Abstract
Understanding how mechanical stress affects erythropoiesis is crucial to produce transfusable erythrocytes in fluid-turbulent bioreactors. We investigated the effects of shear-stress on differentiating CD49d+CD235a+ primary human erythroblasts (EBL) at molecular, cellular, and functional level. Shear-stress, at differentiation onset, enhanced EBL maturation and induced upregulation of genes regulating cholesterol/lipids biosynthesis, causing changes in cell lipid composition. Of note, the osmotic resistance, and the expression of 3-Hydroxy-3-methylglutaryl-CoA reductase (HMGCR), the rate-limiting enzyme of the cholesterol biosynthesis pathway, were higher in dynamic cultures. Inhibition of the S1P-induced proteolytic cleavage, activating SREBPs, led to abrogation of HMCGR expression, and loss of EBL in dynamic cultures, similar to lovastatin administration. This data reveals a role for the S1P-SREBP-HMGCR-axis in the regulation of shear-stress induced adaptation during erythropoiesis, shedding light into mechanisms that will assist the upscaling of erythroid differentiation into bioreactors. Moreover, as shear-stress on hematopoietic cells occurs within the bone-marrow, these results introduce a novel signalling axis in the transduction pathways controlling erythropoiesis.
Similar content being viewed by others
Introduction
Red blood cell (RBC) transfusion is the most common form of cell therapy, yet the practice has remained relatively unchanged for decades1. Although lifesaving, this donor-dependent process carries risks such as alloimmunization and requires prevention of blood-borne diseases spread, while donor-recipient matching limits inventory for certain receiver, based on ABO group or minor/rare antigen mismatch. Thus, the medical need for large-scale production of erythrocytes lacking immunogenic blood-group antigens is evident2. Moreover, cultured RBCs (cRBC) hold great therapeutic potential as carriers for therapeutic molecules like enzymes or inducers of tolerance3. The production of functional, enucleated RBC is feasible in static and turbulent environments45678. Nevertheless, the upscaling of RBC cultures to produce clinically relevant numbers in a cost-effective manner remains a challenge.
Stirred bioreactors are effective for large scale erythroid cultures, similar to upscaling efforts for other mammalian cells, but the turbulent nature of these systems leads the cells to experience a range of distinct mechanical forces4567. These forces vary depending on factors like suspension density, mixing speed, fluid volume, and air sparging strategies8, and may act on mechano-sensors expressed and functional during erythropoiesis9101112. Downstream of these mechano-sensors, specific signal transduction pathways can be induced, including MAPKs, STAT5, and NFATs, which modulate erythroblast (EBL) specification from hematopoietic stem cells11, reticulocyte maturation13, integrin activation9and regulate proliferation potential10. Here we investigated how shear-stress affects terminal erythroblast differentiation, via orbital shakers, commonly used in culture of other mammalian cells, and extensively investigated in terms of fluid dynamics and mechanically-induced biological effects141516. The RBC plasma membrane consists of a lipid bilayer with embedded proteins attached to the underlying membrane skeleton. A variety of lipid and phospholipid species represent the core structure of this membrane, among which cholesterol is the most abundant17. The ratio between cholesterol and phospholipids, controls cell membrane rigidity and fluidity18. Cholesterol concentration in RBC plasma membranes affects cell morphology and functionality: increased plasma membrane cholesterol influences RBC shape and plasma membrane functions such as permeability and transport and impairs RBC flow properties18,19. Cell viability in turbulent conditions is highly dependent on plasma membrane fluidity (PMF). Eukaryotic cells are capable of adaptive responses to changing membrane lipid content ensuring optimal PMF by de novo lipid biosynthesis20. Interestingly, it has been shown that lipid membrane composition changes in shear flow environments are linked to mechanosensing21..
Here we show that erythroid differentiation is accelerated upon orbital shaking. Adaptation to shear-stress is reflected by an altered gene expression profile including upregulation of cholesterol metabolism pathways, confirmed by lipidomic analysis. We uncovered that the rate-limiting enzyme HMGCR (3-Hydroxy-3-methylglutaryl-CoA reductase) of the cholesterol synthesis plays a central role in this phenomenon. Moreover, we described that HMGCR regulation is dependent on the cleavage and subsequent activation of SREBPs (Sterol-regulatory- element-binding-proteins) by S1P (site-1-protease) during erythroblast adaptation to shear-stress.
This data provides the first analysis of the (transcriptional) program induced by culturing erythrocyte precursors in shear-stress environments. Importantly, this response was not detrimental to erythroid differentiation but changed RBC membrane properties and was paramount to the adaptive response to shear environments. Taken together, this study provides new insights in some adaptive molecular mechanisms that will assist in the upscaling of cRBC production, which can be employed for clinical use.
Results
Orbital shaking accelerates erythroid maturation
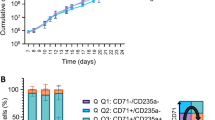
In static conditions, erythroblast (EBL) differentiation to enucleated reticulocytes is accompanied by a progressive loss of CD49d and increase in CD235a expression, taking approximately 10 days (Fig. 1A). We exposed EBL to orbital shaking to generate an average shear-stress of 1.4 Pa, comparable to the shear-stress at the point of the impeller of the bioreactors used4(estimation based on Odeleye22), but also to what can occur in the bone marrow microenvironment, as reviewed before23. Both static and dynamic cultures contained similar CD49d-/CD235a+ population at the end of the differentiation, however dynamic conditions showed significantly faster differentiation kinetics, reaching between 80%–90% CD49d-/CD235a+ cells 4 days earlier compared to static cultures (Fig. 1A, B). A significant increase in enucleated reticulocytes (DRAQ5- cells) was observed at day 4 and 6 in dynamic cultures compared to static cultures, which eventually progressed to similar enucleation percentages at the end of culture (Fig. 1C, D; Supplemental Fig. 1A, B)24. Faster differentiation was further confirmed by morphological analysis (Supplemental Fig. 1C). At the end of differentiation, dynamic cultures demonstrated a slight decrease – yet not significant – in EBL and reticulocytes production (Fig. 1E). Together these results indicate that orbital shaking leads to accelerated differentiation of EBL to enucleated reticulocytes.
