Abstract
Polymorphisms in the MBL2 gene exon 1 can decrease serum levels of mannose-binding lectin (MBL), increasing the risk of infection in immunocompromised individuals. This study evaluated the association between the polymorphism in exon 1 of the MBL2 gene, genotypes, serum MBL levels, and infection in 122 patients with acute lymphoid leukemia (ALL). The MBL*A allele exhibited the highest frequency (0.37) within the study population. The MBL*D (0.32) was the predominant variant. The combined frequency of O polymorphic alleles (either B or D) was 0.63. The frequencies of the A/A, A/O and O/O genotypes were 0.13, 0.49 and 0.38, respectively. All patients exhibited consistently low levels of serum MBL, irrespective of their exon 1 genotype. Parasitic infections (n = 103), bacterial (n = 69) and viral (n = 48). A/O genotype (0.49) had higher infection rates, A/A (0.13) had lower rates, and O/O showed increased viral susceptibility (OR: 0.37; 95% CI 0.13–1.06; p = 0.05). Our findings demonstrated that the study population were MBL-deficient, regardless of their MLB2 genotype. Individuals with the A/O genotype had more infections, while those with the O/O genotype appeared more susceptible to viral infections. These findings highlight the impact of MBL levels and genetic variants on infection susceptibility in ALL patients.
Similar content being viewed by others
Introduction
MBL is produced in the liver and has two domains: the C-terminal carbohydrate recognition domain (CRD) and the N-terminal collagen domain1. It is part of the collectin family and functions to recognize pathogen-associated molecular patterns (PAMPs)2. MBL serves as an important receptor in fighting against harmful microorganisms, mainly by activating the complement system3. Furthermore, this protein is essential for both enhancing the recognition of microbial host cells and orchestrating immune responses against infections caused by viruses, fungi, bacteria, and parasites4.
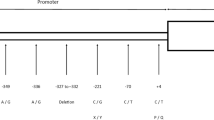
The MBL2 gene comprises 4 exons separated by 3 introns of 600, 1300, and 800 base pairs (bp), respectively1. The MBL2 gene, located on chromosome 10q11.1-q21, has three variations in the wild-type allele in its exon 1. The variants are positioned at codons 52, 54, and 57, resulting in three modifications classified as MBL*D (rs5030737), MBL*B (rs1800450), and MBL*C (rs1800451)1,5. Combinations of these allelic variations can lead to deficiency in serum MBL levels, consequently increasing susceptibility to infections by various pathogens6,7,8,9. These deficiencies may result in heightened susceptibility to infectious diseases, particularly in immunocompromised individuals, such as patients with Acute Lymphoblastic Leukemia (ALL)10.
ALL is characterized by the uncontrolled proliferation of lymphoid progenitor cells in the blood, bone marrow, and other extramedullary sites. Although ALL affects both adults and children, 80% of cases occur in individuals aged 2 to 10 years11. Its symptoms can be nonspecific, presenting a range of signs of bone marrow failure, such as anemia12. In ALL, multiple genetic alterations are observed, including aneuploidy, deletions, insertions, and chromosomal rearrangements13.
Many of these genetic changes interfere with important cellular processes, such as: regulation of the lymphoid lineage; inhibition of tumor suppression through the TP53 tumor suppressor protein pathway; disruption of nucleotide metabolism; and impairment of the regulation of hematopoietic cell development and differentiation6,12,13. Therefore, the paucity of innate immune serum protein such as MBL, attributed to variant genotypes, my significantly accentuate the vulnerability to infectious diseases within patients afflicted by ALL.
Thus, this study analyzed the polymorphic patterns within exon 1 of the MBL2 gene in patients with ALL from the western Brazilian Amazon. Additionally, we explored whether these patterns had any connection with serum MBL levels and susceptibility to infections.
