T., F.G. De Los Santos, and S.H. Phan. The Bleomycin Model of Pulmonary Fibrosis. Methods Mol Biol. 1627, 27–42 (2017).
Han, M. K. et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med 2(7), 548–556 (2014).
Molyneaux, P. L. et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 190(8), 906–913 (2014).
O’Dwyer, D. N. et al. Lung Microbiota Contribute to Pulmonary Inflammation and Disease Progression in Pulmonary Fibrosis. Am J Respir Crit Care Med 199(9), 1127–1138 (2019).
Molyneaux, P. L. et al. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 195(12), 1640–1650 (2017).
Wu, Y. et al. Gut microbiome and metabolites: The potential key roles in pulmonary fibrosis. Front Microbiol 13, 943791 (2022).
Shi, H. et al. The associations between gut microbiota and chronic respiratory diseases: a Mendelian randomization study. Front Microbiol 14, 1200937 (2023).
Quan, Y. et al. The gut-lung axis: Gut microbiota changes associated with pulmonary fibrosis in mouse models induced by bleomycin. Front Pharmacol 13, 985223 (2022).
Zhou, Y. et al. Alterations in the gut microbiota of patients with silica-induced pulmonary fibrosis. J Occup Med Toxicol 14, 5 (2019).
Wei, Y. et al. Astragalus polysaccharide attenuates bleomycin-induced pulmonary fibrosis by inhibiting TLR4/ NF-kappaB signaling pathway and regulating gut microbiota. Eur J Pharmacol 944, 175594 (2023).
Mercader-Barcelo, J., et al. Insights into the Role of Bioactive Food Ingredients and the Microbiome in Idiopathic Pulmonary Fibrosis. Int J Mol Sci. 21(17) (2020).
Chioma, O. S. et al. Gut microbiota modulates lung fibrosis severity following acute lung injury in mice. Commun Biol 5(1), 1401 (2022).
Ashique, S. et al. Short Chain Fatty Acids: Fundamental mediators of the gut-lung axis and their involvement in pulmonary diseases. Chem Biol Interact 368, 110231 (2022).
Gong, G. C., Song, S. R. & Su, J. Pulmonary fibrosis alters gut microbiota and associated metabolites in mice: An integrated 16S and metabolomics analysis. Life Sci 264, 118616 (2021).
Chen, B. et al. Chronic microaspiration of bile acids induces lung fibrosis through multiple mechanisms in rats. Clin Sci (Lond) 131(10), 951–963 (2017).
Bai, L. et al. Glutaminolysis Epigenetically Regulates Antiapoptotic Gene Expression in Idiopathic Pulmonary Fibrosis Fibroblasts. Am J Respir Cell Mol Biol 60(1), 49–57 (2019).
Fang, L. et al. Endogenous tryptophan metabolite 5-Methoxytryptophan inhibits pulmonary fibrosis by downregulating the TGF-beta/SMAD3 and PI3K/AKT signaling pathway. Life Sci 260, 118399 (2020).
Li, J. M. et al. Therapeutic targeting of argininosuccinate synthase 1 (ASS1)-deficient pulmonary fibrosis. Mol Ther 29(4), 1487–1500 (2021).
Saint-Criq, V., Lugo-Villarino, G. & Thomas, M. Dysbiosis, malnutrition and enhanced gut-lung axis contribute to age-related respiratory diseases. Ageing Res Rev 66, 101235 (2021).
Fabbrizzi, A. et al. Microbiota and IPF: hidden and detected relationships. Sarcoidosis Vasc Diffuse Lung Dis 38(3), e2021028 (2021).
Ntolios, P. et al. Mean Platelet Volume as a Surrogate Marker for Platelet Activation in Patients With Idiopathic Pulmonary Fibrosis. Clin Appl Thromb Hemost 22(4), 346–350 (2016).
Xu, W. et al. Association between red blood cell distribution width to albumin ratio and prognosis of patients with sepsis: A retrospective cohort study. Front Nutr 9, 1019502 (2022).
Karampitsakos, T. et al. Increased monocyte count and red cell distribution width as prognostic biomarkers in patients with Idiopathic Pulmonary Fibrosis. Respir Res 22(1), 140 (2021).
Mei, Q. et al. Idiopathic Pulmonary Fibrosis: An Update on Pathogenesis. Front Pharmacol 12, 797292 (2021).
Tsoutsou, P. G. et al. Cytokine levels in the sera of patients with idiopathic pulmonary fibrosis. Respir Med 100(5), 938–945 (2006).
Molyneaux, P. L. et al. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir Res 18(1), 29 (2017).