Orbital shaker accelerates maturation and enucleation compared to static cultures. Erythroblasts (EBL) were differentiated for up to 10 days in dishes (static) or orbital shakers (dynamic). A) Erythroid cells were harvested at indicated days from dishes and flasks and stained with antiCD49d-PB (BD Biosciences, San Jose, CA, US ) (Alexa Fluor 405, y-axis) and antiCD235a-PE (OriGene Technologies, Rockville, MD, USA) (x-axis), and analysed by flow cytometry. B) Maturation states in static vs. dynamic conditions as explained in (A) were averaged for 5 donors (n = 5) by quantifying the percentage of cells within the respective gates Q1: CD49d+/CD235- immature EBL, Q2: CD49+/CD235a+ EBL, and Q3: CD49-/CD235a+ late EBL and reticulocytes. C) Enucleation was quantified by staining cells with the cell-permeable DNA-dye DRAQ5 (1 ng/mL, incubation 5 min at RT; BioLegend, San Diego, CA, USA). (APC, y-axis), forward scatter (FSC, x-axis) on the indicated days. Representative scatter plot displaying the gating for reticulocytes, EBL, and pyrenocytes is shown in Supplemental Fig. 1A. Enucleation percentage is defined as the %reticulocyte/ (%reticulocytes + %nucleated cells) using the gates as defined in Supplemental Fig. 1A. D) Enucleation states in static vs. dynamic conditions as explained in (C) were averaged for 5 donors (n = 5) by quantifying the percentage of cells within the respective gates: reticulocytes, erythroblasts, and pyrenocytes for 5 donors (n = 5). E) Cell-counts for reticulocytes and EBL that were differentiated in static or dynamic conditions for 4 donors (n = 4). The distinction between the two populations was done via DRAQ5 staining. B) 2way ANOVA test was performed, with ***p < 0.001, ** p < 0.01 and *p < 0.05. D and E) Paired two-tailed student t-test was performed, with p** p < 0.01 and *p < 0.05. Unless marked, no significance was observed.
Orbital shaking accelerates erythroblast differentiation immediately after induction
Next, we investigated whether dynamic conditions expedite differentiation at a specific maturation stage. Differentiating EBL were switched between static and dynamic conditions, as described in methods, at different time points and the enucleation rate was measured via flow cytometry. In line with Fig. 1, an inverse correlation between enucleated cells and the initial dynamic culture time was observed (Fig. 2).
Shear-induced enhanced maturation is progressively lost during erythroblast differentiation. A) Enucleation percentage (calculated as described for Fig. 1C) of samples switched from dynamic to static culture on Day 2, 4, or 6 of differentiation, or not switched, for 3 donors (n = 3). B) Enucleation percentage of samples switched from static culture to dynamic culture on Day 2, 4 or 6 of differentiation, as well as dish (static) and flask (dynamic) controls averaged for 3 donors (n = 3). Paired two-tailed student t-test was performed, with ** p < 0.01 and *p < 0.05. Unless marked, no significance was observed.
Cultures switched from orbital shaking to static conditions on day 2 showed slower enucleation kinetic compared to the samples switched on day 4. Importantly, all conditions reached comparable levels of enucleation indicating that the static cultures are not blocked in differentiation but that orbital shaking accelerates the process (Fig. 2A).
In the reverse experiment, transferring static cultures to dynamic conditions on day 2 and day 4 accelerated enucleation compared to the static control, but still lagged compared to cells under continuous orbital shaking. Switching at day 6 to orbital shaking conditions did not accelerate enucleation, suggesting that the regulation of accelerated differentiation occurs before day 6 (Fig. 2B). Together the data indicate that the orbital shaking induced accelerated differentiation occurs during the early phase of EBL maturation, between day 0 and 4.
Shear stress alters the erythroid transcriptome inducing HMGCR expression and cholesterol biosynthesis pathways
To start dissecting the underlying mechanism we performed transcriptomic analysis between static and dynamic conditions during the first 4 days of EBL differentiation.
Principal component analysis (PCA) demonstrated samples cluster per differentiation condition and time, where day 4 demonstrated the biggest difference (Supplemental Fig. 1D). Differential expression analysis between groups demonstrated many differentially expressed genes (DEGs) upon EBL differentiation, with 505 DEGs at day 4 of differentiation (Supplemental Fig. 1E, F; Supplemental table 1–3).
Pearson hierarchical and K-means clustering of these DEGs revealed 3 different gene expression profiles, assigned to 3 major clusters (Fig. 3A). Genes assigned to the first cluster (K1), showed an overall downregulation during differentiation in both static and dynamic cultures, but the downregulation levels were increased in dynamic conditions (Fig. 3A). The second cluster (K2) shows up-regulated genes in dynamic cultures; the third cluster (K3) represents up-regulated genes in static conditions.
RNA analysis between static and dynamic erythroblast differentiation confirms accelerated maturation and characterises the involved processes. Erythroblasts (EBL) of four different donors (n = 4) were differentiated for 4 days as indicated in material and methods. A) Heatmap showing a Pearson hierarchical clustering of z-scores of the 505 differentially expressed RNAs between Day1 and Day4. K-means clustering, reveals 3 clusters indicated as K1, K2, K3. Genes involved in the cholesterol biosynthesis pathway are reported next to their specific coordinates on the heatmap. B) The gene identifiers of the upregulated RNAs of K2 from (A) were extracted and their expression dynamics through complete differentiation of EBL to enucleated reticulocytes, as previously published by Heshusius24, was datamined and determined. Pearson hierarchical clustering was performed on z-scores over time in days as indicated (x-axis). Kmeans clustering (K = 3) was additionally performed and indicated as K2-a, K2-b and K2-c. C) ENRICHR analysis of K2-a to K2-c from (B) shows the top 10 enriched Gene Ontology (GO) term biological processes that are associated with the RNAs in the indicated Kmeans cluster according to the adjusted p-value. D) STRING analysis of the genes within K2-b was performed and genes within the first 3 biological processes according to the false discovery rate highlighted in red, blue, and green. Lines thickness indicates the strength of data support. E) Representation of cholesterol biosynthesis pathway with related z-score of genes up-regulated on day 4 in dynamic (D) compared to static (S) conditions. F) HMGCR (ab242315, Abcam) expression was assessed through western blot analysis, during EBL differentiation of 3 different donors (n = 3) in static and dynamic conditions. GAPDH (MAB374, Millipore) was used as loading control. G) Quantified expression of HMGCR during cell differentiation in dynamic and static conditions. HMGCR expression was normalised according to GAPDH expression. G) 2way ANOVA test was performed, with ** p < 0.01 and *p < 0.05. Unless marked, no significance was observed.
