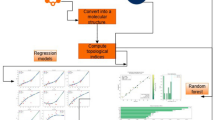
Abstract
This study presents a quantitative read-across structure-property relationship (q-RASPR) approach that integrates the chemical similarity information used in read-across with traditional quantitative structure-property relationship (QSPR) models. This novel framework is applied to predict the physicochemical properties and environmental behaviors of persistent organic pollutants, specifically polychlorinated biphenyls (PCBs) and polybrominated diphenyl ethers (PBDEs). By utilizing a curated dataset and incorporating similarity-based descriptors, the q-RASPR approach improves the accuracy of predictions, particularly for compounds with limited experimental data. The models’ performances were assessed using internal cross-validation and external testing, demonstrating significant enhancements in predictive reliability compared to conventional QSPR models. The findings highlight the potential of q-RASPR for use in regulatory risk assessments and optimizing remediation strategies by providing more precise insights into the environmental fate of these contaminants.
Similar content being viewed by others
Introduction
Polychlorinated biphenyls (PCBs) are persistent environmental pollutants that pose significant risks due to their bioaccumulation and toxicity. Understanding their physicochemical properties, such as partition coefficients and degradation kinetics, is essential for assessing environmental fate and risks. Recent advances in quantitative structure-property relationships (QSPR) models, leveraging computational chemistry and machine learning, have significantly enhanced the quality of prediction of these properties1. Some examples include models for the octanol-air partition coefficient (log KOA) using solvation-free energies2, deep belief networks for comprehensive property predictions3, and extended topological indices for partitioning behaviors4. Novel approaches in predicting degradation kinetics, through multivariate image analysis and quantum chemical calculations combined with machine learning, offer deeper insights into the environmental behavior of PCBs5,6,7. These advancements not only improve our understanding of PCBs but also aid in the development of more effective environmental management strategies.
Understanding the environmental behavior of PCBs, which are persistent organic pollutants, is crucial for their management and regulation. Recent advancements in computational modeling have significantly enhanced our ability to predict the physicochemical properties influencing PCBs’ environmental fate. One key development is a model that elucidates the thermodynamic relationship between the KOA and the solvation-free energy from the air to octanol (ΔGOA), incorporating the effects of dimerization to improve predictions8. Hierarchical Quantitative Structure-Activity Relationship (HQSAR) models have pinpointed the impact of chlorination patterns on PCBs’ bioaccumulation, leading to the design of less bioaccumulative congeners like PCB-2079.
QSPR models, particularly those based on Deep Belief Networks (DBN), have shown superior capability in predicting properties like partition coefficient (log P), octanol-air partition coefficient (log KOA), and bioconcentration factor (log BCF), outperforming earlier models3. Introducing DBN marks a significant advancement in predicting PCBs’ physicochemical properties, showcasing superior accuracy over traditional models. The estimation of the octanol-water partition coefficient (log KOW) using solvation-free energy (ΔGOW) has also emerged as a promising approach, displaying competitive predictive accuracy2. A reassessment of KOA prediction methods, including group contribution and quantum chemical solvation models, has highlighted the advantages of fragment-based approaches10,11. These computational advances are pivotal for improving our understanding of PCBs’ environmental partitioning and for guiding regulatory and remediation efforts. Previous literature explored the predictive capabilities of the Eyring equation for single electron oxidation reactions, with a focus on hydrogen abstraction processes, utilizing kinetic data and Hammett substituent constants to decipher reaction pathways12.
Central to the environmental investigation are PCBs, notorious for their bioaccumulation and biomagnification. A previous study employed molecular simulations alongside spectroscopic techniques and multivariate linear regression to predict the bioconcentration factors (BCFs) of PCBs, based on their molecular structures13. This not only aids in understanding their environmental fate but also in mitigating their ecological impact. Further, previous researchers leveraged Density Functional Theory (DFT) to enhance the precision of BCF predictions in aquatic organisms, using molecular descriptors to bridge quantum mechanics with bioaccumulation phenomena14. The development of Quantitative Structure-Activity Relationship (QSAR) models, both linear and nonlinear, for predicting logarithmic BCFs, highlights the use of large datasets to improve model fit and predictability15. These efforts collectively advance our understanding of chemical kinetics and environmental chemistry, offering novel insights into tackling pollution and preserving ecosystem health.
Previous studies have explored the structural parameters and environmental behavior of various classes of persistent organic pollutants, such as PCBs and diphenyl ethers, employing computational methods to analyze their diverse derivatives16. By utilizing QSAR/QSPR models, researchers have predicted key chemical properties relevant to regulatory requirements, thereby reducing the need for extensive experimental testing. This research builds on these methodologies to extend their applicability across a wider range of structurally similar compounds, underscoring the importance of molecular structure in predicting environmental persistence and toxicity17.
A significant portion of the study focuses on the structural analysis of various diphenyl ethers, highlighting the impact of molecular features on environmental properties18. Additionally, the research examines the photodegradation of PCBs in different media, contributing to a deeper understanding of their environmental transformations19. The investigation extends to the vapor pressure and partitioning behavior of semi-volatile organic compounds (SVOCs), such as polybrominated diphenyl ethers (PBDEs), using temperature-dependent descriptors. This aspect is crucial for assessing the environmental transport and distribution of SVOCs20. This study reviews effective methods for the degradation and removal of PBDEs in environmental settings and employs machine learning to develop QSPR models for predicting BCFs and toxicity in aquatic organisms13,21,22,23. This multifaceted approach not only advances our understanding of diphenyl ethers and related compounds but also supports the principles of reducing animal testing in environmental assessments.
Despite the advances in QSPR modeling, existing approaches often face limitations in terms of predictability and generalizability, particularly when applied to structurally diverse datasets. These models typically do not incorporate chemical similarity information, which can reduce accuracy in predicting complex properties. To address these challenges, this study presents a novel q-RASPR approach that combines QSPR models with read-across techniques, thereby enhancing predictive accuracy by utilizing similarity descriptors and error metrics. This integrated approach not only improves robustness but also reduces overfitting, resulting in models with superior external validation performance.
In the evolving field of chemical informatics, the method of chemical similarity prediction emerges as a critical tool for elucidating the physicochemical characteristics of compounds. This approach, grounded in the principles of QSPR, utilizes descriptors essential for assessing a compound’s properties and for conducting Quantitative Read-Across Structure-Property Relationships (q-RASPR) analysis which combines the merits of supervised QSPR and unsupervised similarity-based read-across24.
