Abstract
The bioluminescent European brittle star Amphiura filiformis produces blue light at the arm-spine level thanks to a biochemical reaction involving coelenterazine as substrate and a Renilla-like luciferase as an enzyme. This echinoderm light production depends on a trophic acquisition of the coelenterazine substrate. Without an exogenous supply of coelenterazine, this species loses its luminous capabilities. Moreover, this species was recently shown not to produce coelenterazine storage forms. As an infaunal suspensive feeder, A. filiformis is assumed to find enough substrate to maintain its bioluminescence capabilities efficiently. To date, no studies have investigated the putative source of coelenterazine in the brittle star diet. A combined analysis using listing based on visual observations and metabarcoding on the planktonic communities highlights planktonic species known as light emitters using coelenterazine. Besides, the A. filiformis stomach content was analyzed seasonally via metabarcoding technique, and coelenterazine-related preys were underlined. Results provide evidence of the presence of preys containing coelenterazine in the fjord environment and within the stomach content of the ophiuroid throughout the year. The results are consistent with the demonstration of the trophic acquisition of luminous capabilities in A. filiformis and give a new step by underlying the constant presence of coelenterazine suppliers throughout the year for the luminescence reaction occurring within this species.
Similar content being viewed by others
Introduction
Bioluminescence is the production of visible light by living organisms thanks to a chemical reaction1. This widespread phenomenon is encountered in many organisms, from bacteria to bony fishes2,3,4. Biochemically, bioluminescence results from a luciferin substrate’s oxidation by a luciferase enzyme’s catalytic action5. The electronically excited oxyluciferin releases light when it relaxes to the ground state5. In some cases, luciferin and luciferase are coupled in a stabilized complex protein called photoprotein, needing external cofactor action to be activated5. To date, 11 different luciferins have been described in various terrestrial and marine taxa2,6,7. Coelenterazine is the most widespread luminous substrate in marine ecosystems. The coelenterazine is a molecule from the imidazopyrazinone class, which oxidizes in the presence of oxygen and a specific luciferase (e.g., Oplophorus, Renilla). This reaction produces an oxyluciferin called coelenteramide and CO28,9,10. Over 75 species used a single luciferase/luciferin system, while only 25 used a photoprotein complex11. Initially thought to be taxon-specific, luciferases are now considered an example of convergent evolution. Recent analysis demonstrates the multiple use of homologous luciferases in various groups of luminous metazoans11. Many assumptions were made about bioluminescence functions that may involve numerous ecological roles, such as defense against predators, help in predation or intra/interspecific communication2,4.
Among echinoderms, luminous species have been observed in four of the five classes: Asteroidea, Ophiuroidea, Holothuroidea, and Crinoidea3,12. No luminous Echinoidea species have been recorded so far as luminous3,12.
The bioluminescent brittle star, Amphiura filiformis, lives burrowed in sublittoral muddy sediments down to 200 m depth in the North Sea and the Mediterranean13. As a suspensive feeder, this species extends two arms at the sediment-water interface to passively capture planktons, the three other arms, and the disc remaining burrowed at 4 to 8 cm down in the sediment13,14. A. filiformis, like other echinoderms, possesses an exceptional ability to regenerate injured or self-induced amputated arms and disc parts (e.g15,16,17). Among other ecological traits, A. filiformis can produce flashes of blue light through specific arm spine-associated photocytes18,19,20 (Fig. 1a,b). The bioluminescent system has been demonstrated to depend on a coelenterazine-based luciferase homologous to the Renilla (Anthozoa) luciferase19,20,21 (Fig. 1c). Mallefet et al., 2020 highlight the dependency of A. filiformis luminous capabilities on a coelenterazine food supply21.
Bioluminescence of the brittle star Amphiura filiformis. (a) Bright light, and (b) bioluminescent pictures of A. filiformis. (c) Schematic illustration of the bioluminescence reaction occurring in A. filiformis in which coelenterazine is oxidized via the enzymatic activity of a luciferase homologous to the Renilla luciferase19,20 to form the oxyluciferin in an excited state. Going down to a stable state, the oxyluciferin (i.e., coelenteramide) releases photons and carbon dioxide8,9. Scale bars: 0.5 cm.
Planktonic communities (i.e., species drifting with the water currents) contain autotrophic and heterotrophic species known as phytoplankton and zooplankton, respectively. Depending on several abiotic factors (e.g., temperature, photoperiod, nutrients), phytoplanktonic rapid, massive expansion of the communities, called blooms, trigger zooplanktonic blooms22. Some zooplanktonic communities spend their entire life cycle as a planktonic species (i.e., holoplankton). Conversely, as for the brittle star A. filiformis, some adult benthic species display first a transitional pelagic stage (i.e., larval stage), followed by a benthic adult stage; they are clustered among the meroplankton zooplanktonic communities22. Seasonal blooms during fall and spring induce a massive increase in planktonic species23, potentially consumed by A. filiformis. Plankton community variations might affect the natural luminous capabilities of the brittle star across seasons. A recent study confirmed that ophiuroid luminous capabilities evolve according to the coelenterazine exogenous acquisition without coelenterazine storage forms synthesis24. Moreover, seasonal, long-term monitoring elicits no variation of A. filiformis luminescence, suggesting a continuous presence and dietary acquisition of prey containing coelenterazine throughout the year24. Nevertheless, the natural dietary origin of coelenterazine within the planktonic communities eaten by A. filiformis remains unknown.
The present work aims to determine the presence of (i) luminous planktonic genera in the fjord where A. filiformis inhabits, (ii) the availability of coelenterazine within the encountered luminous plankton and (iii) the potential presence of remaining DNA traces of these planktonic genera within the brittle star stomach content. Across those different approaches, insights into how the light emission process occurs within a coelenterazine-dietary-dependent model, A. filiformis, are obtained.
Materials and methods
Ophiuroids seasonal sampling
Specimens of the brittle star A. filiformis (n = 50) were collected at each season (summer: August 2021; fall: November 2021; winter: January 2022; spring: May 2022) via an Eckman grab at a depth of 30–40 m in the Gullmarsfjord near the Kristineberg Marine Research Station (Fiskebäckskil, Sweden). The ophiuroids were carefully rinsed out of the mud, and intact specimens were placed in an aquarium with unfiltered running seawater pumped directly from the adjacent fjord.
Plankton seasonal sampling
At each season, plankton communities were collected from the Gullmarsfjord near the monitoring station Släggö (N 58°15.5’, E 11° 26.0’). The communities were sampled with vertical tows from 0 to 30 m using a 200 μm plankton net fitted with a plastic receptacle at the end and resuspended in filtered seawater (0.2 μm; FSW). The plankton mixture from each season was filtered immediately with a 60 μm sieve. For each luminometric measurement, a portion of the plankton is isolated in 2.5 ml Eppendorf tubes (n = 3) and centrifuged at 8000 rpm for 1 min. After removing the supernatant, the plankton mixture was weighed before luminometric assays. For the metabarcoding analysis, the remaining plankton mixture was preserved in a 15 ml Falcon tube filled with 99% undenatured ethanol each season.