Gene ontology analysis revealed that genes in K1 and K3 are enriched for DNA replication and nucleosome organisation processes, respectively (Supplemental Fig. 2A). Moreover, K3 genes during differentiation revealed a cluster of genes upregulated during differentiation (Supplemental Fig. 2C). The differentiation-associated gene expression profile in K1 and K3 is in line with previously reported dataset showing similar gene expression dynamics during differentiation to enucleated reticulocytes (Supplemental Fig. 2B and C24).
In contrast, transcripts within K2 were down-regulated on day 4 in static conditions but up-regulated in dynamic environments. To assess the expression of these genes throughout the complete differentiation to reticulocytes, we compared our data more closely to those from Heshusius24 and re-clustered them based on their gene expression profile, giving rise to 3 additional clusters (K2-a/b/c, Fig. 3B).
The top 10 regulated biological processes according to the adjusted p-value (EnRicHR) revealed that genes in K2-a regulate response to heat and to unfolded protein and chaperone cofactor- dependent protein refolding and are upregulated starting from ~ day 7 of differentiation in static condition. Genes within K2-c are involved in the regulation of cell migration and response to ER stress and are upregulated earlier in differentiation, between day 5 and 6. In contrast, K2-b demonstrated a strong enrichment for genes mostly involved in cholesterol/steroid metabolism being up-regulated in dynamic conditions but downregulated during differentiation in static coltures (Fig. 3C). K2-b STRING analysis confirmed the presence of a cluster of genes involved in secondary alcohol metabolic and cholesterol biosynthetic processes, among which also HMGCR, a rate limiting enzyme of the cholesterol biosynthesis pathway and fundamental coordinator of mevalonate metabolism that assures constant de novo synthesised cholesterol and non-sterol products (Fig. 3D)25. Within the cholesterol biosynthesis pathway, the enzymes HMGCS1 and HMGCR, IDI1, SQLE and HSD17B7 showed a similar trend, being upregulated in dynamic conditions (Fig. 3E). Of note, the protein expression of HMGCR peaked at day 4 and 5 of differentiation, where peak expression during dynamic conditions occurs during day 1 and 2 and decreased on day 3 and 4 (Fig. 3F and Supplemental Fig. 2D), providing further proof of the accelerated maturation in dynamic cultures. Importantly, the high expression of HMGCR detected on day 1 and day 2 of differentiation in dynamic conditions was never reached in static cultures, suggesting an adaptation response of the cells to the shear-stress, and validating the increase in gene expression observed by RNA-sequencing (Fig. 3G). In all, these data confirm accelerated differentiation of EBL upon dynamic conditions and provide a transcriptional and possible functional footprint of downstream effects of shear-induced cholesterol transcriptional program.
Lipidomic analysis of erythroblasts confirms a differential lipid composition between dynamic and static conditions
To assess the effect of the upregulated lipid transcriptional program on day 4 of erythroid differentiation, EBL were subjected to mass spectrometry-based lipidomics analysis. Partial least squares-discriminant analysis (PLS-DA) demonstrated that samples cluster per differentiation condition (Fig. 4A). Clustering of the top 75 lipids selected by adjusted p-value shows that at day 4 of differentiation, static cultures have higher levels of long chain fatty acids and numerous phospholipids (Fig. 4B, cluster A), while dynamic cultures are marked by elevated levels of cholesteryl esters (ChE), free cholesterol (Ch), triglycerides (TG), diglycerides (DG), and phosphatidylcholines (PC) (Fig. 4B, cluster B). Assessment of the contributions of different lipid classes shows that dynamic culturing enriches the EBL lipidome in triglycerides (TG 9%) cholesterol (Ch 16%) at the expense of phosphatidylethanolamine (PE, 7%), fatty acids (FA, 5%) and phosphatidylcholine (PC, 46%), while static cultures show increased percentages of phosphatidylcholine (PC, 51%) phosphatidylethanolamine (PE, 11%) and fatty acids (FA, 11%) (Fig. 4C—outlined in yellow and Fig. 4D). These findings corroborate the upregulation of genes involved in the cholesterol and lipid biosynthesis previously described in dynamic cultures.
Day 4 erythroblasts differentiated in dynamic conditions show higher concentration of cholesterol triglycerides and phosphatidylcholine compared to cells differentiated in static. Day 4 erythroblast (EBL) of three different donors (n = 3) were characterized by global lipidomics to determine their lipid composition. A) Partial Least Squares-Discriminant Analysis (PLS-DA) of samples differentiated in static and dynamic conditions. X-axis represents variations between conditions, y-axis represents variations between donors. B) Heatmap of the top 75 lipids differentially expressed at day 4 of differentiation in static (left- red) and dynamic (right- green) conditions according to the adjusted p-value. Cluster A represents lipids upregulated in static and cluster B lipids upregulated in dynamic. C) Pie-charts representing the lipid composition in percentage of day 4 EBL obtained from static and dynamic cultures. The most expressed lipids in static and dynamic condition are outlined in yellow. D) Comparison of the single lipids detected in reticulocytes obtained from static and dynamic conditions. X-axis represents the abbreviation of lipids name, y-axis the area under the curve. Paired two-tailed student t-test was performed, with ** p < 0.01 and *p < 0.05. Unless marked, no significance was observed.
Reticulocytes generated in shear-stress environment show higher cell stability, but similar lipid composition compared to reticulocytes generated in static conditions
We next investigated whether shear-stress induced transcriptional program can affect erythrocyte homeostasis and reticulocyte plasma membrane stability and deformability (Fig. 5).
Reticulocytes obtained from dynamic cultures show higher cell stability but similar deformability and lipid composition when compared to static. Erythroblasts (EBL) of three different donors (n = 3) were differentiated for 12 days as indicated in material and methods. On day 12 of differentiation, cells were filtered, and the obtained reticulocytes subjected to cell deformability and stability assays. Measure of the reticulocyte’s deformability was detected using the ARCA. Cells were subjected to a shear stress of 3 Pa and 10 Pa. A schematic representation of the ARCA is shown in supplemental Fig. 4D. A) The x-axis represents the ratio between the length and the width of the cell (A/B), the y-axis is the normalised number of cells. A/B ratio directly correlates with reticulocyte deformability. B) Hemolysis percentage (y-axis) measured after incubation with different concentrations of NaCl solution (x-axis) was assessed to determine the stability of reticulocytes obtained from static and dynamic cultures. A and B) Native RBCs were used as control in deformability and stability assays. A) Unpaired two-tailed student t-test of the area under the curve of static condition vs RBC and dynamic condition vs RBC was performed, with *** p < 0.001, ** p < 0.01 and *p < 0.05. B) 2way ANOVA test was performed, with*** p < 0.001, ** p < 0.01 and *p < 0.05. Unless marked, no significance was observed.