Our present study applies the q-RASPR methodology and selectively excludes structurally distinct outlier compounds from similarity assessments within the training set, enhancing the precision of our statistical models. The similarity prediction tool reported here not only forecasts the properties of query compounds but also provides a comprehensive suite of similarity and error metrics, thereby offering a nuanced view of compound behavior.
The current study aims to investigate the enhancement of external predictive capabilities of QSPR models by integrating similarity-based descriptors with conventional structural and physicochemical descriptors. Maintaining consistency with established model frameworks and datasets, this study does not introduce new descriptors or datasets. Instead, it focuses on demonstrating the effectiveness of similarity descriptors in improving model predictive performance using existing data and structural features.
Materials and methods
Central to our q-RASPR analysis is the use of these similarity and error-based measures alongside chosen structural and physicochemical features to generate robust, reproducible, and predictive models. The previous CoMFA (Comparative Molecular Field Analysis) approach, while widely used in QSPR modeling, is known for its sensitivity to molecular alignment and its tendency to overfit data, which can reduce external predictability. By contrast, our proposed q-RASPR method utilizes similarity-based descriptors that do not require molecular alignment and incorporates read-across techniques to enhance robustness and predictive accuracy. This study adheres to the OECD principles for QSPR model validation. The predicted endpoints are clearly defined, and the algorithms are transparent and reproducible. The applicability domain has been determined to ensure reliable predictions. The models demonstrate strong internal and external validation metrics and provide mechanistic interpretations where feasible, thus fulfilling all five OECD principles1.
Data set selection
A comprehensive analysis was conducted on twelve distinct physicochemical data sets to explore a variety of physicochemical properties and environmental fates, including log Koc25, log t1/25, log Koa26,27,28, ln kOH29,30,31, log BCF32, RRT27, log kP33, log k34,35 and log J30 (please see Supplementary Materials SI0). The properties mentioned are key parameters often used in environmental chemistry and ecotoxicology to assess the behavior and impact of chemicals in the environment:
-
1.
log Koc (logarithm of the organic carbon-water partition coefficient): This parameter is a measure of a chemical’s affinity for soil or sediment organic matter. A higher log Koc value indicates a greater tendency of a compound to partition into organic matter, reducing its mobility in the environment. This can influence the chemical’s bioavailability, transport, and ultimate fate in soil and aquatic systems.
-
2.
log t1/2 (logarithm of the half-life of PCB congeners upon exposure to UV radiation (254 nm) in n-hexane solution): The half-life of a substance is the time required for its concentration to reduce to half its initial value. This is crucial for understanding the persistence of chemicals in the environment, in particular in the atmosphere. Substances with long half-lives may accumulate or persist in ecosystems, leading to long-term exposure effects on wildlife and humans.
-
3.
log Koa (logarithm of the octanol-air partition coefficient): This value represents a chemical’s potential to move between the air and organic phases (like fat tissues in organisms or organic matter in soils). It is important for assessing the volatilization of chemicals from soil or water into the atmosphere, and their subsequent deposition or bioaccumulation in terrestrial and aquatic ecosystems.
-
4.
ln kOH (natural logarithm of the gas-phase oxidation rate constant with hydroxyl radicals): This rate constant is crucial for understanding the atmospheric degradation of chemicals by reaction with hydroxyl radicals, a primary atmospheric oxidant. It helps predict the atmospheric lifetime of chemicals and their potential for long-range transport.
-
5.
log BCF (logarithm of the bioconcentration factor): BCF indicates the extent to which a chemical can accumulate in the tissues of organisms from water alone. It is a critical parameter for assessing the potential of substances to bioaccumulate in aquatic organisms, which can lead to biomagnification through food webs and pose risks to predators, including humans.
-
6.
RRT (gas-chromatographic relative retention time): In gas chromatography, RRT is used to identify compounds based on their retention times relative to a reference compound. Although not directly an environmental parameter, it is used in environmental analysis to identify and quantify pollutants in complex environmental samples.
-
7.
log kP (direct photolysis rate constants of PBDEs in hexane and methanol), log k (logarithm of the direct photolysis rate constants for PBDEs dissolved in methanol/water (80:20)), and log J (gas-phase photolysis rate constants): This can refer to various rate constants depending on the degradation rates of PBDEs under specific experimental conditions.
These properties provide a comprehensive picture of how chemicals interact with different components of the environment, their potential for long-term persistence, and their risks to ecosystems and human health. They are used in risk assessments, regulatory decisions, and the design of safer chemicals with minimal environmental impact. These data sets encompass a wide range of compounds, each accompanied by a specific set of structural and physicochemical descriptors, covering diverse chemical and physical property dimensions.
To predict the environmental behavior of persistent organic pollutants, particularly PCBs and PBDEs, we employed a comprehensive computational approach.
The computational tool utilized for descriptor calculation is PaDEL-Descriptor36, which computes a broad spectrum of molecular descriptors. The dataset was divided into training and testing sets using Euclidean Distance-based division at a 70:30 ratio (Table 1). The feature selection process was conducted using a Genetic Algorithm (GA) with the following parameters: a population size of 50 and 100 generations. From an initial pool of 931 potential two-dimensional descriptors, the study systematically refined the selection to develop robust QSPR models37. The GA was facilitated by the tool “GeneticAlgorithm_v4.1_Train,” available from http://teqip.jdvu.ac.in/QSAR_Tools/#GA4, to identify critical descriptors guided by a fitness function based on the Mean Absolute Error (MAE). To constrain the number of descriptors in the final model, a Java-based tool, BestSubsetSelection_v2.1 (available from http://teqip.jdvu.ac.in/QSAR_Tools/), was employed. This tool executed a grid search to identify combinations of descriptors that meet specific criteria for a MLR model.
Partial Least Squares (PLS) regression was then used to develop the QSPR models. PLS is particularly suited for handling datasets with high multicollinearity, a common issue in chemical descriptor data where many variables are interrelated. This method finds latent variables that capture the essential information in the dataset, allowing the model to remain stable even when dealing with complex data structures. By employing PLS, we ensured that the models could make reliable predictions even in the presence of highly correlated descriptors, thereby improving their robustness. The model construction process relied on the Java-based tool “PLS_SingleY_version 1.0” (available from http://teqip.jdvu.ac.in/QSAR_Tools/#PLS) to ensure accuracy and reliability in the analysis.