Materials and methods
Study population and ethic aspects
Between 2015 and 2022, we collected peripheral blood samples from 122 patients with ALL at various stages of the disease and under different treatment regimens at the Hematology and Hemotherapy Foundation of the State of Amazonas (HEMOAM) in Manaus, Amazonas, Brazil. Among them, 76 were male and 46 were female, with ages ranging from 2 to 63 years. We collected all sociodemographic and clinical information, such as sex, age, and infectious diseases, from the FHEMOAM IDOCTOR electronic medical record system. The blood samples were transported to the Laboratory of Virology and Immunology at the National Institute of Amazonian Research, where they were processed and stored at -80 ºC until use. This study was approved by the Research Ethics Committee of HEMOAM under CAAE: 25710819.0.0000.0009, in accordance with the guidelines outlined in Resolution 466/2012 of the National Health Council (CNS) of Brazil.
MBL2 genotyping
A 349 bp fragment of the MBL2 gene (exon 1) was amplified from DNA extracted from the study population (Table 1). The amplification reaction was carried out with a total volume of 50 µL containing: extracted DNA (500 ng); dNTPs (225 µM each); primers (5 µM each); KCl (50 mM); MgCl2 (2.5 mM); Tris-HCl (pH 8.3; 10 mM); and Taq DNA polymerase (0.5 U)14.
The amplification reaction proceeded with an initial denaturation cycle at 94 °C for 2 min, followed by 35 cycles with the following configuration: 94 °C for 30 s; 58 °C for 60 s; and 72 °C for 120 s14. The oligonucleotides utilized in the PCR are detailed in Table 1.
The MBL*D (rs5030737) was detected using a semi-nested PCR technique, which involves two rounds of amplification. In the second round, one of the primers from the initial round is combined with a new internal primer to increase specificity, the amplification. Reaction was conducted in a total volume of 25 µL, comprising extracted DNA (500 ng); dNTPs (225 µM each); primers (5 µM); KCl (50 mM); MgCl2 (1.1 mM); Tris-HCl (pH 8.3; 10 mM); and 0.5 U of Taq DNA polymerase.
Following an initial denaturation cycle at 94 °C for 2 min, 30 cycles ensued with the following configuration: 94 °C for 20 s; 62 °C for 20 s; 72 °C for 30 s. Subsequently, a final extension cycle at 72 °C for 5 min was conducted. The amplified product determines the presence of the MBL*D variant. A second PCR was performed using different internal primer as mentioned above14.
The MBL*A, MBL*B, and MBL*C alleles were discerned employing enzymatic cleavage by BanI for MBL*A/B (rs1800450), resulting in two fragments of 260 bp and 89 bp, and by MboII for MBL*C (rs1800451), yielding two fragments of 279 bp and 70 bp within the amplified product (Table 2)15.
Measurement of serum MBL levels
Serum MBL protein concentration was measured in patient plasma via a commercial ELISA (Invitrogen, USA). A “sandwich” assay format with anti-MBL antibody sensitization was used. Samples were diluted 1:800, followed by reagent and standard dilutions according to manufacturer instructions. The optical density (OD) of samples was measured using a spectrophotometer with a 450 nm filter. The intensity of OD signal was directly linked to the concentration of MBL. The concentration per ng was determined through interpolation using the standards provided by the manufacturer.
Statistical analysis
Allele frequencies of the MBL2 gene were estimated using the maximum likelihood method and compared using the unpaired Student’s t-test. Associations between alleles and susceptibility to infections were assessed using Odds Ratio (OR) analysis. Chi-square test with Yates correction was employed for comparisons of the frequency of each allele and studied genotype. The Hardy-Weinberg equilibrium was tested for goodness of fit of the genotype distribution using the chi-squared test. Results with p-values ≤ 0.05 were considered statistically significant. All analyses were performed using GraphPad Prism version 8 software.
Results
Allelic and genotypic frequency of the MBL2 gene in the Study Population
For the codon 54 (rs1800450), the allelic frequency of the wild-type allele MBL*A (0.37) was higher compared to the MBL*B allele (0.31), as shown in Table 3. Regarding codon 52 (rs5030737), the frequency of MBL*D (rs5030737) was 0.32, representing the more prevalent allelic variant. The MBL*C allele (rs1800451) was not detected in the study population. The allele distributions were consistent with Hardy-Weinberg equilibrium.