We measured the deformability of the cultured reticulocytes using an automated rheoscope and cell analyzer (ARCA) subjecting the cells to a shear-stress of 3 and 10 Pa (Supplemental Fig. 4D). We detected lower, but yet not significant, deformability in reticulocytes obtained from static and dynamic cultures compared to native RBC, no significant difference was detected between static and dynamic conditions (Fig. 5A). In contrast, the osmotic resistance of reticulocytes derived from dynamic cultures was comparable to RBC controls where static cultures showed significant lower osmotic resistance (Fig. 5B).
Interestingly, global lipidomics analysis of reticulocytes showed minimal to no differences between static and dynamic cultures, aside from a slight increase in fatty acids in static cultures (Supplemental Fig. 3). The data suggest that the transcriptional changes detected at the onset of EBL differentiation in dynamic conditions, is associated with higher osmotic resistance of reticulocytes generated in shaker, but similar lipid composition compared to static conditions.
The HMGCR inhibitor lovastatin leads to a failure in adaptation to orbital shaking conditions and a severe reduction of reticulocyte formation during erythroblast differentiation
Increased production of cholesterol in dynamic conditions poses the question whether the increased HMGCR expression is essential for the adaptive response of differentiating EBL during dynamic conditions. Statins are well-described HMGCR inhibitors used in the clinic to reduce cholesterol levels26,27.
Lovastatin treatment at different concentrations did not affect cell yield and general hemoglobinization during EBL differentiation in static cultures, with the exception of 1 µM treatment. In contrast, dynamic cultures treated with the same lovastatin concentrations showed a significant reduction of cell viability and hemoglobinization (Fig. 6A, C, E and Supplemental Fig. 4). CD49d/CD235a marker expression was not affected in lovastatin-treated EBL differentiated in static conditions, except for a reduction of CD235a+/CD49d- cells with 1 µM lovastatin concentration (Fig. 6B, D). We were unable to characterize differentiation markers for dynamic cultures in presence of lovastatin because most of the cells did not survive the HMGCR inhibition (Fig. 6B, D, E).
Lovastatin inhibition of HMGCR prevents cells adaptation to shear stress environment. Erythroblasts (EBL) of three different donors (n = 3) were incubated with different concentrations of lovastatin on day 0 of differentiation and cultured in static and dynamic conditions. Erythroid cells were harvested on day 12 of differentiation and analysed by flow cytometry. A) Representative example of living gate strategy plotting forward scatter (FSC x-axis) vs side scatter (SSC y-axis) in cells differentiated in static and dynamic conditions treated with different concentrations of lovastatin. B) Maturation states of cells differentiated in static and dynamic conditions incubated with different concentrations of lovastatin according to antiCD49d-PB (BD Biosciences, San Jose, CA, US) (Alexa Fluor 405, y-axis) and antiCD235a-PE (OriGene Technologies, Rockville, MD, USA) (x-axis), staining. C) Quantified percentage of cells in the living gate, averaged for 3 donors (n = 3). Gating strategy to identify the living cells gate is described in Supplemental Fig. 4). D) Quantified maturation states of cells differentiate in static and dynamic incubated with different concentrations of lovastatin were measured as described in Fig. 1A. Percentage of cells in gates was averaged for 3 donors (n = 3). ND indicates not-detectable measurement due to the low percentage of cells in the living gate. E) Representative picture of pelleted cells on day 12 of differentiation incubated with different concentration of lovastatin. C and D) 2way Anova test was performed, with *** p < 0.001 ** p < 0.01 and *p < 0.05. Unless marked, no significance was observed.
Overall, the data shows that HMGCR enzymatic inhibition was not tolerated during EBL differentiation under turbulent culture conditions and thus it is required for the adaptive response of EBL to orbital shaking.
S1P inhibition prevents SREBP cleavage and activation reducing HMGCR expression and inducing cell death in dynamic-differentiated cells
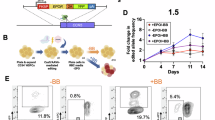
SREBPs (Sterol-regulatory- element-binding-proteins) are master regulators of cholesterol metabolism in many cells. The precursor of SREBPs is located in the ER and when the cellular steroids level is reduced, a cascade of cleavage processes, initiated by the site-1 protease (S1P), allows SREBPs translocation into the nucleus, where they regulate the expression of genes involved in cholesterol metabolism, such as HMGCR28. To explore if the increase of HMGCR expression detected during erythroid differentiation in dynamic conditions, was due to the cleavage and subsequent activation of SREBPs, we inhibited S1P via the administration of its inhibitor, PF-429242 (1 µM). PF-429242 effects were evaluated 24 and 48 h after its administration (Fig. 7). PF-429249 did not alter the total cell count on day 1 of differentiation, with a slight reduction of EBL differentiated in static conditions on day 2. Of note, the cell count of EBL differentiated in dynamic environment was similar between the untreated-controls and the samples subjected to S1P inhibition, during the 2 days of erythroid maturation (Fig. 7A). The reduction in hemoglobinization after PF-429242 treatment suggests an increase in cell death in dynamically differentiated cells after S1P inhibition (Fig. 7B). In line with this, flow cytometry data showed that the inhibition of S1P induced a reduction of cells in the living gate in samples differentiated in dynamic condition on day 1. The reduction in living cells was also detected in static cultures on day 2, but it did not reach the levels observed in dynamic conditions on the same day (Fig. 7C, D and Supplemental Fig. 4). Moreover, an increase of more mature cells, CD235a+/CD49d-, was observed on day 1 and 2, in both static and dynamic cultures, with a stronger effect in the latter one, reaching ~ 80% of CD235a+/CD49- EBL on day 2, possibly related to a selection of the most mature cells after the S1P inhibition (Fig. 7E). HMGCR expression, evaluated by Western blot, was severely reduced in both static and dynamic cultures treated with PF-429242 on day 1 and 2 of differentiation. BAND 3 was used as loading control. (Fig. 7F, G and Supplemental Fig. 5). All together this data suggests a novel role for the S1P-induced cleavage of SREBPs to transcriptionally induce HMGCR expression and increase cholesterol biosynthesis in dynamically differentiated EBL.