Yang, D., et al. Dysregulated Lung Commensal Bacteria Drive Interleukin-17B Production to Promote Pulmonary Fibrosis through Their Outer Membrane Vesicles. Immunity. 50(3), 692–706 e7 (2019).
Nakashima, S. et al. Identification of Helicobacter pylori VacA in human lung and its effects on lung cells. Biochem Biophys Res Commun 460(3), 721–726 (2015).
Li, K. J. et al. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir Res 20(1), 272 (2019).
Li, W. et al. Effects of phycocyanin on pulmonary and gut microbiota in a radiation-induced pulmonary fibrosis model. Biomed Pharmacother 132, 110826 (2020).
Tong, X. et al. Alterations to the Lung Microbiome in Idiopathic Pulmonary Fibrosis Patients. Front Cell Infect Microbiol 9, 149 (2019).
Sencio, V. et al. Alteration of the gut microbiota’s composition and metabolic output correlates with COVID-19-like severity in obese NASH hamsters. Gut Microbes 14(1), 2100200 (2022).
Trandafir, L.M., et al. Can Bioactive Food Substances Contribute to Cystic Fibrosis-Related Cardiovascular Disease Prevention? Nutrients. 15(2) (2023).
Hu, H. et al. Bu-Fei-Huo-Xue capsule alleviates bleomycin-induced pulmonary fibrosis in mice through modulating gut microbiota. Front Pharmacol 14, 1084617 (2023).
Ren, Y. et al. Simple determination of L-hydroxyproline in idiopathic pulmonary fibrosis lung tissues of rats using non-extractive high-performance liquid chromatography coupled with fluorescence detection after pre-column derivatization with novel synthetic 9-acetylimidazol-carbazole. J Pharm Biomed Anal 142, 1–6 (2017).
Huang, Y.Y., A. Martinez-Del Campo, and E.P. Balskus, Anaerobic 4-hydroxyproline utilization: Discovery of a new glycyl radical enzyme in the human gut microbiome uncovers a widespread microbial metabolic activity. Gut Microbes. 9(5), 437–451 (2018).
Brzezniakiewicz-Janus, K. et al. Red Blood Cells Mean Corpuscular Volume (MCV) and Red Blood Distribution Width (RDW) Parameters as Potential Indicators of Regenerative Potential in Older Patients and Predictors of Acute Mortality - Preliminary Report. Stem Cell Rev Rep 16(4), 711–717 (2020).
Enaud, R. et al. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front Cell Infect Microbiol 10, 9 (2020).
Gao, H. G. et al. Increased serum and musculotendinous fibrogenic proteins following persistent low-grade inflammation in a rat model of long-term upper extremity overuse. PLoS One 8(8), e71875 (2013).
Pourgholamhossein, F. et al. Thymoquinone effectively alleviates lung fibrosis induced by paraquat herbicide through down-regulation of pro-fibrotic genes and inhibition of oxidative stress. Environ Toxicol Pharmacol 45, 340–345 (2016).
Ji, Y. et al. Hydroxyproline Attenuates Dextran Sulfate Sodium-Induced Colitis in Mice: Involvment of the NF-kappaB Signaling and Oxidative Stress. Mol Nutr Food Res 62(21), e1800494 (2018).
Li, N., Zhou, H. & Tang, Q. Red Blood Cell Distribution Width: A Novel Predictive Indicator for Cardiovascular and Cerebrovascular Diseases. Dis Markers 2017, 7089493 (2017).
Byndloss, M. X. et al. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357(6351), 570–575 (2017).
Pral, L. P. et al. Hypoxia and HIF-1 as key regulators of gut microbiota and host interactions. Trends Immunol 42(7), 604–621 (2021).
Funding
The study was funded by the foundation of the Key Laboratory of Ethnomedicine (Minzu University of China), the Ministry of Education (KLEM-ZZ2020GD01), Ningxia Natural Science Foundation Project (2021AAC03358) and Ningxia Autonomous Region Key Research and Development Program (2023BEG02025).
Author information
Authors and Affiliations
Contributions
Sihan Hou conducted experiments and wrote the manuscript, Xueer Wang participated in writing the manuscript, and all authors participated in reviewing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was conducted with approval from the Ethics Committee of Minzu University of China (ECMUC2023006AO).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hou, S., Wang, X., Guo, J. et al. Triangle correlations of lung microbiome, host physiology and gut microbiome in a rat model of idiopathic pulmonary fibrosis. Sci Rep 14, 28743 (2024). https://doi.org/10.1038/s41598-024-80023-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-80023-y