The frequency of the O polymorphic allele (B or D) was 0.63 (Table 4). Genotype analysis revealed that A/O was more frequent (0.49), followed by O/O (0.38). The sum of A/O and O/O genotypes resulted in a frequency of 0.87. The MBL*D allele occurred in heterozygosity with a frequency of 0.63 and it was not found in homozygous (Table 4). The MBL*B allele had a frequency of 0.45 in heterozygosity and 0.08 in homozygous.
When the results were stratified based on sex, the MBL*A, MBL*B, and MBL*D allelic frequency observed between male (0.22, 0.18, and 0.21, respectively) and female (37.7, 0.26, and 0.20, respectively) were similar (Table 5). When we examined the allele frequencies based on age groups, we found that the MBL*A allele had a higher frequency in the 19–30 years age group (0.30), while the MBL*B allele was more common in the 2–7 years age group (0.21). Similarly, the MBL*D allele was more prevalent in the 19–30 years age group (0.22).
Regarding genotype frequency, heterozygous genotypes (A/O) were more common among the study population. The A/B genotype had a higher frequency in females (0.22) compared to males (0.12), while the A/D genotype was more prevalent in males (0.35) than in females (0.30). The B/D genotype was more prevalent in individuals aged 2–7 years (0.30) and 8–18 years (0.35). On the other hand, the A/D genotype showed a higher frequency in individuals aged 19–30 years (0.56). Among participants aged over 30 years, there was a higher frequency of the B/D genotype (0.35) (Table 5). However, it’s important to emphasize that the occurrence of an allele or genotype is not related to age, as it is genetically determined. Our stratification based on age was solely for descriptive purposes.
Serum levels of MBL according to genotypes
The serum levels of MBL were notably low within the study population, particularly among individuals with the O/O genotype, which suggest that patients with ALL are MBL-deficient. No statically significant difference was observed in MBL serum levels between the A/A and A/O (p = 0.577), A/O and O/O (p = 0.668), and A/A and O/O genotypes (p = 0.420) (p > 0.05) (Fig. 1). Despite this, the A/A genotype exhibited the highest mean serum concentration among the groups analyzed (377.7 ± 257.4).
Association of Exon 1 polymorphisms in the MBL2 gene and susceptibility to infections
Between 2015 and 2022, a total of 239 infection cases were documented within the study population (Table 6). These cases comprised 103 parasitic, 69 bacterial, 48 viral, and 19 fungal infections. An association between the O/O genotype and susceptibility to viral infections was found, but it was not strongly evident (OD 0.37; 95% IC: 0.13–1.06, p = 0.05).
A higher prevalence of parasitic infections (37.86%) was observed in patients with the A/O genotype compared to other genotypes, particularly among patients with the A/D genotype (66.67%) (Table 6). Similarly, increased numbers of parasitic infections (33.33%) were observed in patients with the A/B genotype. Descriptions of bacterial, viral, and fungal infections are detailed in supplementary Table 1.
The potential association between the overall genotypes (A/A, A/O and O/O) of exon 1 of the MBL2 gene and the risk of acquiring infection (regardless of type of infection) was not statistically significant (p > 0.05) (Fig. 2). The OR analysis showed that possessing the variant genotype (A/O or O/O) did not result in a higher susceptibility to infections within the study population when compared to the wild type. Nevertheless, the consistently low levels of serum MLB in all patients with ALL may increase their susceptibility to infections. This is supported by the finding that these patients had a total of 239 infection episodes over a seven-year period.
Although no statistically significant association was found between polymorphisms and infections, certain pathogens were more prevalent in the study population. Klebsiella pneumoniae was the most common bacterium, accounting for 20.29% of bacterial infections. Among viral infections, hepatitis B (HBV) was the leading cause with 15 cases (31.25%). For protozoan infections, Endolimax nana was identified in 25 cases (24.27%). Finally, Candida parapsilosis was the predominant fungal pathogen, affecting 8 individuals (42.11%). (Supplementary Table 1)
Discussion
This study provides, for the first time, the characterization of exon 1 polymorphism of the MBL2 gene and investigates its potential association with serum MBL levels and susceptibility to infections in individuals living with ALL in the state of Amazonas, Brazil.