S1P inhibition in the first 2 days of differentiation, reduced cell viability and HMGCR expression and led to a loss of EBL viability in dynamic cultures. Erythroblasts (EBL) of three different donors (n = 3) were incubate with the S1P inhibitor PF-429242 on day 0 of differentiation and cultured in static and dynamic conditions for 2 days. The effects of S1P inhibition were evaluated every 24 h. A) Cell-counts for EBL that were cultured in static or dynamic conditions in presence or absence of the inhibitor. B) Representative picture of pelleted cells of 3 donors (n = 3) on day 2 of differentiation in static or dynamic conditions, incubated with PF-429242. C) Representative example of living gate strategy plotting forward scatter (FSC x-axis) vs side scatter (SSC y-axis) in cells differentiated in static and dynamic conditions treated with PF-429242. D) Quantified percentage of cells in the living gate, averaged for 3 donors (n = 3). Gating strategy to identify the living cells gate is described in Supplemental Fig. 4. E) Maturation states in static vs. dynamic conditions as explained in Fig. 1A were averaged for 3 donors (n = 3) by quantifying the percentage of cells within the respective gates Q1: CD49d-/CD235a-, Q2: CD49d+/CD235- immature EBL, Q3: CD49-/CD235a+ late EBL and reticulocytes, Q4 CD49+/CD235a+ EBL. Representative example of gating strategy corresponding to the described conditions are reported below. F) HMGCR expression assessed through western blot analysis, during 2 days of EBL differentiation in static and dynamic conditions treated with PF-429242 and controls. BAND 3 was used as loading control. G) Quantified expression of HMGCR during cell differentiation in dynamic and static conditions. HMGCR expression was normalised according to BAND3 expression. (A, D, and G 2way Anova test was performed, with **** p < 0.0001, *** p < 0.001 **, p < 0.01 and *p < 0.05. Unless marked, no significance was observed.
Discussion
EBL can be expanded to large numbers, sufficient to produce transfusion products4(for review, see Di buduo29). However, cost-effective upscaling is crucial for the implementation of such products. To this end, advances in high cellular density cultures, media components and stirred tank bioreactor perfusion systems are required. As most upscaling systems introduce mixing by stirring, shaking and/or sparging of air/oxygen, the effects of mechanical stress on erythroid cells need to be evaluated.
Here we show that shear-stress during CD49d+/CD235low EBL maturation to enucleated CD49-/CD235highreticulocytes accelerates erythroid differentiation without effects on cell viability. We used a maximum shear-stress of 1.4 Pa through orbital shaking at 0.7 g, mimicking the shear-stress experienced at the impeller of our stirred-tank-reactors4(based on Odeleye22) and the possible mechanical forces within the bone marrow23. Orbital shaking altered the EBL transcriptional program, indicating that erythroid cells adapt to turbulent environments. The highest effect of mechanical forces on the acceleration of cell differentiation was detected within the first 4 days of erythroid maturation.
Interestingly, similar transcriptional responses to shear-stress were also observed in other cell-types30,31. Characterization of the RNA expression throughout the first 4 days of differentiation revealed > 500 DEGs between dynamic and static cultures. Of note, no differences in the expression of mechanosensory genes, like PIEZO1 and TRPV2 between static and dynamic conditions were detected32,33. Among the biological processes upregulated in dynamic cultures, the cholesterol biosynthesis pathway was highly enriched. This process may represent one of the adaptation mechanisms, enabling cells to withstand the shear-stress, ensuring a more rigid membrane by the increase of cholesterol content.
We found the upregulation of several members of the cholesterol biosynthesis pathways in dynamic conditions, such as SQLE, HMGCS1 and HMGCR. The latter is a key rate limiting catalyst in this process34(for review, see Sharpe35). Increased HMGCR expression correlates with higher cholesterol concentration in day 4 EBL differentiated in dynamic compared to static conditions. This was associated with higher stability of reticulocytes derived from dynamic cultures, comparable to RBC controls. In contrast, cells differentiated in static environment showed lower stability compared to dynamically differentiated reticulocytes and RBC controls. Nevertheless, RBC deformability and stability depend on a multitude of factors such as the surface area to volume ratio, the viscosity of the cytoplasm and the plasma membrane structure (e.g. spectrin based skeletal structure)36,37 More insights in the variability of the mentioned factors are required to determine their involvement in the deformability and stability characteristics of cultured reticulocytes.
The increase of cholesterol, triglycerides, and phosphatidylcholine in day 4 EBL cultured in dynamic compared to static conditions suggests an adaptation process to the shear-stress environment, possibly involving changes in the lipid plasma membrane composition and as consequence in plasma-membrane fluidity (PMF), and a probable increase in cholesterol storage via lipid droplet formation. Nevertheless, the reduced difference in the enucleation rate, differentiation markers and lipid composition between reticulocytes obtained from static and dynamic cultures, indicates a loss of the adaptation phenotype towards the end of differentiation.
HMGCR converts HMG-coenzyme A into mevalonic acid, and it is a known pharmacological target of cholesterol-decreasing therapeutics like statins26. Of note, the mevalonate pathway is also suppressed by the tumor protein p53, by mechanism of up-regulation of the cholesterol transporter ABCA138.The node of p53—a central transcriptional regulatory node in several stress responses39– has long been associated with physiological or stress erythropoiesis both in the primitive40and adult41 settings. The evidence that cells differentiated in dynamic conditions do not survive the inhibition of HMGCR, confirms the involvement of genes involved in the cholesterol biosynthetic pathway in the cellular adaptation mechanism to shear-stress.
It is known that mechanosensing proteins are present and functional in the erythroid membrane9,11,12. EBL differentiate on central macrophages in erythroid islands in the bone marrow, where they may experience shear forces through crawling and dynamic contacts with the central macrophages42,43. In addition, it has been shown that the mechano-sensor channel Piezo1 plays a fundamental role in regulating cholesterol biosynthesis in other tissues, specifically in neural development where its activity influences neural stem cells fate44. In light of this, there may be a specific role for mechano-sensors on hematopoietic progenitors and EBL during migration, sensing of tissue stiffness or in cell–cell contacts. Investigation of the putative mechanisms that regulate cell adaptation to mechanical forces could be useful to understand how cells regulate their transcriptional program upon shear-stress, also in the bone marrow environment. HMGCR is a target gene of Steroid Responsive Element Binding Proteins (SREBPs)45. Some SREBPs regulating proteins such as HSD17B7, AACS and INSIG1 (for review, see Dong46) are also upregulated in dynamic conditions. SREBPs maintain cellular lipid homeostasis by regulation of cholesterol, fatty acids, and triglyceride biosynthesis47. In sterol-depleted conditions, SREBPs are firstly cleaved by site-1-protease (S1P) and then by site-2-protease (S2P) and translocated from the ER into the nucleus where they regulate the expression of genes involved in the lipid metabolism, such as HMGCR.28.