To further enhance the model’s predictions, especially for compounds with limited experimental data, we integrated regression with a read-across technique. This method uses structural similarity to predict the properties of compounds by comparing them with structurally related chemicals that have known properties. By incorporating similarity-based descriptors, the read-across approach refines the QSPR model’s ability to make accurate predictions, particularly for compounds where direct experimental data are scarce. This integration addresses the limitations of traditional QSPR models, which often struggle with predictions for under-represented compounds. The primary tool employed was Read-Across-v4.1 (available from https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home), which predicts outcomes based on a similarity approach; that is, by comparing the new query compounds with a known set of source compounds in the source set. To quantify similarity, three distinct algorithms were utilized: similarity measurement based on Euclidean distance, Gaussian kernel similarity, and Laplacian kernel similarity. These methods involved several hyperparameters, such as sigma (σ) for the Gaussian kernel and gamma (γ) for the Laplacian kernel, as well as the number of similar source compounds selected. This study adopted the basic configurations of these methods (σ = 1, γ = 1, and 10 similar source compounds) for predictions, to ensure fair comparisons without additional optimization of hyperparameters. To develop the q-RASPR models, the study integrated aspects based on similarity and employed the Java-based tool RASAR-Desc-Calc-v2.0 (available from https://sites.google.com/jadavpuruniversity.in/dtc-lab-software/home) to calculate the necessary q-RASPR descriptors.
Comprehensive internal and external validation was conducted to assess the reliability of the models. Internal validation included cross-validation techniques to ensure the model’s stability as well as Y-randomization tests to confirm that the predictive power was not due to random correlations. These rigorous validation procedures are crucial to demonstrate the robustness and reliability of the models for practical applications, particularly in regulatory settings where accurate predictions of environmental behavior are essential for risk assessment and management. Model validation was carried out using various internal and external validation metrics, such as the Coefficient of Determination (R²), Adjusted R², Root Mean Square Error (RMSE), MAE, External Validation metrics (Q2F1 and Q2F2), and the Concordance Correlation Coefficient (CCC). These metrics demonstrate the model’s robustness, fitness, and predictive capabilities.
This study adheres to the OECD principles for QSPR model validation. The endpoints predicted are clearly defined, and the algorithms used are transparent and reproducible. The applicability domain has been defined to ensure reliable predictions. The models demonstrate satisfactory values of internal and external validation metrics and provide mechanistic interpretations where feasible, thus fulfilling all five OECD principles.
The Y-randomization test
To further substantiate the statistical significance of the models, a Y-randomization test was conducted, involving 100 random permutations of the response values. The intercepts of the trend lines in the R2 and Q2 plots were determined by contrasting the Q2 and R2 values of the original model against those derived from models generated randomly. This process verified the model’s correlation was not due to random chance.
Descriptor analysis
The importance of descriptors in the q-RASPR model was presented through bubble plots, illustrating the relationship between standardized regression coefficients and Variable importance in Projections (VIP), represented by the size of the bubbles. Furthermore, heat maps showcased low inter-correlation among descriptors within the model, ensuring the diversity and reliability of the model.
Predictive accuracy
The predictive accuracy of the model was demonstrated through scatter plots of observed versus predicted data for both training and test datasets, showcasing an even distribution of data points around the trend line, thus illustrating the model’s accuracy in fitting and predicting values.
Results and discussion
This research undertook modeling across various chemical datasets for twelve distinct physicochemical properties to assess the external predictive capability of the q-RASPR models, and compare them with the corresponding QSPR models (Tables 1 and 2). The developed q-RASPR models showed comparable internal validation metrics to traditional models but outperformed in external validation metrics (Table 3). In most of the cases, the reported models are obtained from PLS regression except a few where the final model is a multiple linear regression (MLR) equation (Table 3). As shown in Table 3, the models consistently achieved R² values above 0.90 for the training set, indicating a good fit. Additionally, the Q²F1 and Q²F2 metrics for the test set exceeded the threshold of 0.7, demonstrating strong external predictability and model robustness. We retained the combination of structural and physicochemical descriptors consistent with previous QSPR studies, employing the same training and query sets. Additionally, this study includes metrics such as the composite index RA function and the consistency measure gm, along with other similarity-based RASPR descriptors (e.g., Avg. Sim, MaxPos, MaxNeg, AbsDiff), which are less common in traditional QSPR models. The scatter plots depict the proximity of observed and predicted values for the modeled dataset compounds. The variable importance approach was employed to assess the relative significance of descriptors, illustrating the relationship between standardized regression coefficients and Variable importance in projections (VIP), as illustrated in the bubble plots. Moreover, heat maps showcased low inter-correlation among descriptors within the model, confirming the diversity and reliability of the model. Additionally, a Y-randomization test was implemented using the SIMCA-P 10.0 (available from https://umetrics.com/products/simca) and MLRPlusValidation v1.3 (available from https://sites.google.com/site/mlrplusvalidation/) software, reshuffling the dependent variable 100 times to ensure that the developed models were not obtained by chance (see sheets 1–5 of Supplementary Material SI-1 and Figures S1-S7 in Supplementary Material SI-2). In this investigation, we have verified that the trends observed in our models are consistent with established chemical principles. For instance, the relationship between the log Kow (octanol-water partition coefficient) and environmental persistence aligns with well-known concepts in environmental chemistry. Additionally, we explore how molecular weight, functional groups, and aromatic rings influence properties like the log Koc (organic carbon-water partition coefficient), demonstrating coherence with widely accepted chemical theories. Our models indicate that increased branching in aliphatic compounds leads to lower log Kow values, which is consistent with the known behavior of branched versus linear compounds. Similarly, we explain how the presence of electronegative substituents results in reduced log Koc values, in line with established chemical behaviors. Furthermore, our models identify a trend where increasing molecular weight corresponds to higher log Koc values, which aligns with the understanding that larger molecules tend to exhibit stronger sorption to organic matter due to enhanced hydrophobic interactions.
Polychlorinated biphenyls (PCBs)
Modeling of logKoc
Following a rigorous feature selection process, four key descriptors were identified to construct a PLS QSPR model with a single latent variable. The model equation is listed in Table 1. This model demonstrated exceptional performance in both internal and external validation metrics (Table 3), thereby attesting to its statistical reliability and predictive prowess. The model’s predictive accuracy was proven by comparing observed and predicted data in scatter plots (Fig. 1) for both training and test datasets. Figure 1 shows apparently discontinuous distribution for logKoc of PCBs, which may be due to isomeric PCBs with the same chlorine counts (such as compounds 2,4-dichloro-1-(3-chlorophenyl)benzene, 2,4-dichloro-1-(2-chlorophenyl)benzene, 1,3,5-trichloro-2-phenylbenzene, 1,3,5-trichloro-2-(3-chlorophenyl)benzene, 1,2,4,5-tetrachloro-3-(4-chlorophenyl)benzene, 1,2,4,5-tetrachloro-3-(3,5-dichlorophenyl)benzene, etc.).