Our findings showed a higher frequency of the MBL*A allele and a lower frequency of the MBL*B allele within the study population. A recent study conducted in Istanbul, Turkey, found frequencies of MBL*A alleles (0.82) and MBL*B alleles (0.17) in ALL patients, but no statistically significant differences were observed compared to healthy individuals16. Another study that analyzed the allelic frequency of the MBL2 gene in Swedish adults with febrile neutropenia who were undergoing chemotherapy also found a higher frequency of the MBL*A allele (0.64). However, unlike our data, they observed a low frequency of the variant MBL*D allele (0.11)17. Despite the variation in genetic backgrounds and geographical contexts among the study populations, both our study and the two previously referenced studies have identified a consistent pattern: a higher prevalence of the MBL*A allele and a lower prevalence of the MBL*B allele in patients. However, it is important to note that this shared trend is more likely due to population genetic background than to ALL itself.
The MBL*C variant was not identified in this study, similar to previous research conducted in the northern region of Brazil14. The absence or low frequency of the MBL*C variant in the Amazonian region could be due to specific genetic and evolutionary factors, such as the historical admixture between European, African, and indigenous ethnic groups18,19. However, further studies are necessary to comprehensively assess this situation. Our findings provide important insights into the frequencies of the MBL*A, MBL*B, and MBL*D alleles in the study population. These findings have important implications for future studies of host genetic variation in hematological diseases and could lead to improved clinical management of patients, especially those with genotypes associated with low MBL levels. This association with variants of the MBL2 gene could be particularly relevant in the context of ALL patients undergoing chemotherapy treatments20,21. Our findings revealed the O polymorphic allele (B or D) were more frequent in young patients (1–10 years old). While the frequency of an allele is not inherently linked to age, the presence of MBL structural gene mutation in homozygous or heterozygous form may directly impact disease course and prognosis in young patients by amplifying existing immunosuppression. Further research is needed to better understand the direct influence of MBL gene variants on ALL development.
Upon analyzing the genotype frequencies, it was observed that the A/O genotype exhibited the highest prevalence within the study population (0.49), whereas 63% of individuals carried the A/D genotype and 45% carried the A/B genotype. The A/A genotype had a frequency of 0.13, while the O/O genotype occurred in 38% of the individuals. The genotype frequencies found in our study differ from those reported in previous studies conducted with individuals suffering from different neoplastic hematological disease. For example, a study of patients with myeloid leukemia in Poland found that the A/A genotype was more frequent (64%) than the O/O genotype (17,7%)22.Similarly, in another study with patients with ALL, it was observed that the A/A genotype (73.3%) was more frequent than the B/B genotype (8.1%)23. This variations in genotype frequencies across regions and populations, including those seen in our study, could be attributed to a number of factors, including the genetic composition of the population, the specific characteristics of our study group, and the overall diversity within the study population.
MBL exon 1 genotypes have been associated with increased susceptibility to infections throughout the years. Patients carrying the A/O genotype, particularly A/D, exhibited higher infection frequencies. Pana et al. (2014) identified a statically significant association between MBL2 gene A/O variants and an increased susceptibility to bacterial infections in children with B-cell acute lymphoblastic leukemia24. In another study, a link was demonstrated between the A/O and O/O genotypes and an increased risk of febrile neutropenia or infections in oncohematological patients undergoing chemotherapy25. In a study carried out in South Africa, it was observed that MBL2 gene polymorphisms were associated with increased susceptibility to infection and humoral IgG response to C. trachomatis infection (p = 0.048)26. In this other study, an association of polymorphisms in codon 54 (A/B or B/B) was observed with the development of more severe symptoms of COVID-19 in children in a Turkish city27. Molle et al. (2006) found that multiple myeloma (MM) patients with the A/A genotype had a reduced risk of septicemia after autologous stem cell transplantation28. Although our study did not establish a statistically significant association between the A/O genotype and infection susceptibility, the higher number of infections observed in patients carrying this genotype suggests its clinical significancy in increasing susceptibility to infection in the context of ALL.