To dive more into the mechanisms regulating the cholesterol and lipid metabolism during erythropoiesis in dynamic conditions, we inhibited S1P via the administration of PF-429242 on day 0 of differentiation. The inhibition of the emergence of the transcriptional active form of SREBPs, was accompanied by reduction in cell numbers and viability and increase of the more mature cells (CD235a+/CD49d-) in both static and dynamic conditions, but with a more evident effect in dynamic setups, proving again the dependency of orbital-shaken EBL on lipid biosynthesis. The decrease in cell number and the cell viability impairment and the increase in the more mature cells could be related to a loss of the most immature EBL, unable to adapt their membrane to the shear-stress environment, leaving space to more mature cells, whose plasma membrane composition was defined and able to tolerate mechanical stresses. In line with this, HMGCR expression was severely reduced after PF-429242 treatment in both static and dynamic conditions. Although the reduction in HMGCR expression could be related to the increase of mature cells after S1P inhibition, the evidence of its reduction on day 1 in static samples (subjected to a lower variation in differentiation after S1P inhibition), suggests a role for PF-429242 in HMGCR downregulation. Of note, recently Wang et al., showed that lipid homeostasis was critically important at the early onset of erythroblast differentiation48.
In conclusion, we show that erythroid precursors withstand and adapt to significant shear-stress during the early stages of differentiation to enucleated reticulocytes. The metabolic rewiring underlying this essential adaptation centers on cholesterol metabolism, via SREBPs. Such adaptation was reduced nearing the end of maturation, leading to phenotypically similar reticulocytes from both static and dynamic cultures. Shear-stress does not affect cell viability but accelerates erythroid differentiation, allowing a faster and possibly more cost-effective protocol and permitting the upscaling of EBL differentiation into stirred-bioreactors. Moreover, these data give insights into the biological processes regulating erythropoiesis due to mechanosensing and mechanisms of cellular adaptation to external stresses.
Material and methods
All methods were carried out in accordance with relevant guidelines and regulations at Sanquin. All experimental protocols were approved by Stichting Sanquin Bloedbank, Department of Research and Sanquin Ethical Advisory Board. Written informed consent was obtained from all subjects.
Erythroblast expansion and differentiation
Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll density separation according to the manufacturer’s protocol (GE Healthcare, Chicago, Illinois, USA). Informed consent was given in accordance with the Declaration of Helsinki, the Dutch National and Sanquin Internal Ethic Boards. PBMC were cultured to proerythroblasts in IMDM medium (PAN P04-20,250) supplemented with EPO (2 U/mL; ProSpec, East Brunswick, NJ, USA), human recombinant Stem-Cell Factor (100 ng/mL, supernatant SCF-producing cell line) and dexamethasone (1 μM; Sigma, St. Louis, MO, USA). Interleukin 3 -IL3 (1 ng/mL STEMCELL technologies) was added on day0 of culture24. During the expansion phase, EBL were kept at a density of 0.7–1.5 million/mL. To differentiate EBL to reticulocytes, cells were washed twice in PBS and resuspended in differentiation media (1 million/mL): IMDM supplemented with EPO (10 U/mL), 5% Omniplasma (Octopharma, Wien, Austria), holotransferrin (700 µg/mL; Sanquin), and heparin (5 U/mL; LEO Pharma BV, Breda, The Netherlands)24. Cells were kept in differentiation medium for 10–12 days, without further dilutions, sampled daily, pelleted, washed twice in PBS and subjected to cell count and flow cytometry analysis. Cell counts were obtained using a CASY (CASY®Model TTC, Schärfe System GmbH, Reutlingen, Germany). The day of change to differentiation media is referred to as Day 0 of differentiation.
Orbital shaking cultures
Dynamic cultures were performed using 125 mL Corning Erlenmeyer flasks with ventilated caps (Sigma Aldrich) on a Thermo Scientific™ CO2 Resistant Shaker (#15,341,105). 250 revolutions-per-minute (RPM) was used, yielding to a 1.4 Pa maximum shear-stress. The starting volumes were 12 mL in dishes and 25 mL in flasks.
Static and dynamic conditions were switched at indicated days by moving samples cultured in dish to flask in orbital shaker and vice versa, without media change.
Flow-Cytometry
Samples were collected daily for 10 days of differentiation washed twice in PBS and resuspended in HEPES buffer (132 mM NaCl, 20 mM HEPES, 6 mM KCl, 1 mM MgSO4, 1.2 mM K2HPO4) supplemented with 0.5% bovine serum albumin (BSA) and stained for 30 min as indicated in the figure legends. Samples were measured with FACS Canto II (BD Biosciences, Oxford, UK) and results analysed using FlowJo software (FlowJo v10; Tree Star, Inc., Ashland, OR USA).
Cytospins
Cytospins were performed with Shandon Cytospin-II (Thermo Scientific), stained with benzidine and Differential Quik Stain Kit (PolySciences, Warrington, PA) as described before24. Stained cytospins were imaged with 50 × objective on Leica DM-2500 microscope (Leica, Germany).
Western blotting
EBL were pelleted, washed 3 times with PBS and lysed in CARIN buffer (20 mM Tris–HCl pH 8.0, 138 mM NaCl, 10 mM EDTA, 100 mM NaF, 1% NonidetP-40, 10% glycerol). Lysates were heated in Laemmli sample buffer (2% sodiumdodecylsulfate [SDS] wt/vol, 10% glycerol, 1% 2-mercaptoethanol, 60 mM Tris–HCl pH6.8, bromophenolblue) for 5 min at 95 °C, subjected to SDS–polyacrylamide gel electrophoresis, blotted using iBlot-PVDF blotting system (ThermoFisher Scientific, Bleiswijk, The Netherlands), and stained as indicated in figure legends.
Stability and deformability assays
Day 12 differentiating EBL from static and dynamic cultures were filtered (Spuitfilters-Acrodisc® WBC) to remove nucleated cells and pyrenocytes. Purified reticulocytes were subjected to osmotic shock by incubation in 0% to 0.7% NaCl for 5 min at RT, centrifuged and supernatants’ absorbance measured at 415 nm. Reticulocytes’ deformability was assessed using ARCA (ARCA-Linkam CSS450) as described before24,49..
HMGCR and S1P inhibition
Day 0 differentiating-EBL were incubated with the HMGCR inhibitor Lovastatin (Ab120614) or the S1P inhibitor PF-429242 (Bio Techne) and cultured in static or dynamic condition. Cells differentiation, enucleation, stability, and deformability were assessed as described above.