Coelenterazine content and Renilla luciferase activity seasonal monitoring
For each season, coelenterazine content and Renilla luciferase activity of plankton mixtures (n = 3) were directly measured with coelenterazine and luciferase assays following the protocol described in Coubris et al., 202421.
An FB12 tube luminometer (Tirtertek-Berthold, Pforzheim, Germany) was kept in the dark room and calibrated using a standard 470 nm light source (Beta light, Saunders Technology, Hayes, UK) following Deheyn et al.25. Light responses were recorded using FB12-Sirius PC Software (Tirtertek-Berthold). All data were standardized per unit of mass (g).
For coelenterazine detection, 1 ml of cold methanol was added to the plankton mixture Eppendorf tube. Plankton was crushed with micro-pestle. Then, 5 µl of the methanolic extract was placed into a tube filled with 195 µl of Tris buffer (20 mM Tris, 0.5 M NaCl; pH 7.4) and inserted in the luminometer. Afterward, 200 µl of Renilla luciferase solution with 4 µl of Renilla luciferase (Prolume Ltd., working dilution of 0.2 g l–1 in a Tris-HCl buffer 10 mM, NaCl 0.5 M, BSA 1%; pH 7.4) diluted in 196 µl of Tris buffer was injected into the luminometer tube. The Ltot was recorded to calculate the coelenterazine content per gram of arm tissue (ng g− 1), assuming that 1 ng of pure coelenterazine coupled with Renilla luciferase emits 2.52 × 1011 photons5.
For the luciferase assay, 1 ml of Tris buffer was added to the plankton mixture Eppendorf tube. Plankton was crushed with micro-pestle until a homogenized extract was obtained. Then, 20 and 40 µl of the extract were placed in two tubes with 180 and 160 µl Tris buffer, respectively. Each tube with the diluted luciferase solutions was put in the luminometer, and a solution with 5 µl of 1/200 stock of coelenterazine (1OD in cold methanol at 430 nm; Prolume Ltd, USA) diluted in 195 µl of Tris buffer was injected. Two measures of maximum light emission (Lmax) were recorded and averaged to calculate the maximal light decay rate corresponding to the luciferase activity5, expressed in 109 quanta g− 1 s− 1.
Plankton list analysis
A planktonic species list was extracted from the public database SHARKweb (https://sharkweb.smhi.se/hamta-data/), maintained by the Swedish Meteorological and Hydrological Institute (SMHI). The plankton data set includes abundance measurements of phytoplanktonic and zooplanktonic communities from station Släggö between August 2021 and May 2022. After a first analysis of the planktonic communities, the percent frequency of occurrence of phytoplankton and zooplankton and the luminous species from each community was evaluated. Then, a trimming was performed only to keep the zooplankton since no phytoplanktonic species are known to use coelenterazine as a luminous substrate. Based on the recent list of eukaryotic marine bioluminescent species and subsequent information on the luminescence of those species, luminous planktonic species using coelenterazine or another substrate for light production were retrieved4.
Stomach content extraction
At each season, directly after sampling, ophiuroid specimens (n = 50) were immersed in 99% ethanol and microdissected to collect the stomach content. The microdissection was carried out under a binocular microscope to carefully remove the oral jaws with a micro scalpel and forceps and access to the stomach cavity. This procedure was used to avoid excessive contamination from the disk’s surrounding tissues (e.g., gonads, coelomic fluid, and stomach walls). All the stomach content was carefully collected with a Pasteur pipette and pooled in an Eppendorf tube.
DNA extractions
DNA from plankton mixture samples obtained at each season was isolated following the manufacturer’s protocols of the E.Z.N.A Tissue DNA kit (Omega Biotek, Georgia, USA). In parallel, following the manufacturer’s protocols, DNA from stomach content samples obtained seasonally was isolated using the DNeasy Blood and Tissue Extraction Kit (Qiagen, Germany). For both extraction protocols, the DNA concentration was quantified for each extract using the Qubit High Sensitivity dsDNA Assay (Thermo Fisher Scientific). Then, all the DNA samples were stored at -20 °C.
Metabarcoding protocol
To obtain an identification of the different species occurring in the planktonic mixture from the Gullmarsfjord along the season, a 313 base-pair fragment of the mitochondrial cytochrome oxidase subunit I (COI) gene and around 150 base-pair fragment of the ribosomal subunit 18 (18S) gene were amplified through thermal cycler (Veriti, Thermal cycler, Thermo Fisher Scientific) using the Supreme NZYTaq 2x Green Master Mix DNA polymerase (NZYtech, Portugal). PCRs were performed in a final volume of 12.5 µL containing 1.25 µL of template DNA, 0.5 µM of the primers, 3.125 µL of Supreme NZYTaq 2x Green Master Mix DNA polymerase, and ultrapure water up to 12.5 µL. Polymerase chain reaction (PCR) was performed with conserved consensus primers: mlCOIintF (Forward: 5’ GGWACWGGWTGAACWGTWTAYCCYCC 3’)26 and LoboR1 (Reverse 5’ TAAACYTCWGGRTGWCCRAARAAYCA 3’)27 for COI, and (forward: 5’ TGGTGCATGGCCGTTCTTAGT 3’) and (reverse: 5’ CATCTAAGGGCATCACAGACC 3’) for 18 S28. The PCR conditions were: for the COI, 95 °C for 5 min, 35 cycles of 95 °C for 30 s, 56 °C for 45 s, 52 °C for 45 s, 72 °C for 45 s, and a final step at 72 °C for 7 min; for the 18 S, 95 °C for 5 min, 30 cycles of 95 °C for 30 s, 61 °C for 45 s, 54 °C for 45 s, 72 °C for 45 s, and a final step at 72 °C for 7 min. PCR products were electrophoresed in 2% agarose and visualized after staining with GreenSafe (NZYtech). Following the manufacturer’s protocol, positive PCR products were purified using Mag-Bind RXNPure Plus magnetic beads (Omega Biotek). Finished libraries were pooled in equimolar amounts according to the quantification results of the Qubit dsDNA HS Assay (Thermo Fisher Scientific, USA). The pool was sequenced in a fraction of 1/16 of a MiSeq flow cell (Illumina).