Our findings suggested that the O/O genotype might be associated with susceptibility to viral infections. Similar findings were found in research conducted in Morocco, which revealed a significant increase in susceptibility to HIV-1 infection in individuals carrying the O/O genotype29. Another study evaluating the influence of MBL2 gene on susceptibility to Dengue (DENV) infection in a Vietnamese population, observed that the frequency of the O/O genotype was significantly higher among dengue patients, suggesting that this genotype is associate with the susceptibility to DENV infection30. Although our results showed a significant association between the O/O genotype and susceptibility to viral infections, this association was not statistically strong. Further studies with larger numbers of individuals are necessary to better elucidate the role of the O/O genotype on infection susceptibility in the context of ALL.
While no correlation was found between serum MBL levels and MBL2 gene polymorphisms in this study, the observed serum MBL levels in the study group (mostly < 500 ng/mL) were considerably lower than the typical levels found in the general population (> 2000 ng/mL)31,32,33. This suggests a serum MBL deficiency among ALL patients, regardless of their genotype.
Previous studies have reported a direct association between serum MBL levels and increased susceptibility to infections. A Merlen et al. (2015) demonstrated that the use of L-asparaginase therapy resulted in a reduction of serum MBL protein levels. This decrease was associated with a higher risk of thrombotic and infectious events in ALL patients34. Furthermore, a study carried out in Brussels, Belgium involving 255 oncohematological patients undergoing 569 chemotherapy cycles unveiled a heightened prevalence of infectious manifestations in comparison with healthy individuals35. These results highlight the complex relationship between MBL protein levels and susceptibility to infections. Our findings underscore the importance of monitoring MBL levels as a potential predictor of infection risk in this clinical setting, especially in ALL patients, who have many factors that can affect MBL levels.
This study presents some limitations. For example, the study population size may have affected the correlation between the MBL2 gene polymorphism and susceptibility to infections. However, the number of individuals included in this study is relatively high, considering that we are working with ALL patients, who are difficult to recruit in large numbers. It is also important to mention that access to patient clinical records was limited. This included detailed information on medical history, immune status, and treatments. In many cases, the information was incomplete, which hindered further analyses. Furthermore, it would be valuable to assess serum MBL levels across different stages of the disease and treatment to more clearly determine how these factors may influence MBL levels. Unfortunately, this was not possible to accomplish in the present study. Despite these limitations, this study contributes to the scientific knowledge of the immunogenetic profile of the studied population and may provide valuable insights for future investigations on the interaction between the innate immune system and clinical aspects of ALL.
Conclusions
This study marks the pioneering effort to elucidate the genetic variability within exon 1 of the MBL2 gene and its relationship with infections among patients residing in the Amazonas state of Brazil, who are affected by ALL. The MBL*D allele was the allele variant more frequent. The frequency of infections episodes was higher among patients carrying variant heterozygous genotypes (A/O), especially the A/D genotype. The presence of genotype O/O seemed to be associated with an increased risk to viral infections. Interestingly, our findings showed that ALL patients were MLB-deficient and this was not related to exon 1 polymorphism.
While further investigations with larger sample sizes are needed to thoroughly characterize the link between exon 1 polymorphism and susceptibility to infection in the context of ALL, this study offers valuable insights into three key aspects: the genetic diversity within the MBL2 gene, its potential connection with infections, and the intricate interplay of MBL levels and their possible role in the prognosis of ALL patients.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Taylor, M. E., Brickell, P. M., Craig, R. K. & Summerfield, J. A. Structure and evolutionary origin of the gene encoding a human serum mannose-binding protein. Biochem. J. 262, 763–771 (1989).
Eppa, Ł. et al. Deposition of mannose-binding lectin and ficolins and activation of the lectin pathway of complement on the surface of polyurethane tubing used for cardiopulmonary bypass. J. Biomed. Mater. Res. B Appl. Biomater. 106, 1202–1208 (2018).
Giang, J. et al. Complement activation in inflammatory skin diseases. Front. Immunol. 9, 639 (2018).