RNA sequencing and analysis
EBL differentiated in static or dynamic conditions were sampled at days 1, 2 and 4 following differentiation induction. RNA was isolated from cell pellets using Trizol as indicated by the manufacturer’s protocol (Invitrogen Technologies, CA, USA). DNA remnants were removed by RNA clean up kits (Zymo Research, CA, USA). RT-reactions and RNA sequencing was performed by GENEWIZ (NJ, USA) using random hexamers, with on average 30 million reads deep. After sequencing, quality control and adapter trimming were performed using FastQC v0.11.9 (Babaham Bioinformatics) and fastp 0.23.250with additional arguments –length_required 36, –cut_right, –cut_right_window_size 4, –cut_right_mean_quality 15. Next, pair-ended trimmed fastq files were mapped to GRCh38.p13 (Ensembl 87) using STAR 2.7.0f51with –quantMode GeneCounts. Deferential expression analysis was performed using second stranded reads in EdgeR 3.40.252. In short, low expressed reads were filtered out, all samples were TMM normalised, and dispersion was calculated using the estimateDisp function. Samples were categorised in two different groups (dish and flask) for the different differentiation days. Pairwise comparisons between the different groups were carried out for the different days using the glmLRT/ glmLRT functions; p-values were Bonferroni adjusted and genes with adjpval \(\le\) 0.05 were considered differentially expressed between groups. Analyses were performed in Ubuntu 22.04.2 LTS and R version 4.2.3. Heatmaps were plotted using Morpheus (https://software.broadinstitute.org/morpheus). Gene ontology analysis was performed using EnRicHR535455..
Lipidomic analysis
Day 4 EBL and Day12 filtered reticulocytes were washed twice in ice-cold PBS then pelleted and residual PBS aspirated before snap-freezing. Lipids were extracted from frozen cell pellets at 1.07 cells/mL with ice cold methanol, vortexed vigorously for 30 min at 4 °C, then centrifuged for 10 min at 18,213 rcf. Non-polar lipids were resolved using UHPLC coupled to mass spectrometry with data-dependent fragmentation (ddMS2). 5 uL injection volumes were used with a 5-min gradient as previously described. Briefly, samples were resolved over an Kinetex C18 column (2.1 × 30 mm, 1.7 µm; Phenomenex, Torrance, CA) at 50ºC using a 5 min gradient at 300 µL/min for negative and positive ESI. The negative mode gradient was from 10%−100% B and positive mode was from 30%−100% B (A: 75% water, 25% acetonitrile, 5 mM ammonium acetate; B: 90% isopropanol, 10% acetonitrile, 5 mM ammonium acetate). Sample were randomized and quality control was assessed using technical replicates throughout the sequence as previously described.56 Lipidomics data were analyzed using LipidSearch 5.0 (Thermo Scientific), which provides lipid identification on the basis of accurate intact mass, isotopic pattern, and fragmentation pattern to determine lipid class and acyl chain composition.
Data Availability
Sequencing data are available at GEO under accession number GSE247121. Upon request, the raw data supporting the conclusion of this article will be made available by the authors without undue reservation.
References
Learoyd, P. The history of blood transfusion prior to the 20th century–part 1. Transfusion Medicine 22, 308–314 (2012).
Shah, S., Huang, X. & Cheng, L. Concise review: stem cell-based approaches to red blood cell production for transfusion. Stem Cells Transl Med 3, 346–355 (2014).
Koleva, L., Bovt, E., Ataullakhanov, F. & Sinauridze, E. Erythrocytes as carriers: From drug delivery to biosensors. Pharmaceutics 12, 276 (2020).
Gallego-Murillo, J. S. et al. Expansion and differentiation of ex vivo cultured erythroblasts in scalable stirred bioreactors. Biotechnol Bioeng 119, 3096–3116 (2022).
Chu, L. & Robinson, D. K. Industrial choices for protein production by large-scale cell culture. Curr Opin Biotechnol 12, 180–187 (2001).
Rodrigues, M. E., Costa, A. R., Henriques, M., Azeredo, J. & Oliveira, R. Technological progresses in monoclonal antibody production systems. Biotechnol Prog 26, 332–351 (2010).
Tapia, F., Vázquez-Ramírez, D., Genzel, Y. & Reichl, U. Bioreactors for high cell density and continuous multi-stage cultivations: options for process intensification in cell culture-based viral vaccine production. Appl Microbiol Biotechnol 100, 2121–2132 (2016).
Han, S. Y. et al. Red cell manufacturing using parallel stirred-tank bioreactors at the final stages of differentiation enhances reticulocyte maturation. Biotechnol Bioeng 118, 1763–1778 (2021).
Aglialoro, F., Hofsink, N., Hofman, M., Brandhorst, N. & Van den Akker, E. Inside out integrin activation mediated by PIEZO1 signaling in erythroblasts. Front Physiol 11, 958 (2020).
Aglialoro, F. et al. Mechanical stress induces Ca2+-dependent signal transduction in erythroblasts and modulates erythropoiesis. Int J Mol Sci 22, 955 (2021).
Caulier, A. et al. PIEZO1 activation delays erythroid differentiation of normal and hereditary xerocytosis-derived human progenitor cells. Haematologica 105, 610 (2020).
Flormann, D. et al. Transient receptor potential channel vanilloid type 2 in red cells of cannabis consumer. Am J Hematol 97, E180–E183 (2022).
Moura, P. L. et al. Reticulocyte and red blood cell deformation triggers specific phosphorylation events. Blood Adv 3, 2653–2663 (2019).
Clincke, M. et al. Very high density of CHO cells in perfusion by ATF or TFF in WAVE bioreactorTM. Part I. Effect of the cell density on the process. Biotechnol Prog 29, 754–767 (2013).
Zamani, L. et al. High cell density perfusion culture has a maintained exoproteome and metabolome. Biotechnol J 13, 1800036 (2018).
Karst, D. J., Serra, E., Villiger, T. K., Soos, M. & Morbidelli, M. Characterization and comparison of ATF and TFF in stirred bioreactors for continuous mammalian cell culture processes. Biochem Eng J 110, 17–26 (2016).
Mohandas, N. & Gallagher, P. G. Red cell membrane: past, present, and future. Blood, The Journal of the American Society of Hematology 112, 3939–3948 (2008).
Cooper, R. A. Influence of increased membrane cholesterol on membrane fluidity and cell function in human red blood cells. J Supramol Struct 8, 413–430 (1978).
Basford, J. M., Glover, J. & Green, C. Exchange of cholesterol between human β-lipoproteins and erythrocytes. Biochimica et Biophysica Acta (BBA)-Specialized Section on Lipids and Related Subjects 84, 764–766 (1964).