For metazoan library preparation, the same COI primers as above were used. These primers included the Illumina sequencing primer sequences attached to their 5’ ends. To prevent the amplification of host Amphiura filiformis DNA, a blocking primer (5’ AATCAGAAGAGRTGTTGGTAGAGAATAGGG 3’) with a C3 CPG spacer added to the 3’ end was designed and used in the library preparation29. In the first amplification step, PCRs were performed in a final volume of 25 µL, containing 2 µL of template DNA, 1 µM of the primers, 10 µM of the blocking primer, 5 µL of 5x GoTaq Green Master Mix (Promega, USA), and ultrapure water up to 25 µL. The reaction mixture was incubated as follows: an initial denaturation step at 95 ºC for 5 min, followed by 35 cycles of 95 ºC for 30 s, 56 ºC for 45 s, 52 ºC for 45 s, 72 ºC for 45 s, and a final extension step at 72 ºC for 7 min. The two primer annealing steps (56 ºC for 45 s and 52 ºC for 45 s) were included to enhance the binding of the blocking primer to the target DNA. The amplified PCR products were verified on 2% agarose gels stained with ethidium bromide and imaged under UV light. After cleaning up the PCR products with Sera magnetic beads, the index PCR was performed with the Illumina Nextera Index. Finished libraries were pooled in equimolar amounts according to the quantification results of the Qubit dsDNA HS Assay (Thermo Fisher Scientific, USA). The pool was sequenced on a NovaSeq machine by Genomics Core Leuven.
In silico analysis
The quality of the FASTQ files was assessed using FastQC software (Version 0.12.0)30. Then, the tool DADA2 (Version 1.26) was used to remove the PCR primers, filter the reads according to their quality, denoise, merge the forward and reverse reads, cluster the resulting sequences into Amplicon Sequence Variants (ASVs) and remove chimeric sequences31. The taxonomic assignment was conducted using the MetaCOXI reference database32 based on the European Nucleotide Archive (ENA)33 and the Barcode of Life Data System (BOLD)34. The database was completed by targeting Dinophyceae, Hexanauplia, Cnidaria, Polychaeta, Ctenophora, and Tunicata species. The MetaCOXI reference database was settled with COI sequences obtained from BOLD for the following taxa: Pyrrophycophyta, Rhodophyta, Haptophyta, Bacillariophyta, Chlorarachniophyta, Ciliophora, Heterokontophyta, and Copepoda. In addition, for the planktonic mixture DNA pool 18 S taxonomic assignment, a pre-trained classifier of the Protist Ribosomal database (PR2) reference database was used35. All recorded COI barcode sequences were then matched to the obtained custom reference library with > 97% bootstrap support using the ‘assignTaxonomy’ command in DADA2. All recorded 18 S barcode sequences were then compared against the reference database using the feature-classifier approach ‘classify-sklearn’ implemented in QIIME 2 (Version 2024.10)36. A second taxonomic assignment method was optionally implemented in the pipeline, allowing assigning recorded barcode sequences using the basic local alignment search tool BLAST+ (Version 2.6.0) based on minimum similarity and minimum coverage (-perc_identity 70 and –qcov_hsp 80). An initial test implementing BLAST + assigned taxonomy only to the COI data set using a 96% percent identity threshold led to the exclusion of most clusters. After a first analysis of the obtained planktonic sequences through COI and 18 S protocol, a trimming was performed only to keep the zooplankton in the next steps of the analysis since no phytoplanktonic species are known to use coelenterazine as a luminous substrate.
Statistical analysis
All the statistical analyses were performed with R Studio (Version 2023.06.1, 2022, Posit Software, USA). Variance normality and equality were tested using the Shapiro–Wilk test and Levene’s test, respectively. When these parametric assumptions were met, with or without a log transformation, an ANOVA was coupled with the Tuckey test. Each difference was considered to be significant at a minimum P-value < 0.05. Values were graphically illustrated with mean and standard error of the mean (s.e.m). Additionally, Principal Component Analysis (PCA) was performed to assess correlations between luminometric parameters.
Results
Seasonal coelenterazine content and Renilla luciferase activity of plankton mixture
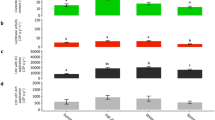
The plankton mixture’s luminous components (i.e., coelenterazine content and luciferase activity) were measured in Kristineberg Marine Research Station (Fiskebäckskil, Sweden) at each season (n = 3). Firstly, coelenterazine was detected throughout the year in the plankton mixture, significantly rising in spring 2022 (2.13 ± 0.6 ng g− 1) (Fig. 2a) (P-value = 0.093; F-value = 7.7852; Df = 3). Then, no statistical differences in luciferase activities were observed over the season (P-value = 0.3094; F-value = 1.4096; Df = 3), displaying a maximal value of 0.022 ± 0.009 109 q g− 1s− 1, following the same pattern as the coelenterazine content measurements (Fig. 2b). These results are consistent with the PCA analysis (Fig. 2c), highlighting positive correlations between the two luminometric parameters (i.e., coelenterazine content and luciferase activity). Supplementary Table S1 summarizes the mean and s.e.m values from coelenterazine and luciferase assays and recaps the statistical tests and the associated p-values for each season.
Seasonal luminometric measurements on the plankton mixture collected in the Gullmarsfjord (a) coelenterazine content, and (b) luciferase activity. (c) Principal component analysis (PCA) showing correlations between luminometric measurements. Different lettering indicates statistical differences. Error bars correspond to s.e.m.
Seasonal characterization of planktonic communities
Analysis of the total plankton list revealed a percent frequency of occurrence over the year of 68.20% of phytoplanktonic genera compared to 31.80% of zooplanktonic ones. Globally, phytoplankton communities dominated the plankton at each season (Fig. 3a; Supplementary Tables S2 and S3). Comparatively, zooplankton increased during spring (Fig. 3a; Supplementary Tables S2 and S3). Analysis of the zooplankton list indexing the species collected in the fjord from summer 2021 to spring 2022 revealed that Cyclopoida and Calanoida species represent the largest proportion among all seasons (Fig. 4a; Supplementary Table S3). This preponderance of Cyclopoida and Calanoida orders is visible across seasons (Supplementary Fig. S1a; Table S3). The plankton list analysis reveals a percent frequency of occurrence over the year of 21.06% of luminous plankton compared to total plankton abundance.
Zooplankton and phytoplankton percent frequency of occurrence in the Gullmarsfjord evaluated seasonally on the online plankton list (https://sharkweb.smhi.se/hamta-data/). Percent frequency of occurrence over the seasons of (a) the total phyto- and zooplankton, (b) luminous genus only, (c) retrieved phytoplankton luminous, and (d) retrieved zooplankton luminous genera.
Zooplankton percent frequency of occurrence in the Gullmarsfjord. Zooplanktonic communities evaluated on (a–c) the plankton list available online (https://sharkweb.smhi.se/hamta-data/) and through (d–f) metabarcoding method using COI and 18 S sequences. Percent frequency of occurrence over the year of (a,d) zooplankton, (b,e) luminous genus only and (c,f) luminous genus using coelenterazine only.