Hammad, N. M., El Badawy, N. E., Nasr, A. M., Ghramh, H. A. & Al Kady, L. M. Mannose-binding lectin gene polymorphism and its association with susceptibility to recurrent vulvovaginal candidiasis. Biomed. Res. Int. https://doi.org/10.1155/2018/7648152 (2018).
Sumiya, M. et al. Molecular basis of opsonic defect in immunodeficient children. Lancet 337, 1569–1570 (1991).
Litzman, J. et al. Mannose-binding lectin gene polymorphic variants predispose to the development of bronchopulmonary complications but have no influence on other clinical and laboratory symptoms or signs of common variable immunodeficiency. Clin. Exp. Immunol. 153, 324–330 (2008).
Kilpatrick, D. C. Mannan-binding lectin and its role in innate immunity. Transfus. Med. 12, 335–352 (2002).
Goeldner, I. et al. Mannose binding lectin and susceptibility to rheumatoid arthritis in Brazilian patients and their relatives. PLoS One. https://doi.org/10.1371/journal.pone.0095519 (2014).
OrnelasAMDM et al. Association between MBL2 haplotypes and dengue severity in children from Rio De Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. https://doi.org/10.1590/0074-02760190004 (2019).
Lausen, B., Schmiegelow, K., Andreassen, B., Madsen, H. O. & Garred, P. Infections during induction therapy of childhood acute lymphoblastic leukemia–no association to mannose-binding lectin deficiency. Eur. J. Haematol. 76, 481–487 (2006).
Rafei, H., Kantarjian, H. M. & Jabbour, E. J. Targeted therapy paves the way for the cure of acute lymphoblastic leukaemia. Br. J. Haematol. 188, 207–223 (2020).
Greaves, M. F., Maia, A. T., Wiemels, J. L. & Ford, A. M. Leukemia in twins: lessons in natural history. Blood 102, 2321–2333 (2003).
Greaves, M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat. Rev. Cancer 18, 471–484 (2018).
Pontes, G. S. et al. Characterization of mannose-binding lectin gene polymorphism among human T-cell lymphotropic virus 1 and 2-infected asymptomatic subjects. Hum. Immunol. 66, 892–896 (2005).
Carlos, A. et al. Characterization of mannose-binding lectin plasma levels and genetic polymorphisms in HIV-1-infected individuals. Rev. Soc. Bras. Med. Trop. 44, 1–3 (2011).
Oguz, R. et al. The association of HLA-DRB1 alleles and MBL2 gene variant in pediatric acute lymphoblastic leukemia patients. Hematol. Transfus. Cell. Ther. https://doi.org/10.1016/j.htct.2023.02.002 (2023).
Wong, M. et al. Mannose-binding lectin 2 polymorphisms do not influence frequency or type of infection in adults with chemotherapy induced neutropaenia. PLoS One. https://doi.org/10.1371/journal.pone.0030819 (2012).
Esteves Agostinho, M. Black American colonization in the Brazilian Amazon. Sæculum – Revista De História 25, 164–179 (2020).
Edge, M. D. & Rosenberg, N. A. Implications of the apportionment of human genetic diversity for the apportionment of human phenotypic diversity. Studies in History and Philosophy of Science Part C:Studies in History and Philosophy of Biological and Biomedical Sciences 52:32–45 (2015).
Cedzyński, M. & Świerzko, A. S. Components of the lectin pathway of complement in Haematologic Malignancies. Cancers (Basel). https://doi.org/10.3390/cancers12071792 (2020).
Yen, H-J. et al. Pediatric acute lymphoblastic leukemia with t(1;19)/TCF3-PBX1 in Taiwan. Pediatr. Blood Cancer. https://doi.org/10.1002/pbc.26557 (2017).
Sokołowska, A. et al. Associations of ficolins and mannose-binding lectin with acute myeloid leukaemia in adults. Sci. Rep. https://doi.org/10.1038/s41598-020-67516-2 (2020).
Oguz, R. et al. The association of HLA-DRB1 alleles and MBL2 gene variant in pediatric acute lymphoblastic leukemia patients. Hematol. Transfus. Cell. Ther. 46, 327–334 (2024).