We employed a read-across (RA) prediction approach, utilizing the structural and physicochemical parameters from the developed QSPR model. The RA predictions were executed by using the default setting of the hyperparameters and employing various similarity metrics such as Euclidean distance, Gaussian kernel, and Laplacian kernel similarity. The predictions, based on these methodologies, indicated superior performance (Table 3).
Furthermore, we developed a novel q-RASPR model, integrating chemical structure attributes with RA-based similarity information. By amalgamating these features within the training set, four significant descriptors were selected for model construction. The PLS model with 2 LVs is expressed as:
The q-RASPR model for predicting log Koc demonstrated strong predictive performance with an R² of 0.976 and a Q² (LOO) of 0.971, indicating a robust fit to the training data. Additionally, the model achieved Q²F1 and Q²F2 values of 0.974 and 0.970, respectively, confirming its external predictive power. The test set MAE was 0.074, and RMSE was 0.092, showing minimal prediction errors. A high Q²F1 value of 0.974 indicates that the q-RASPR model generalizes well to new compounds, suggesting it is suitable for use in regulatory assessments. The lower RMSE value for the q-RASPR model compared to the QSPR model indicates fewer deviations in predictions, highlighting the effectiveness of incorporating similarity-based descriptors.
A comparative analysis of the q-RASPR model with the initial QSPR model revealed that despite similar internal validation metrics, the q-RASPR model exhibited superior performance in test set predictions, particularly in terms of MAE (q-RASPR model’s MAE for the test set = 0.074 versus QSPR model’s MAE for the test set = 0.140). This demonstrates that the integration of RA similarity information into the q-RASPR model enables more precise log KOC predictions with the same amount of chemical information.
For predicting the water-organic carbon partition coefficient (Log KOC) of PCBs, the q-RASPR model (Eq. (1)) integrates four descriptors: average molecular weight (AMW), the maximum topological distance (MAXDP2), standard error of the experimental response values of close source neighbors based on Laplacian kernel similarity (SE(LK)), and the absolute difference in the similarity values of the closest positive and negative source compounds (Abs MaxPos-MaxNeg), unveiling the intricate relationship between compound structural characteristics and their distribution behavior in the environment (Figs. 2 and 3).
AMW, as a fundamental physicochemical property, directly reflects the size and mass of the compound. Its positive contribution (Fig. 2) to log KOC may indicate that larger molecules tend to have a higher enrichment capacity in the organic carbon phase. The square of the MAXDP2 (Fig. 2), as an indicator of molecular structural complexity, underscores the significance of molecular shape and size in determining the distribution in the organic phase.
The SE(LK) descriptor with its positive impact (Fig. 2) suggests the essential role of molecular similarity and interaction in the distribution process. The absolute difference in the similarity values of the closest positive and negative close congeners (Abs MaxPos-MaxNeg) has a negative contribution (Fig. 2); therefore in most cases, the similarity value to the closest negative congener is higher than the similarity value to the closest positive congener.
Our research illustrates the efficacy of meticulously designed QSPR and q-RASPR models, coupled with RA prediction methodologies, in accurately forecasting the water-organic carbon partition coefficients of PCBs. The success of these models highlights the potential of incorporating similarity information into QSPR model development. Through these steps, the study aimed to ensure that the developed q-RASAR model not only performed well in the internal validation test but also maintained high accuracy and reliability in external predictions.
Modeling of log t1/2 and log KOA
In the present section, we successfully developed two distinct QSPR models based on MLR regression targeting the log t1/2 and log KOA datasets, respectively. The development of these models involved the identification of chemically significant descriptors through an optimal subset selection process to optimize model performance. Through an optimal subset selection approach, we identified two sets of key descriptors for the prediction of log t1/2 and logKOA. Based on these identified descriptors, we formulated the following model equations:
To assess the performance of these models, rigorous internal and external validation exercises were conducted, adhering to the fourth principle of the OECD guidelines. The results of the validation metrics demonstrated that both models achieved computed R2 and/or R2Adj values exceeding the threshold of 0.8, confirming good model fit. Additionally, the correlation coefficients obtained through leave-one-out cross-validation (Q2LOO) also surpassed the 0.7 benchmark, further affirming model robustness. The external validation correlation coefficients (Q2F1 and Q2F2) were also within acceptable ranges, indicating that our preliminary QSPR models possess commendable external predictive capabilities.
The provided equation (Eq. (2)) represents a q-RASPR model (Fig. 1) for predicting the logarithm of the half-life (log t1/2) of PCBs based on its molecular descriptors and a specific consistency measure known as the Banerjee-Roy coefficient (gm). Here, Log t1/2 is the logarithm of the half-life of the PCB, indicating how long it takes for half of the compound to undergo a chemical change or degradation. ATS6i is a molecular descriptor, representing a 2D autocorrelation of a specific property calculated over six bonds in the molecule. The negative coefficient (Fig. 2) suggests that an increase in this descriptor value leads to a decrease in the half-life of the PCB. MATS1m is another molecular descriptor, indicating Moran autocorrelation of a molecular property calculated over one bond. The significant negative coefficient (Fig. 2) implies a strong inverse relationship with the PCB’s half-life, meaning that as the value of this descriptor increases, the half-life decreases. gm(LK) represents the Banerjee-Roy concordance coefficient, calculated using the Laplacian kernel (LK) method, which is a measure of similarity or consistency among compounds. The negative coefficient (Fig. 2) indicates that higher consistency or similarity values are associated with shorter half-lives. This model (Eq. (2)) essentially correlates specific molecular features and a similarity measure (Fig. 3) with the degradation rate of a compound, providing insights into how structural characteristics influence its stability and persistence in an environment.