Analysis of the total plankton list focused on the luminous genera highlighted a percent frequency of occurrence over the year of 36.05% and 63.95% of bioluminescence genera for phytoplankton and zooplankton, respectively. At each season, luminous phytoplankton genera presented a higher proportion than luminous zooplankton (Fig. 3b; Supplementary Tables S2 and S3). Among the plankton list analysis, 9 phytoplankton genera were identified as luminous (Fig. 3c; Supplementary Tables S2). Consistently with the total plankton analysis, an increase of luminous zooplanktonic genera occurred in spring (Fig. 3b; Supplementary Tables S2 and S3).
By refining the data analysis and restricting to the luminous genera, 14 different luminous zooplanktonic genera were found to occur in the fjord water with a prevalence of Oithona, Paracalanus and Centropages genera (Figs. 3d and 4b; Supplementary Table S3). The plankton list analysis highlights a percent frequency of occurrence of 42.36% of luminous zooplankton over the total zooplankton abundance. According to the seasonal monitoring, the summer presented the highest species richness (Supplementary Fig. S1b; Table S3). At each season, Oithona, Paracalanus and Centropages genera represented the highest percent frequency of occurrence of luminous zooplanktonic genera monitored in the fjord (Supplementary Fig. S1b; Table S3). Species known to use coelenterazine as the substrate for light production were retrieved throughout the year in the zooplankton list (Fig. 4c; Supplementary Table S3). The luminous zooplanktonic species, using coelenterazine, percent of occurrence represents 3.89% of the total abundance of the zooplankton recorded in the Gullmarsfjord. The genera Triconia, Oikopleura, Metridia, and Mnemiopsis were, in order of frequency of occurrence, the major representatives retrieved in the luminous zooplankton using coelenterazine for light production (Fig. 4c; Supplementary Table S3). While Triconia, Oikopleura, and Mnemiopsis genera dominated during summer, Triconia appeared to be the almost exclusive coelenterazine users during winter (Supplementary Fig. S1c; Table S3). Finally, the Metridia genus proportion increased considerably during the spring compared to the other seasons (Supplementary Fig. S1c; Table S3).
The metabarcoding analysis (i.e., through COI and 18 S sequences analysis) revealed the presence of both phytoplankton and zooplankton in the Gullmarsfjord over the year (Supplementary Tables S4, S5, and S6). Sequences of two phytoplanktonic genera were retrieved (i.e., Tripos and Protoperidinium; Supplementary Tables S4) before trimming the plankton metabarcoding dataset.
Consistently with the zooplankton list analysis, the metabarcoding performed on the zooplankton seasonally collected either using 18 S and COI sequences presented a preponderance of the Cyclopoida and Calanoidea orders over the year (Fig. 4d; Supplementary Tables S5 and S6). For each season, the metabarcoding analysis performed with 18 S and COI primers highlighted that five genera belonging to the former orders were the most abundant in the fjord waters (Supplementary Fig. S2a and S3a; Tables S5 and S6). The COI sequence’s percent frequency of occurrence analysis revealed a similar pattern between the Paracalanus and Microcalanus genera (Supplementary Fig. S2a; Table S5). The Paracalanus genus was dominant from the summer to winter, while the Microcalanus percentage appeared lower during these seasons and drastically higher during spring. The COI analysis displayed a high percentage of Acartia and Oithona genera during summer compared to the three other seasons (Supplementary Fig. S2a; Table S5). The 18 S analysis provided evidence of a community switch between Paracalanus and Pseudocalanus genera over seasons. The Paracalanus genus appeared prevalent during the summer, fall, and winter seasons while drastically decreasing in fall and being replaced by Pseudocalanus genus communities (Supplementary Fig. S3a; Table S6). Among the luminous genera retrieved through the metabarcoding analysis, the Paracalanus genus widely dominates the luminous zooplankton communities (Fig. 4e; Supplementary Fig. S2b and S3b; Tables S5 and S6). Whatever primer was used, the Oithona, Centropages and Metridia genera appeared in the luminous zooplankton communities (Fig. 4e). Conversely, the Ctenophora (i.e., Mnemiopsis genus) and the Appendicularia (i.e., Oikopleura genus) occurred respectively in the COI and 18 S analysis (Fig. 4f).
The seasonal metabarcoding analysis spotted differences in the occurrence of zooplanktonic genera that use coelenterazine as a luminous substrate. According to the COI assay, the Oikopleura genus appears in higher proportion in summer and fall. On the contrary, Clytia and Metridia genera appear exclusively during the fall and winter seasons, respectively (Supplementary Fig. S2c; Table S5). The COI analysis did not detect any zooplanktonic genus containing coelenterazine during spring (Supplementary Fig. S2c; Table S5). Consistently, the 18 S approach pinpointed a high occurrence of Clytia and Metridia genera in fall and winter. Concerning the cnidarians and the ctenophora containing coelenterazine, the Mmemiopsis proportion rose drastically in summer, while the Obelia genus popped up only during the spring (Supplementary Fig. S3c; Table S6).
Seasonal characterization of Amphiura filiformis diet
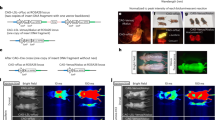
The stomach content of A. filiformis was analyzed with a metabarcoding technique based on COI primers. Despite the use of blocking primer to limit the detection of A. filiformis sequences, the metabarcoding analysis reveals 99.12% of brittle star sequences within its stomach content (Fig. 4a; Supplementary Table S7). This high proportion of A. filiformis sequences in the stomach content can be linked to cannibalism behavior observed in this species (Fig. 5b, c, d). Besides, the remaining 0.88% of sequences found in the stomach content encompassed 99.62% of sequences from the Cyclopoida species (Fig. 5a; Supplementary Table S7). Cyclopoida order contains multiple species known to be luminous, some using coelenterazine as a luminous substrate. As mentioned in previous studies37, A. filiformis feeds from planktonic pelagic species by extending its arms in the water column (Fig. 5e).
Analysis of Amphiura filiformis stomach content. (a) Percent frequency of occurrence in the stomach content DNA samples across all seasons; bold species name indicates the luminous status, and asterisk signifies the use of coelenterazine for light production. (b–d) Time-lapse pictures of cannibalism behavior observed in A. filiformis; interval time of 2 min between each picture. (e) Picture showing arms extending in the water column to catch suspended materials. G: gonad, M: mouth, sp: spine. Scale bar: 0.5 cm.
Finally, the exclusion of the Cyclopoida species from the metabarcoding analysis revealed the presence of seven benthic species or genera in extremely low proportion (0.38%), in A. filiformis stomach content (Fig. 5a; Supplementary Table S7). Among these species, genera, or orders, the luminous cnidaria Pennatula phosphorea was identified (Fig. 5a; Supplementary Table S7). The seasonal analysis of the stomach content highlighted the presence of luminous organisms using coelenterazine as substrate at each season (Supplementary Table S7).