Levental, K. R. et al. Lipidomic and biophysical homeostasis of mammalian membranes counteracts dietary lipid perturbations to maintain cellular fitness. Nat Commun 11, 1339 (2020).
Yamamoto, K. & Ando, J. Endothelial cell and model membranes respond to shear stress by rapidly decreasing the order of their lipid phases. J Cell Sci 126, 1227–1234 (2013).
Odeleye, A. O. O., Marsh, D. T. J., Osborne, M. D., Lye, G. J. & Micheletti, M. On the fluid dynamics of a laboratory scale single-use stirred bioreactor. Chem Eng Sci 111, 299–312 (2014).
Zhang, P. et al. The physical microenvironment of hematopoietic stem cells and its emerging roles in engineering applications. https://doi.org/10.1186/s13287-019-1422-7.
Heshusius, S. et al. Large-scale in vitro production of red blood cells from human peripheral blood mononuclear cells. Blood Adv 3, 3337–3350 (2019).
Brown, M. S. & Goldstein, J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res 21, 505–517 (1980).
Stancu, C. & Sima, A. Statins: mechanism of action and effects. J Cell Mol Med 5, 378–387 (2001).
Forsyth, A. M., Braunmüller, S., Wan, J., Franke, T. & Stone, H. A. The effects of membrane cholesterol and simvastatin on red blood cell deformability and ATP release. Microvasc Res 83, 347–351 (2012).
Sakai, J. et al. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85, 1037–1046 (1996).
Di Buduo, C. A. et al. Latest culture techniques: cracking the secrets of bone marrow to mass-produce erythrocytes and platelets ex vivo. Haematologica 106, 947 (2021).
Zhan, C. et al. Low shear stress increases recombinant protein production and high shear stress increases apoptosis in human cells. iScience 23, (2020).
Fledderus, J. O. et al. Prolonged shear stress and KLF2 suppress constitutive proinflammatory transcription through inhibition of ATF2. Blood 109, 4249–4257 (2007).
Cahalan, S. M. et al. Piezo1 links mechanical forces to red blood cell volume. Elife 4, (2015).
Istvan, E. S. & Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 1979(292), 1160–1164 (2001).
Sharpe, L. J. & Brown, A. J. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). Journal of Biological Chemistry 288, 18707–18715 (2013).
Mohandas, N. & Chasis, J. Red blood cell deformability, membrane material properties and shape: regulation by transmembrane, skeletal and cytosolic proteins and lipids. Semin Hematol (1993).
Mohandas, N. & Evans, E. Mechanical properties of the red cell membrane in relation to molecular structure and genetic defects. Annu Rev Biophys Biomol Struct 23, 787–818 (1994).
Moon, S.-H. et al. p53 Represses the Mevalonate Pathway to Mediate Tumor Suppression Article p53 Represses the Mevalonate Pathway to Mediate Tumor Suppression. Cell 176, 564–580 (2019).
Haronikova, L. et al. SURVEY AND SUMMARY The p53 mRNA: an integral part of the cellular stress response. Nucleic Acids Res 47, 3257–3271 (2019).
Nemkov, T. et al. Circulating primitive murine erythroblasts undergo complex proteomic and metabolomic changes during terminal maturation. Blood Adv 6, 3072–3089 (2022).
Pant, V., Quintás-Cardama, A. & Lozano, G. The p53 pathway in hematopoiesis: lessons from mouse models, implications for humans. Blood 120, 5118 (2012).
Heideveld, E. & van den Akker, E. Digesting the role of bone marrow macrophages on hematopoiesis. Immunobiology 222, 814–822 (2017).
Heideveld, E. et al. Glucocorticoids induce differentiation of monocytes towards macrophages that share functional and phenotypical aspects with erythroblastic island macrophages. Haematologica 103, 395 (2018).
Nourse, J. L. et al. Mechanotransduction by Membrane Proteins Piezo1 regulates cholesterol biosynthesis to influence neural stem cell fate during brain development. Journal of General Physiology 154, (2022).
Vergnes, L. et al. SREBP-2-deficient and hypomorphic mice reveal roles for SREBP-2 in embryonic development and SREBP-1c expression [S]. J Lipid Res 57, 410–421 (2016).
Dong, X.-Y., Tang, S.-Q. & Chen, J.-D. Dual functions of Insig proteins in cholesterol homeostasis. Lipids Health Dis 11, 1–8 (2012).
Brown, M. S. & Goldstein, J. L. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res 50, S15–S27 (2009).
Wang, E. et al. An Optimized Human Erythroblast Differentiation System Reveals Cholesterol-Dependency of Robust Production of Cultured Red Blood Cells Ex Vivo. Adv Sci (Weinh) https://doi.org/10.1002/ADVS.202303471 (2024).
van Zwieten, R. et al. Partial pyruvate kinase deficiency aggravates the phenotypic expression of band 3 deficiency in a family with hereditary spherocytosis. Am J Hematol 90, E35–E39 (2015).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 1–14 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–W97 (2016).
Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr Protoc 1, e90 (2021).
Nemkov, T., Hansen, K. C. & D’alessandro, A. A Three-Minute Method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. https://doi.org/10.1002/rcm.7834.
Aknowledgment
This study was supported by research funding from Horizon 2020 European Union International Training Network grants “RELEVANCE” (AB, FA) (H2020-MSCA ITN-2015, grant agreement N.675115) and “EVIDENCE” (GI) (H2020-MSCA ITN-2015, grant agreement N.860436), by the Dutch Ministry of Health, Welfare and Sports (JGM, NY; PPOC 17-28), and the Dutch Science Organisation NWO (12583). We thank the Sanquin Central Facility for Flow cytometry support.
Author information
Authors and Affiliations
Contributions
GI and AB performed most of the experiments with help of FA, NY, EV, TB, and JS. JGM provided help with orbital shaking fluid dynamics calculations. KF performed the deformability assay. JR, AA, ADA performed and helped with the lipidomic analysis. AFS performed the RNAseq analysis. GI, AB, MvL and EvdA designed, interpreted, and wrote the manuscript. All authors critically read and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests:
The author(s) declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Iacono, G., Abay, A., Murillo, J.S.G. et al. Differentiating erythroblasts adapt to mechanical stimulation by upregulation of cholesterol biosynthesis via S1P/SREBP-induced HMGCR expression. Sci Rep 14, 30157 (2024). https://doi.org/10.1038/s41598-024-81746-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-81746-8