Equation (3) represents a q-RASPR model (Fig. 1) for predicting the Log KOA of PCBs. In QSPR modeling, Log KOA is a critical parameter as it indicates the compound’s potential to partition between the octanol and air phases, which is essential in understanding its environmental fate and behavior, particularly in terms of volatility. It is a measure of a substance’s potential to undergo long-range atmospheric transport and bioaccumulation. The descriptor ATS7s (Fig. 2) represents a topological descriptor related to the molecule’s structure. The descriptor CVsim(GK) (Fig. 2) is related to the coefficient of variation in the similarity measures, derived from a Gaussian Kernel (GK) similarity assessment among molecules. The model (Eq. (3)) indicates that the Log KOA value increases with an increase in the ATS7s descriptor value. Conversely, the Log KOA value decreases with an increase in the CVsim(GK) descriptor value, implying that greater variability in the similarity of the compound to others, as measured by the GK method, is associated with a lower propensity of PCBs to partition into octanol. Additionally, heat maps (Fig. 3) demonstrated low inter-correlation among descriptors, affirming the diversity and reliability of the model.
Modeling of log kOH and log BCF
In this section, we meticulously developed two distinct QSPR models (Table 1) using PLS regression for the ln kOH and log BCF datasets, and compared the performance of these models with that of q-RASPR models (Eqs. (4) and (5)). For each dataset, optimal subsets were carefully selected, identifying three descriptors for modeling the experimental values of ln kOH and log BCF datasets.
Rigorous internal and external validation tests were conducted, adhering to the OECD’s Principle 4 for model performance assessment; the calculated validation metrics are listed in Table 3. The computed R2 and/or R2Adj values for the datasets under investigation exceeded the threshold of 0.8. However, R2 and R2Adj are not the primary criteria for model selection; the cross-validated correlation coefficients (Q2LOO) for both models also surpassed the threshold of 0.7. The external correlation coefficients (Q2F1 and Q2F2) for these two PLS models fell within acceptable ranges, indicating that the preliminary QSPR models possess commendable external predictive capabilities.
The most significant and statistically robust q-RASPR PLS models for ln kOH and log BCF (Fig. 1) along with their quality across various internal and external validation metrics, are showcased in Table 3. These models, tailored specifically for ln kOH and log BCF, utilized three descriptors and incorporated RASPR and two-dimensional structural descriptors. Based on the leave-one-out (LOO) Q2 values, these models were developed using 2 and 1 LVs, respectively. All developed models met the threshold requirements for robustness, reliability, and good predictability.
It was observed that the q-RASPR models generally performed better on the test set than on the training set, attributed to the computational algorithm of RASPR descriptors (refer to Table 2). For the computation of RASPR descriptors on the training set, the algorithm operates on a Leave-Same-Out (LSO) basis, avoiding the consideration of identical compounds in the search for close analogs to prevent overfitting. In any QSPR modeling study, the chemical or physicochemical descriptors of training compounds are calculated based on the specific structure or properties of that compound. However, the RASPR descriptor of a specific training compound is not derived from that specific compound but is computed based on similarity features from its close analogs. Thus, a predictive aspect is inherently built into the computation of RASPR descriptors. QSPR models are fitted based on the descriptor data of the training set, whereas RASPR models are fitted based on training set descriptor data as per the leave-same-out (LSO) computation. Moreover, the number of components (LVs) in the PLS models during development is selected based on Leave-One-Out cross-validation (LOO-CV). Due to the combined effect of LSO descriptor computation and LOO cross-validation, the q-RASPR models exhibit comparable or slightly lower performance on training data compared to test data.
Equation (4) developed in this study aims to predict the hydroxyl radical (•OH) reaction rate constants (ln kOH) of compounds through a series of computational chemistry descriptors. The model reveals the relationship between three key descriptors—ATSC1c, GGI2, and SE(LK)—and ln kOH, each representing different chemical properties that influence the reaction rate. The descriptor ATSC1c is a distance matrix descriptor based on atomic type and second-order assignment, reflecting the electronic properties of the carbon atom environment within the compound. A positive coefficient indicates that enhanced electronic characteristics of carbon atoms (such as increased electronegativity or decreased electron cloud density) will enhance the reaction rate with the hydroxyl radical. This may relate to the carbon atoms’ increased efficiency in stabilizing the transition state. The GGI2 descriptor, a topological index, relates to the molecule’s geometric shape and connectivity. Its negative coefficient suggests that higher geometric connectivity complexity in a molecule leads to lower reaction rates with the hydroxyl radical. This could be due to increased structural complexity introducing spatial hindrance, making the attack by •OH more difficult. The SE(LK) descriptor represents standard error of observed response values of close congeners, measured by the LK. A positive coefficient implies that higher dispersion among similar close source neighbors correlates with higher reaction rates with the hydroxyl radical. This might indicate common structural features or functional groups that make compounds more susceptible to reaction with •OH.
The model (Eq. (4)) provides a theoretical framework for predicting and understanding the reactivity of different compounds with hydroxyl radicals. By revealing the contributions of different descriptors to ln kOH, this study not only aids in understanding the reaction mechanisms of organic molecules but also offers valuable information for designing new compounds and predicting their environmental behavior.
In Eq. (5), the logarithm of the BCF (Log BCF) is predicted through four key descriptors: GGI5, Avg.Sim(LK), SD similarity(LK), and their corresponding coefficients (Figs. 2 and 3). The GGI5 descriptor, multiplied by the coefficient 2.375, indicates the positive impact (Fig. 2) of the compound’s topological geometric characteristics on its bioconcentration potential. Specifically, this suggests that the geometric configuration and topological structure of compounds play a key role in their accumulation process within organisms. The Avg.Sim(LK) descriptor, multiplied by the coefficient 2.535, emphasizes the positive contribution (Fig. 2) of a compound’s average similarity to known high-BCF compounds in predicting BCF. This indicates that if a compound structurally resembles those known to accumulate easily within organisms, it is also more likely to accumulate. The SD similarity(LK) descriptor coefficient, -1.958, indicates the negative impact (Fig. 2) of the standard deviation of a compound’s similarity to high-BCF compounds on BCF values. This may mean that if the close source compounds of a query compound show a high dispersion in their BCF values, the compound is less likely to accumulate within organisms.
Equation (5) provides a quantitative framework for understanding and predicting the bioconcentration potential of compounds, emphasizing the importance of compounds’ geometric, topological characteristics, and their similarity to known high-BCF compounds in the bioconcentration process.