Discussion
The burrowing brittle star, A. filiformis, is known to depend on an exogenous supply of coelenterazine to emit blue light at the basis of its arm spine21,24. However, the main question is identifying prey within the planktonic communities that provide the coelenterazine to the brittle star.
Through seasonal monitoring, fluctuations of phytoplanktonic and zooplanktonic genera were highlighted. Similarly, the abundance of luminous zooplanktonic genera varied throughout the year. Based on the first luminometric measurements performed on the plankton from Swedish waters, the plankton mixtures were demonstrated to contain coelenterazine and Renilla luciferase-based luminous system at each season. Finally, the zooplanktonic genera using a coelenterazine-based luminous system are found annually. This finding confirms the recent hypothesis suggesting a nonrestricted amount of coelenterazine in the fjord that allows a continuous exogenous supply of the luminous substrate24. This acquisition is required to maintain the brittle star’s luminous capabilities across seasons24. Besides, DNA traces of coelenterazine-dependent luminous organisms were retrieved within the stomach content of A. filiformis, adding evidence of an external acquisition of the coelenterazine substrate from the diet.
As observed in most marine environments, the Gullmarsfjord had a larger yearly proportion of phytoplankton than zooplankton38,39,40. Concerning the zooplankton communities’ abundance variation, whatever the technique used, the present results highlighted a dominancy of the Calanoida and Cyclopoidea over the season. This result is consistent with the literature on the zooplankton communities (e.g., Pseudocalanus, Microcalanus, Acartia, and Oithona genera) within the Gullmarsfjord23,41.
The analysis shows a lower proportion of Calanus in winter, likely due to changing environmental conditions (e.g., hydrodynamic). Our study sampled plankton at 25–30 m (i.e., depth of A. filiformis sampling), while some zooplankton genera (e.g., Calanus, Pseudocalanus, and Microcalanus) may be trapped in the deep layer of the fjord (i.e., below 60 m) due to winter physicochemical conditions42,43. Therefore, the observed proportions may not fully represent the plankton communities in the Gullmarsfjord. Some biotic factors might also impact the zooplanktonic communities, such as predation by other planktonic species (e.g., Ctenophora and Cnidaria) through top-down effect44.
The spring season displays both phyto- and zooplanktonic blooms with a rise in abundance and diversity of the plankton. A similar increase was already mentioned in various Swedish fjords and could be related to an increasing number of small development stages of copepods (e.g., Acartia, Centropages, Temora genera)45,46. Moreover, this spring bloom is commonly followed by higher sedimentation rates that impact the benthic community as a source of organic matter47,48,49.
Finally, the current results identify light-emitting plankton species in the Gullmarsfjord water across seasons through plankton list analysis and the metabarcoding method. First, several phytoplanktonic genera were identified as luminous. Some retrieved luminous genera from the current analysis (i.e., Alexandrium, Gonyaulax, Lingulodinium, Noctiluca) displayed a well-characterized luminous system50,51,52,53. Up to now, all dinoflagellate species produce light upon a tetrapyrrole substrate, the dinoflagellate luciferin, and a specific luciferase4,11,54,55,56.
Then, our results underlined different zooplanktonic genera known to be luminous2,4. Some listed species are considered dubious luminous, such as Oithona, Paracalanus, Centropages, Corycaeus or Pleurobrachia, even if mentions or data exists on their luminescence. Several reports were made on the luminescence of Oithona species57,58,59,60. Similarly, Lapota and Losee, 1984 performed a listing of the luminous plankton occurring in the Sea of Cortez and reported Paracalanus, Centropages, and Corycaeus as displaying luminous species61. A conflictual status is recorded for the ctenophore Pleurobrachia pileus with a study mentioning the absence of light production62, against others stating and arguing with recorded luminometric measurements on the bioluminescence of this species63,64. Some of these retrieved luminous genera use coelenterazine as a luminous substrate (e.g., Mnemiopsis, Metridia, Oikopleura)2,4. It should be noted that Triconia conifera is the only luminous Oncaeidae copepod, and it is strongly suggested that a coelenterazine-based system be used to produce light65,66,67. This result was confirmed by the luminometric measurements performed in the plankton mixture. Coelenterazine is the most widespread phylogenetic luciferin substrate found in at least ten taxa2,4,68,69. Zooplanktonic genera using coelenterazine were retrieved during all seasons, with the lowest species diversity in winter. However, it should be noted that the luminous substrate is known only for a limited number of bioluminescent zooplankton species4,6,65. The luminometric assays performed on the plankton supported these results with an increase in coelenterazine content and luciferase activity during spring. It should be noted that coelenterazine is retrieved in small amounts in non-luminous organisms70. Additionally, some research has highlighted the presence of coelenterazine in non-luminous tissues (e.g., gonads) and larval stages of luminous organisms18,70,71,72,73. The luciferase activity measurements in the present study are essential to evaluate the presence of plankton using a functional coelenterazine-Renilla luciferase system to produce light.
Our results bring to light several bioluminescent pelagic genera or species using coelenterazine that live seasonally in sympatry with the brittle star A. filiformis. These zooplankton genera and larval stages are potential prey and sources of coelenterazine for the brittle stars. The fact that they were detected seasonally may support the constant supply of coelenterazine to maintain the stable luminous capabilities of A. filiformis over the seasons24. Consistently, the present data underlined the presence of Cyclopoida in the brittle star stomach content. The most represented coelenterazine-dependent luminous genera found in the plankton list is the Cyclopoida Triconia65, which could represent a source of substrate acquisition for the brittle star species. Other luminous Cyclopoida are abundant in the retrieved zooplanktonic fjord communities, such as Oithona. According to Herring, 1985, 1988 and Temnykh et al., 2022, Oithona similis is reported as a luminous species without any clues on its luminous system, which could be dependent on coelenterazine as Triconia57,58,59,60. Another luminous species using coelenterazine was found in the stomach content analysis, the sea pen Pennatula phosphorea74. This species is known to live in sympatry with A. filiformis in the muddy sediment of the fjord; hence, this meroplanktonic species could represent a source of coelenterazine. Finally, as visually observed and pinpointed by the metabarcoding analysis, A. filiformis is a cannibalism species that might snatch the coelenterazine substrate from conspecifics. Without being able to discriminate ontogenic stages (i.e., meroplankton) with the metabarcoding analysis, parent-offspring cannibalism can not be excluded75,76. Similar observations have been made for the luminous midshipman fish, Porichthys notatus, which depends on Cypridina-type luciferin exogenous supply77. The young fishes were easily eaten by older conspecifics78. Consequently, the exclusive suspensive feeding mode of A. filiformis needs to be redefined since our data have highlighted a mix of feeding modes with a potential shift toward the suspension and detritivore, cannibalism, and deposit feeding modes that have already been suggested in the literature37,79.