Pana, Z. D. et al. Mannose binding lectin and ficolin-2 polymorphisms are associated with increased risk for bacterial infections in children with B acute lymphoblastic leukemia. Pediatr. Blood Cancer 61, 1017–1022 (2014).
Frakking, F. N. J. et al. Mannose-binding lectin (MBL) and the risk for febrile neutropenia and infection in pediatric oncology patients with chemotherapy. Pediatr. Blood Cancer 57, 89–96 (2011).
Verweij, S. P. et al. Genetic variation in the MBL2 gene is Associated with Chlamydia trachomatis infection and Host Humoral Response to Chlamydia trachomatis infection. Int. J. Mol. Sci. https://doi.org/10.3390/IJMS23169292 (2022).
Yilmaz, D. et al. Association between mannose binding lectin gene polymorphisms and clinical severity of COVID-19 in children. Mol. Biol. Rep. 50, 1 (2023).
Mølle, I., Peterslund, N. A., Thiel, S. & Steffensen, R. MBL2 polymorphism and risk of severe infections in multiple myeloma patients receiving high-dose melphalan and autologous stem cell transplantation. Bone Marrow Transpl. 38, 555–560 (2006).
Bouqdayr, M. et al. MBL2 gene polymorphisms related to HIV-1 infection susceptibility and treatment response. Hum. Immunol. 84, 80–88 (2023).
Giang, N. T. et al. Complement protein levels and MBL2 polymorphisms are associated with dengue and disease severity. Sci. Rep. 10, 14923 (2020).
Pehlivan, M. et al. Role of MIF-173G/C and Mbl2 Codon 54A/B variants in the risk of multiple myeloma: an Association study. Endocr. Metab. Immune Disord Drug Targets 21, 925–931 (2021).
Izakovicova, P. et al. Gene polymorphisms and serum levels of mannose-binding lectin in Czech patients with recurrent aphthous stomatitis: a case–control study. J. Oral Pathol. Med. 52, 81–90 (2023).
Baioumy, S. A. et al. Mannose-binding lectin serum levels and (Gly54asp) gene polymorphism in recurrent aphthous stomatitis: a case-control study. Int. J. Immunopathol. Pharmacol. https://doi.org/10.1177/20587384211064454 (2021).
Merlen, C. et al. L-Asparaginase lowers plasma antithrombin and mannan-binding-lectin levels: impact on thrombotic and infectious events in children with acute lymphoblastic leukemia. Pediatr. Blood Cancer 62, 1381–1387 (2015).
Vekemans, M. et al. Low mannose-binding lectin concentration is associated with severe infection in patients with hematological cancer who are undergoing chemotherapy. Clin. Infect. Dis. 44, 1593–1601 (2007).
Acknowledgements
Acknowledgments: I thank the University of the State of Amazonas (UEA), the foundation of Hematology and Hemotherapy of the State of Amazonas (FHEMOAM), and the laboratory of Virology and Immunology of the National Institute of Amazonian Research (INPA) for the support provided throughout this study.
Author information
Authors and Affiliations
Contributions
Author Contributions: Conceptualization, G.S.P. , L. C. d. O and P. H. R. d. S; methodology, L. C. d., L. S. M and P. H. R. d. S.; investigation, L. C. d. O., P. H. R. d. S. and L. S. M; formal analysis, L. C. d. O and A. N. B.; writing—original draft preparation, G.S.P. , L. C. d. O. and P. H. R. d. S ; supervision, G.S.P. and A. N. B; project administration, G.S.P., L. C. d. O, and P. H. R. d. S. ; funding acquisition/resources, G.S.P. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
Informed consent was obtained from all subjects involved in the study.
Institutional review board statement
The study was approved by the Research Ethics Committee of HEMOAM under CAAE: 25710819.0.0000.0009, in accordance with the guidelines outlined in Resolution 466/2012 of the National Health Council (CNS) of Brazil.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
de Oliveira, L.C., de Souza, P.H.R., Barbosa, A.N. et al. Impact of Exon 1 polymorphism in the MBL2 gene on MBL serum levels and infection susceptibility in acute lymphoid leukemia. Sci Rep 15, 244 (2025). https://doi.org/10.1038/s41598-024-81971-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-81971-1