Polybrominated diphenyl ethers (PBDEs)
Modeling of RRT
We have developed an innovative QSPR model aimed at predicting the relative retention time (RRT) for PDBEs during chromatographic processes. The model is developed based on PLS regression and incorporates an optimal subset selection method to identify the most influential latent variable from a pool of multiple features (Tables 1 and 2). The performance of the model was rigorously evaluated through a series of internal and external validation metrics, all of which adhere to the internationally recognized standards set by the OECD, thereby ensuring its high accuracy and reliability in prediction (Table 3).
The results of internal validation unequivocally demonstrate the model’s fitting accuracy and robustness, as evidenced by high coefficients of determination (R²) and leave-one-out cross-validation correlation coefficients (Q²LOO), coupled with a minimal mean absolute error for the training set (MAEtrain), indicative of the model’s precision in replicating known data. External validation metrics, such as external predictive R² (Q²F1) and the mean absolute error for the test set (MAEtest), further underscore the model’s predictive capability on unseen data, showcasing its high reliability when applied to novel samples.
We explored predictions based on chemical analogs to enhance the reliability of similarity-based predictions. Through the evaluation of different similarity metrics, GK similarity was found to provide the best prediction quality for the test set under default hyperparameters. This not only validates the effectiveness of the chosen similarity metric but also serves as a crucial reference for future predictions based on chemical analogs.
By leveraging structural and physicochemical features in conjunction with similarity-based and error metrics, a new descriptor matrix was developed, encompassing structural attributes and RA-based similarity information. Through internal validation metrics, four key descriptors were identified, and a PLS model with two latent variables was developed based on these descriptors. This model’s external predictive capability significantly surpasses that of the initial QSPR model, as reflected by improved external predictive R² (Q²F1) and a lower MAEtest, highlighting its advantage in providing accurate predictions. The model’s predictive accuracy was proven by comparing observed and predicted data in scatter plot (Fig. 1) for both training and test datasets.
Equation (6) delineates the relationship between a PBDE’s relative retention time (RRT) and several descriptors (Fig. 3). RRT is a common chromatographic parameter employed to characterize the mobility speed of a compound in a chromatographic system relative to a reference compound. This specific model (Eq. (6)) predicts RRT by integrating various descriptors, including ATSC2e, an autocorrelation topological descriptor considering the distribution of electronegativity-related properties within the molecule. In Eq. (6), the coefficient of ATSC2e indicates a negative correlation with RRT, implying that PBDEs with higher ATSC2e values have comparatively shorter retention times in the chromatographic system. CVact(LK): This descriptor is the coefficient of variation of the response of close congeners of a PBDE. Its coefficient also indicates a negative relationship, that is, the greater the chemical variation or activity of close congeners of a compound, the smaller its RRT. MaxPos(LK): It represents the maximum positive similarity among the close source congeners. Its positive coefficient suggests a positive correlation with RRT, meaning that higher values of this characteristic result in longer retention times for compounds in the chromatographic system. Abs MaxPos-MaxNeg: A descriptor representing the absolute value of the difference between positive and negative characteristics within a compound. Its coefficient indicates that a larger difference in these characteristics correlates with shorter retention times for the PBDE.
These descriptors provide a multiparametric model (Eq. (6)) for estimating the behavior of PBDEs in chromatographic analysis. This study not only developed a QSPR model with high accuracy and reliability but also enhanced its predictive performance through chemical analogy (RA) approaches. These achievements provide a powerful tool for predicting RRT in chromatographic processes and also offer new perspectives on the application of similarity metrics in predictions based on chemical analogs.
Modeling of Log kP (hexane), Log kP (methanol), Log J, and Log k.
In this section, we developed three QSPR models using PLS regression, tailored for the datasets Log kP (methanol), Log J, and Log k, respectively, while the Log kP (hexane) model was developed by using MLR regression technique. Through optimal subset selection, we identified sets of descriptors - two for each of the Log kP (hexane) and Log kP (methanol) datasets, and three and two descriptors for the Log J and Log k datasets, respectively (Table 1).
In an endeavor to enhance the external predictive power of the related QSPR models, q-RASPR descriptors were computed. These descriptors, derived from the determined structural features, encompass read-across similarity, error, consistency, and predictive functions. The mathematical representations of the MLR-derived q-RASPR models for the Log kP (hexane), Log kP (methanol), Log k, and Log J (PLS-derived) datasets are articulated in the following equations (Table 2) respectively.
The finalized q-RASPR models underwent stringent validation, with the internal and external validation metrics computed and listed in Table 3. The Q²LOO values for all three PLS-based q-RASPR models (Log kP (hexane) dataset = 0.862; Log kP (methanol) dataset = 0.828; Log J = 0.914; Log k = 0.914) surpassed the threshold of 0.8, attesting to the models’ acceptable fitness and robustness. The satisfactory Q²F1 values further underscored the developed models’ external predictive capabilities.
A comparative analysis of the computational validation metrics for QSPR and q-RASPR models across all studied datasets revealed a superior external predictivity for q-RASPR models, evident from higher Q²F1 (Log kP (hexane) dataset: QSPR = 0.899, q-RASPR = 0.922; Log kP (methanol) dataset: QSPR = 0.863, q-RASPR = 0.903; Log J: QSPR = 0.879, q-RASPR = 0.919; Log k: QSPR = 0.959, q-RASPR = 0.990) and Q²F2 values, along with lower MAETest values. The close proximity of R², Q²LOO, and MAETrain values between QSPR and q-RASPR models also attested to the similar degree of fit and robustness offered by both algorithms. Hence, it can be asserted that the q-RASPR modeling approach achieved enhanced external predictivity across all four datasets without compromising the fit and robustness inherent to QSPR (Fig. 4).
Model (7) reveals the contributions of two main factors to the Log kP (hexane) value (Fig. 5). The maxwHBa descriptor reflects the PBDE’s ability as a hydrogen bond acceptor, significantly affecting its transmembrane transport properties. A higher maxwHBa value indicates a stronger hydrogen bond accepting capacity. The positive coefficient (Fig. 5) of maxwHBa in the model suggests that an increase in hydrogen bond acceptor strength leads to an increase in the Log kP (hexane) value. On the other hand, the SD Activity(GK) descriptor (Fig. 5), based on a similarity approach, reflects the variability of property values of close congeners of a query compound within a given set of PBDEs. The introduction of this descriptor considers the distribution of PBDEs in similarity space and how this distribution affects the activation energy of the photodegradation reaction. The positive coefficient (Fig. 5) of SD Activity(GK) indicates that an increase in the variability of property values (i.e., a greater diversity in PBDE property) could lead to higher Log kP (hexane) values.