To sum up, the present study is the first to seasonally identify planktonic luminous species depending on coelenterazine in the Gullmarsfjord. Moreover, the stomach content metabarcoding analysis added the first milestone concerning the A. filiformis diet and confirmed the coelenterazine trophic acquisition hypothesis.
To resume, A. filiformis reproduces during summer, producing non-luminous ophiopluteus pelagic larvae. As the brittle star gonads, these larvae contain coelenterazine. The brittle star can emit light after the metamorphosis and the settlement of the juvenile. The newly benthic juvenile produces light when luciferase is expressed in the developing arm’s spines. The fast arm growth is essential for the feeding process and the capture of prey containing coelenterazine. The continuous presence of coelenterazine across seasons in the Gullmarsfjord allows the permanent luciferin supply for trophic acquisition and maintenance of A. filiformis luminous capabilities throughout the year. Nevertheless, the ingestion rate of coelenterazine remains to be determined. To complete the trophic acquisition story of luminous capabilities in A. filiformis, ongoing works focused on the physiological transfer of the coelenterazine from the stomach content to the photogenic sites.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Harvey, E. N. Bioluminescence (Academic, 1952).
Haddock, S. H. D., Moline, M. A. & Case, J. F. Bioluminescence in the sea. Annu. Rev. Mar. Sci. 2, 443–493. https://doi.org/10.1146/annurev-marine-120308-081028 (2010).
Claes, J. M., Haddock, S. H. D., Coubris, C. & Mallefet, J. Systematic distribution of bioluminescence in marine animals: a species-Level Inventory. Life 14, 432. https://doi.org/10.3390/life14040432 (2024).
Duchatelet, L. & Dupont, S. Marine eucaryotes bioluminescence: a review of species and their functional biology. Mar. Life Sci. Technol. https://doi.org/10.1007/s42995-024-00250-0 (2024).
Shimomura, O. Bioluminescence: Chemical Principles and Methods (World Scientific, 2012).
Kaskova, Z. M., Tsarkova, A. S. & Yampolsky, I. V. 1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 45, 6048–6077. https://doi.org/10.1039/C6CS00296J (2016).
Lau, E. S. & Oakley, T. H. Multi-level convergence of complex traits and the evolution of bioluminescence. Biol. Rev. 96, 673–691. https://doi.org/10.1111/brv.12672 (2021).
Schramm, S. & Weiß, D. Bioluminescence – the vibrant glow of nature and its chemical mechanisms. ChemBioChem 25 (e202400106). https://doi.org/10.1002/cbic.202400106 (2024).
Markova, S. V. & Vysotski, E. S. Coelenterazine-dependent luciferases. Biochem. Mosc. 80, 714–732. https://doi.org/10.1134/S0006297915060073 (2015).
Shimomura, O. & Teranishi, K. Light-emitters involved in the luminescence of coelenterazine. Luminescence 15, 51–58 (2000).
Delroisse, J., Duchatelet, L., Flammang, P. & Mallefet, J. Leaving the dark side? Insights into the evolution of luciferases. Front. Mar. Sci. 8, 673620. https://doi.org/10.3389/fmars.2021.673620 (2021).
Mallefet, J. Echinoderm bioluminescence: where, how and why do so many ophiuroids glow? (2009).
Rosenberg, R. & Lundberg, L. Photoperiodic activity pattern in the brittle star Amphiura filiformis. Mar. Biol. 1 https://doi.org/10.1007/s00227-004-1365-z (2004).
Sköld, M., Loo, L. O. & Rosenberg, R. Production, dynamics and demography of an Amphiura filiformis population. Mar. Ecol. Prog. Ser., 81–90. https://doi.org/10.3354/meps103081 (1994).
Burns, G. et al. Dynamic gene expression profiles during arm regeneration in the brittle star Amphiura filiformis. J. Exp. Mar. Biol. Ecol. 407, 315–322. https://doi.org/10.1016/j.jembe.2011.06.032 (2011).
Czarkwiani, A., Ferrario, C., Dylus, D. V., Sugni, M. & Oliveri, P. Skeletal regeneration in the brittle star Amphiura filiformis. Front. Zool. 13, 18. https://doi.org/10.1186/s12983-016-0149-x (2016).
Dupont, S. & Thorndyke, M. C. Growth or differentiation? Adaptive regeneration in the brittlestar Amphiura filiformis. J. Exp. Biol. 209, 3873–3881. https://doi.org/10.1242/jeb.02445 (2006).
Coubris, C., Duchatelet, L., Dupont, S. & Mallefet, J. A brittle star is born: Ontogeny of luminous capabilities in Amphiura filiformis. PLoS One 19, e0298185. https://doi.org/10.1371/journal.pone.0298185 (2024).
Delroisse, J. et al. A puzzling homology: a brittle star using a putative cnidarian-type luciferase for bioluminescence. Open Biol. 7, 160300. https://doi.org/10.1098/rsob.160300 (2017).
Lau, E. S. et al. Functional characterization of luciferase in a brittle star indicates parallel evolution influenced by genomic availability of haloalkane dehalogenase. Preprint at https://doi.org/10.1101/2024.10.14.618359 (2024).
Mallefet, J., Duchatelet, L. & Coubris, C. Bioluminescence induction in the ophiuroid Amphiura filiformis (Echinodermata). J. Exp. Biol., jeb.218719. https://doi.org/10.1242/jeb.218719 (2020).
Larink, O. & Westheide, W. Coastal Plankton: Photo Guide for European Seas (Verlag Dr. Friedrich Pfeil, 2011).
Lindahl, O. & Hernroth, L. Phyto-Zooplankton community in coastal waters of western Sweden -An ecosystem off balance? Mar. Ecol. Prog. Ser. 10, 119–126. https://doi.org/10.3354/meps010119 (1983).
Coubris, C. et al. Maintain the light, long-term seasonal monitoring of luminous capabilities in the brittle star Amphiura filiformis. Sci. Rep. 14, 13238. https://doi.org/10.1038/s41598-024-64010-x (2024).
Deheyn, D., Mallefet, J. & Jangoux, M. Intraspecific variations of bioluminescence in a polychromatic population of Amphipholis squamata (Echinodermata: Ophiuroidea). JMBA 77, 1213–1222. https://doi.org/10.1017/S0025315400038728 (1997).
Leray, M. et al. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 10, 34. https://doi.org/10.1186/1742-9994-10-34 (2013).
Lobo, J. et al. Enhanced primers for amplification of DNA barcodes from a broad range of marine metazoans. BMC Ecol. 13, 34. https://doi.org/10.1186/1472-6785-13-34 (2013).
Hardy, C. M., Krull, E. S., Hartley, D. M. & Oliver, R. L. Carbon source accounting for fish using combined DNA and stable isotope analyses in a regulated lowland river weir pool. Mol. Ecol. 19, 197–212. https://doi.org/10.1111/j.1365-294X.2009.04411.x (2010).