In model (8), Log kP (methanol) is predicted through two descriptors, maxaasC and Pos.Avg.Sim (Fig. 5). The maxaasC descriptor represents the maximum surface area of specific atoms or molecular structures within a compound, while the Pos.Avg.Sim descriptor indicates the average similarity to positive proximal compounds. The coefficients in Eq. (8) signify the contributions of these descriptors to Log kP (methanol). Specifically, a higher value of maxaasC, meaning a larger maximum surface area, significantly positively impacts (Fig. 5) Log kP (methanol), suggesting that compounds with larger surface areas are likely to have higher lipophilicity or a greater tendency to degradation in organic phases. The positive coefficient (Fig. 5) of Pos.Avg.Sim implies that an increase in similarity to positively situated proximal compounds leads to an increase in Log kP (methanol), possibly reflecting the similarity in partitioning behavior among structurally similar compounds.
For models (9) and (10) (Fig. 6), the ATSC2c descriptor represents the weighted sum of electronic state indices of secondary carbon atoms in the molecule, reflecting the charge distribution and polarity of the molecule. Its negative coefficient (Fig. 6) indicates that an increase in ATSC2c value, i.e., an enhanced electronegativity of secondary carbon atoms in the molecule, leads to a decrease in log J and log k values. The descriptor maxsBr represents the maximum value of a specific property related to bromine atoms within the PBDEs being studied. The positive coefficient (Fig. 6) suggests that an increase in this property is associated with a significant increase in the value of Log J. The descriptor SD Activity(GK) could represent the standard deviation of a certain activity or property of the PBDEs, calculated using a GK method, which emphasizes the influence of similar PBDEs in the dataset. The positive coefficient (Fig. 6) implies that higher variability in this property among similar PBDEs correlates with an increase in Log J. ATSC2c is a topological descriptor related to the second-order connectivity of carbon atoms, reflecting the influence of molecular topology on the property being modeled. The negative coefficients (Fig. 6) indicate that an increase in this topological feature leads to a decrease in the values of Log J and Log k. The gm*Avg.Sim descriptor is a composite descriptor combining the model-based consistency measure (gm) and the average similarity (Avg.Sim) of a query compound with a set of reference compounds. The positive coefficient (Fig. 6) of this descriptor suggests that when a PBDE is more structurally or property-wise similar to PBDEs in the reference set, and these similarities positively correlate with the gm, its log k value tends to increase, indicating an increase in the PBDE’s similarity. These observations not only showcase the efficacy of employing PLS methodology for QSPR model development but also highlight an innovative approach to augmenting model external predictivity through the incorporation of RASPR descriptors. These findings offer valuable insights for the development of efficient, robust, and highly predictive models in the field of environmental remediation. Modeling of Log KOA and Ln kOH.
Modeling of log KOA and ln kOH
In the final section, we developed MLR-QSPR (Table 1) and PLS-q-RASPR (Table 2) models for the datasets of ln kOH and MLR-q-RASPR log KOA for PBDEs. The model’s predictive accuracy was showcased by comparing observed and predicted data in scatter plots (Fig. 4) for both the training and test datasets. The MLR and PLS-q-RASPR models constructed for the Log KOA and Ln kOH datasets are as follows:
Equation (11) appears to represent a model predicting the octanol-air partition coefficient (Log KOA) of a compound based on its molecular properties and similarities to other compounds. This model (Eq. (11)) (Fig. 6) employs the following descriptors: AMW (atomic molecular weight) reflects the total mass of the atoms in a molecule, suggesting that as the size of the molecule increases, there is a tendency for the Log KOA to increase, indicated by the positive coefficient. GATS1e (Geary Autocorrelation - lag 1 weighted by atomic Sanderson electronegativities) is a 2D autocorrelation descriptor reflecting the distribution of electronegative atoms across the molecule. The negative coefficient (Fig. 6) implies that as the electronegativity distribution becomes more uniform or extensive, the Log KOA decreases, potentially indicating less volatility or lower affinity for the air phase. The descriptor gm*SD Similarity involves a combination of a similarity measure (SD Similarity) adjusted by a consistency or concordance measure (gm). The negative coefficient (Fig. 6) suggests that as the variability in similarity to a reference set of PBDEs increases (or as the PBDEs become less consistent in their similarity to the reference set), the Log KOA decreases. This could imply that PBDEs with less consistent structural or property similarities to known reference compounds tend to have lower octanol-air partition coefficients.
Equation (12) represents a q-RASPR model (Fig. 6) that predicts the natural logarithm of the rate constant for the reaction between PBDEs and hydroxyl radicals. Here, GGI2 represents a geometric or topological feature of the molecule, possibly related to its shape or the arrangement of atoms. Abs MaxPos-MaxNeg measures the absolute difference between the maximum positive and maximum negative similarity values, which could be related to electronic properties or polarities. Pos.Avg.Sim denotes the average similarity of the PBDE to other PBDEs with positive characteristics within a dataset, reflecting a measure of chemical likeness. Each term in the equation modifies the rate constant based on the molecular characteristics they represent, with the coefficients indicating the strength and direction of each descriptor’s effect. The negative coefficients (Fig. 6) suggest that increases in the corresponding descriptor values lead to a decrease in the reaction rate constant with hydroxyl radicals, indicating less reactivity or slower degradation in the presence of hydroxyl radicals. This model (Eq. (12)) provides insights into how specific molecular features influence the reactivity of PBDEs in oxidative environments, such as atmospheric chemistry or environmental degradation processes. The inter-correlation among different descriptors is shown in Fig. 7.
Conclusions
The results of this study demonstrate the effectiveness of the integrated q-RASPR approach in enhancing the predictive accuracy of QSPR models. By utilizing similarity-based descriptors and read-across techniques, our models exhibit strong external validation performance. This method not only improves robustness but also provides reliable predictions in line with OECD principles, making it a powerful tool for future applications in chemical property prediction. The study’s findings have profound implications for environmental remediation strategies. The predictive models offer insights into the environmental fate and bioaccumulation potential of hazardous compounds, facilitating the design of safer chemicals and contributing to the reduction of environmental and health risks associated with persistent organic pollutants. Tables S1 to S12 in the Supplementary Materials show how the current models outperform previously reported models at the same endpoints.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Roy, K. et al. Is it possible to improve the quality of predictions from an intelligent use of multiple QSAR/QSPR/QSTR models? J. Chemom. 32(4), e2992 (2018).