Vestheim, H. & Jarman, S. N. Blocking primers to enhance PCR amplification of rare sequences in mixed samples – a case study on prey DNA in Antarctic krill stomachs. Front. Zool. 5, 12. https://doi.org/10.1186/1742-9994-5-12 (2008).
Andrews, S. FastQC A quality control tool for high throughput sequence data (2010).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. https://doi.org/10.1038/nmeth.3869 (2016).
Balech, B., Sandionigi, A., Marzano, M., Pesole, G. & Santamaria, M. MetaCOXI: an integrated collection of metazoan mitochondrial cytochrome oxidase subunit-I DNA sequences. Database 2022, baab084. https://doi.org/10.1093/database/baab084 (2022).
Leinonen, R. et al. The European nucleotide archive. Nucleic Acids Res. 39, D28–D31. https://doi.org/10.1093/nar/gkq967 (2011).
Ratnasingham, S. & Hebert, P. D. N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364 https://doi.org/10.1111/j.1471-8286.2007.01678.x (2007).
Guillou, L. et al. The Protist Ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. https://doi.org/10.1093/nar/gks1160 (2012).
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. https://doi.org/10.1186/s40168-018-0470-z (2018).
Loo, L., Jonsson, P., Sköld, M. & Karlsson, Ö. Passive suspension feeding in Amphiura filiformis (Echinodermata: Ophiuroidea): feeding behaviour in flume flow and potential feeding rate of field populations. Mar. Ecol. Prog. Ser. 139, 143–155. https://doi.org/10.3354/meps139143 (1996).
Nielsen, E. S. The balance between phytoplankton and zooplankton in the sea. ICES J. Mar. Sci. 23, 178–188. https://doi.org/10.1093/icesjms/23.2.178 (1958).
Longhurst, A. R. Interactions between zooplankton and phytoplankton profiles in the eastern tropical Pacific Ocean. Deep Sea Res. Oceanogr. Abstr. 23, 729–754. https://doi.org/10.1016/S0011-7471(76)80017-4 (1976).
Le Fèvre, J. Aspects of the biology of frontal systems. Adv. Mar. Biol., 163–299. https://doi.org/10.1016/S0065-2881(08)60109-1 (1987).
Langer, J. A. F. et al. Community barcoding reveals little effect of ocean acidification on the composition of coastal plankton communities: evidence from a long-term mesocosm study in the Gullmar fjord, Skagerrak. PLoS One 12, e0175808. https://doi.org/10.1371/journal.pone.0175808 (2017).
Lindahl, O. & Perissinotto, R. Short-term variations in the zooplankton community related to water exchange processes in the Gullmar fjord, Sweden. J. Plankton Res. 9, 1113–1132. https://doi.org/10.1093/plankt/9.6.1113 (1987).
Gao, S. et al. Overwintering distribution, inflow patterns and sustainability of Calanus finmarchicus in the North Sea. Prog. Oceanogr. 194, 102567. https://doi.org/10.1016/j.pocean.2021.102567 (2021).
Tiselius, P. & Møller, L. F. Community cascades in a marine pelagic food web controlled by the non-visual apex predator Mnemiopsis leidyi. J. Plankton Res., fbw096v1. https://doi.org/10.1093/plankt/fbw096 (2017).
Olsson, I. & Lundh, E. On plankton production in Kungsbacka fjord, an estuary on the Swedish west coast. Mar. Biol. 24, 17–28. https://doi.org/10.1007/BF00402843 (1974).
Hernroth, L. Marine pelagic rotifers and tintinnids – important trophic links in the spring plankton community of the Gullmar Fjord, Sweden. J. Plankton Res. 5, 835–846. https://doi.org/10.1093/plankt/5.6.835 (1983).
Båmstedt, U. Spring-bloom dynamics in Kosterfjorden, western Sweden: variation in phytoplankton production and macrozooplankton characteristics. Sarsia 70, 69–82. https://doi.org/10.1080/00364827.1985.10420619 (1985).
Graf, G., Schulz, R., Peinert, R. & Meyer-Reil, L. A. Benthic response to sedimentation events during autumn to spring at a shallow-water station in the Western Kiel Bight: I. Analysis of processes on a community level. Mar. Biol. 77, 235–246. https://doi.org/10.1007/BF00395812 (1983).
Tiselius, P. & Kuylenstierna, M. Growth and decline of a diatom spring bloom phytoplankton species composition, formation of marine snow and the role of heterotrophic dinoflagellates. J. Plankton Res. 18, 133–155. https://doi.org/10.1093/plankt/18.2.133 (1996).
Li, L. & Woodland Hastings, J. The structure and organization of the luciferase gene in the photosynthetic dinoflagellate Gonyaulax polyedra. Plant. Mol. Biol. 36, 275–284. https://doi.org/10.1023/A:1005941421474 (1998).
Liu, L. & Hastings, J. W. Two different domains of the luciferase gene in the heterotrophic dinoflagellate Noctiluca scintillans occur as two separate genes in photosynthetic species. Proc. Natl. Acad. Sci. U. S. A. 104, 696–701 (2007). https://doi.org/10.1073/pnas.0607816103
Fajardo, C. et al. New perspectives related to the bioluminescent system in dinoflagellates: Pyrocystis lunula a case study. Int. J. Mol. Sci. 21, 1784. https://doi.org/10.3390/ijms21051784 (2020).
Park, S. A. et al. Bioluminescence capability and intensity in the dinoflagellate Alexandrium species. Algae 36, 299–314. https://doi.org/10.4490/algae.2021.36.12.6 (2021).
Liu, L., Wilson, T. & Hastings, J. W. Molecular evolution of dinoflagellate luciferases, enzymes with three catalytic domains in a single polypeptide. Proc. Natl. Acad. Sci. U. S. A. 101, 16555–16560. https://doi.org/10.1073/pnas.0407597101 (2004).
Marcinko, C. L. J., Painter, S. C., Martin, A. P. & Allen, J. T. A review of the measurement and modelling of dinoflagellate bioluminescence. Prog. Oceanogr. 109, 117–129. https://doi.org/10.1016/j.pocean.2012.10.008 (2013).
Nicolas, M. T., Nicolas, G., Johnson, C. H., Bassot, J. M. & Hastings, J. W. Characterization of the bioluminescent organelles in Gonyaulax polyedra (dinoflagellates) after fast-freeze fixation and antiluciferase immunogold staining. J. Cell. Biol. 105, 723–735. https://doi.org/10.1083/jcb.105.2.723 (1987).
Herring, P. J. Bioluminescence in the Crustacea. J. Crust Biol. 5 (4), 557–573. https://doi.org/10.2307/1548235 (1985).
Herring, P. J. Copepod luminescence. Hydrobiologia 167–168, 183–195. https://doi.org/10.1007/BF00026304 (1988).
Tokarev, Y. N. Fundamentals of Biophysics Ecology of Hydrobionts (EKOSI-Gidrofizika, 2006).
Temnykh, A. V., Silakov, M. I. & Melnik, A. V. Large luminous plankton in bioluminescence peaks in the Black Sea. Russ. J. Mar. Biol. 48, 247–255. https://doi.org/10.1134/S1063074022040113 (2022).
Lapota, D. & Losee, J. R. Observations of bioluminescence in marine plankton from the Sea of Cortez. J. Exp. Mar. Biol. Ecol. 77(3), 209–239. https://doi.org/10.1016/0022-0981(84)90121-7
Haddock, S. H. D. & Case, J. F. Not all ctenophores are bioluminescent: Pleurobrachia. Biol. Bull. 189(3), 356–362. https://doi.org/10.2307/1542153
Temnykh, A. V., Silakov, M. I. & Mashukova, O. V. Bioluminescence of Ctenophore Pleurobrachia pileus (O. F. Müller, 1776) in the summer period. Biophysics 68, 596–606. https://doi.org/10.1134/S000635092304022X
Melnik, A., Silakov, M. I., Mashukova, O. V. & Melnik, L. Research into bioluminescence of the Black Sea ctenophores Pleurobrachia pileus O.F. Müller, 1776. Luminescence 38(8), 1477–1484. https://doi.org/10.1002/bio.4529
Thomson, C. M., Herring, P. J. & Campbell, A. K. The widespread occurrence and tissue distribution of the imidazolopyrazine luciferins. J. Biolumin. Chemilumin. 12, 87–91 (1997).
Herring, P. J., Latz, M. I., Bannister, N. J. & Widder, E. A. Bioluminescence of the poecilostomatoid copepod Oncaea conifera. Mar. Ecol. Prog Ser., 297–309 (1993).
Takenaka, Y., Yamaguchi, A. & Shigeri, Y. A light in the dark: ecology, evolution and molecular basis of copepod bioluminescence. J. Plankton Res. 39, 369–378. https://doi.org/10.1093/plankt/fbx016 (2017).
Widder, E. A. Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science 328, 704–708. https://doi.org/10.1126/science.1174269 (2010).
Shimomura, O., Inoue, S., Johnson, F. H. & Haneda, Y. Widespread occurrence of coelenterazine in marine bioluminescence. Comp. Biochem. Physiol. B. 65, 435–437. https://doi.org/10.1016/0305-0491(80)90044-9 (1980).
Shimomura, O. Presence of coelenterazine in non-bioluminescent marine organisms. Comp. Biochem. Physiol. B. 86, 361–363. https://doi.org/10.1016/0305-0491(87)90306-3 (1987).
Mallefet, J. & Shimomura, O. Presence of coelenterazine in mesopelagic fishes from the Strait of Messina. Mar. Biol. 124, 381–385. https://doi.org/10.1007/BF00363911 (1995).
Thomson, C. M., Herring, P. J. & Campbell, A. K. Evidence for de novo biosynthesis of coelenterazine in the bioluminescent midwater shrimp, Systellaspis debilis. J. Mar. Biol. Assoc. UK. 75, 165–171. https://doi.org/10.1017/S0025315400015277 (1995).
Duchatelet, L. et al. Coelenterazine detection in five myctophid species from the Kerguelen Plateau. In The Kerguelen Plateau: Marine Ecosystem + Fisheries: Proceedings of the Second Symposium (2019).
Duchatelet, L. et al. Waves of light at the bottom of the ocean: insights into the luminous systems of three Pennatuloidea (Anthozoa). Preprint at https://doi.org/10.1101/2024.04.30.591678 (2024).
Crowe, W., Josefson, A. & Svane, I. Influence of adult density on recruitment into soft sediments: a short-term in situ sublittoral experiment. Mar. Ecol. Prog. Ser. 41, 61–69. https://doi.org/10.3354/meps041061 (1987).
Sköld, M., Josefson, A. & Loo, L. O. Sigmoidal growth in the brittle star Amphiura filiformis (Echinodermata: Ophiuroidea). Mar. Biol. 139, 519–526. https://doi.org/10.1007/s002270100600 (2001).
Warner, J. A. & Case, J. F. The zoogeography and dietary induction of bioluminescence in the midshipman fish, Porichthys notatus. Biol. Bull. 159, 231–246. https://doi.org/10.2307/1541021 (1980).
Mensinger, A. F. & Case, J. F. Bioluminescence maintenance in juvenile Porichthys notatus. Biol. Bull. 181, 181–188. https://doi.org/10.2307/1542501 (1991).
Buchanan, J. B. A comparative study of some features of the biology of Amphiura filiformis and Amphiura chiajei [Ophiuroidea] considered in relation to their distribution. J. Mar. Biol. Assoc. UK. 44, 565–576. https://doi.org/10.1017/S0025315400027776 (1964).
Acknowledgements
The authors acknowledge Ursula Schwarz, captain of the Alice vessel, and the skillful members of the Kristineberg Center (Goteborg University, Sweden) for their help during the Amphiura filiformis collections. The authors acknowledge Isabel Casties, Linda Svanberg, and Peter Tiselius during the seasonal plankton collection and for providing the plankton monitoring list. The authors also want to thank Sam Dupont for the continuous support during the study and Adeline Tauran for all the advice and helpful skills during the planktonologist list analysis. The authors acknowledge Linda Dhondt for her technical support during the stomach DNA extraction. CC is a Ph.D. student under an FRIA fellowship, KM is a bioinformatician working at the Katholieke Universiteit Leuven (KU Leuven) Genomics Core, LD is a postdoctoral researcher at the Université catholique de Louvain - UCLouvain, and JM is a research associate FRS-FNRS. This study is the contribution of BRC 423 of the Biodiversity Research Center (UCLouvain) from the Earth and Life Institute Biodiversity (ELIV) and the “Centre Interuniversitaire de Biologie Marine” (CIBIM).
Funding
This work was supported by a FRIA grant (40014530) awarded to CC and an FRS-FNRS grant (T.0169.20) awarded to the Université de Louvain – UCLouvain Marine Biology Laboratory and the Université de Mons Biology of Marine Organisms and Biomimetics Laboratory. The research leading to these results also received funding from the European Union’s Horizon 2018 research and innovation program under grant agreement No 231, ASSEMBLE Plus project.
Author information
Authors and Affiliations
Contributions
CC, LD, and JM collected samples. LD and JM performed the microdissections to analyze the stomach content. KM and CC performed all the stomach content metabarcoding analyses. CC and LD performed, analyzed, and interpreted the experiments and were major contributors to figure creation and writing. LD and JM supervised the work and contributed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Coubris, C., Mirzaei, K., Duchatelet, L. et al. Availability and occurrence of coelenterazine in a Swedish fjord to maintain Amphiura filiformis bioluminescence. Sci Rep 14, 31803 (2024). https://doi.org/10.1038/s41598-024-82811-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-82811-y