Li, W. et al. Estimation of octanol-water partition coefficients of PCBs based on the solvation free energy. Comput. Theor. Chem. 1202, 113324 (2021).
Safder, U. et al. Quantitative structure-property relationship (QSPR) models for predicting the physicochemical properties of polychlorinated biphenyls (PCBs) using deep belief network. Ecotoxicol. Environ. Saf. 162, 17–28 (2018).
Pandey, S. K. & Roy, K. QSPR modeling of octanol-water partition coefficient and organic carbon normalized sorption coefficient of diverse organic chemicals using extended Topochemical Atom (ETA) indices. Ecotoxicol. Environ. Saf. 208, 111411 (2021).
Jalili-Jahani, N., Fatehi, A. & Zeraatkar, E. PLS and N-PLS based MIA-QSPR modeling of the photodegradation half-lives for polychlorinated biphenyl congeners. RSC Adv. 10(56), 33753–33761 (2020).
González-Mariño, I. et al. Photodegradation of nitenpyram under UV and solar radiation: kinetics, transformation products identification and toxicity prediction. Sci. Total Environ. 644, 995–1005 (2018).
Terzaghi, E. et al. Rhizoremediation half-lives of PCBs: role of congener composition, organic carbon forms, bioavailability, microbial activity, plant species and soil conditions, on the prediction of fate and persistence in soil. Sci. Total Environ. 612, 544–560 (2018).
Li, W. et al. Prediction of octanol-air partition coefficients for PCBs at different ambient temperatures based on the solvation free energy and the dimer ratio. Chemosphere 242, 125246 (2020).
Yang, J., Gu, W. & Li, Y. Biological enrichment prediction of polychlorinated biphenyls and novel molecular design based on 3D-QSAR/HQSAR associated with molecule docking. Biosci. Rep., 39(5). (2019).
Mathieu, D. QSPR versus fragment-based methods to predict octanol-air partition coefficients: revisiting a recent comparison of both approaches. Chemosphere 245, 125584 (2020).
Ebert, R. U., Kühne, R. & Schüürmann, G. Octanol/Air Partition CoefficientA General-Purpose Fragment Model to Predict Log Koa from Molecular Structure57p. 976–984 (Environmental Science & Technology, 2023). 2.
Nolte, T. M. et al. Thermochemical unification of molecular descriptors to predict radical hydrogen abstraction with low computational cost. Phys. Chem. Chem. Phys. 22(40), 23215–23225 (2020).
Kobayashi, Y. & Yoshida, K. Development of QSAR models for prediction of fish bioconcentration factors using physicochemical properties and molecular descriptors with machine learning algorithms. Ecol. Inf. 63, 101285 (2021).
Zhang, X. et al. QSPR modeling of the logKow and logKoc of polymethoxylated, polyhydroxylated diphenyl ethers and methoxylated-, hydroxylated-polychlorinated diphenyl ethers. J. Hazard. Mater. 353, 542–551 (2018).
Bertato, L., Chirico, N. & Papa, E. Predicting the Bioconcentration factor in Fish from Molecular structures. Toxics 10(10), 581 (2022).
Yang, M. et al. Estimating subcooled liquid vapor pressures and octanol-air partition coefficients of polybrominated diphenyl ethers and their temperature dependence. Sci. Total Environ., 628–629 : pp. 329–337. (2018).
Mansouri, K. et al. OPERA models for predicting physicochemical properties and environmental fate endpoints. J. Cheminform. 10(1), 10 (2018).
Kundi, V. & Ho, J. Predicting Octanol–Water partition coefficients: are Quantum Mechanical Implicit Solvent models better than empirical fragment-based methods? J. Phys. Chem. B. 123(31), 6810–6822 (2019).
Huang, C. et al. Comprehensive exploration of the ultraviolet degradation of polychlorinated biphenyls in different media. Sci. Total Environ. 755, 142590 (2021).
Hu, P. T. et al. New equation to predict size-resolved gas-particle partitioning quotients for polybrominated diphenyl ethers. J. Hazard. Mater. 400, 123245 (2020).
Yao, B. et al. Current progress in degradation and removal methods of polybrominated diphenyl ethers from water and soil: a review. J. Hazard. Mater. 403, 123674 (2021).
Ai, H. et al. QSAR modelling study of the bioconcentration factor and toxicity of organic compounds to aquatic organisms using machine learning and ensemble methods. Ecotoxicol. Environ. Saf. 179, 71–78 (2019).
Nendza, M. et al. PBT assessment under REACH: screening for low aquatic bioaccumulation with QSAR classifications based on physicochemical properties to replace BCF in vivo testing on fish. Sci. Total Environ. 616-617, 97–106 (2018).
Banerjee, A., Gajewicz-Skretna, A. & Roy, K. A machine learning q-RASPR approach for efficient predictions of the specific surface area of perovskites**. Mol. Inf. 42(4), 2200261 (2023).
Yu, S. et al. QSAR models for predicting octanol/water and organic carbon/water partition coefficients of polychlorinated biphenyls. SAR QSAR Environ. Res. 27(4), 249–263 (2016).
Chen, Y. et al. Prediction of octanol-air partition coefficients for polychlorinated biphenyls (PCBs) using 3D-QSAR models. Ecotoxicol. Environ. Saf. 124, 202–212 (2016).
Xu, H. Y. et al. QSPR/QSAR models for prediction of the physicochemical properties and biological activity of polybrominated diphenyl ethers. Chemosphere 66(10), 1998–2010 (2007).
Wang, Z. Y., Zeng, X. L. & Zhai, Z. C. Prediction of supercooled liquid vapor pressures and n-octanol/air partition coefficients for polybrominated diphenyl ethers by means of molecular descriptors from DFT method. Sci. Total Environ. 389(2), 296–305 (2008).
Luo, S. et al. A novel model to predict gas–phase hydroxyl radical oxidation kinetics of polychlorinated compounds. Chemosphere 172, 333–340 (2017).
Raff, J. D. & Hites, R. A. Deposition versus photochemical removal of PBDEs from lake superior air. Environ. Sci. Tech. 41, 6725–6731 (2007).
Fei, J. et al. The Internal relation between Quantum Chemical descriptors and empirical constants of Polychlorinated compounds. Molecules 23(11), 2935 (2018).
Liu, H. et al. QSAR studies of bioconcentration factors of polychlorinated biphenyls (PCBs) using DFT, PCS and CoMFA. Chemosphere 114, 101–105 (2014).
Fang, L. et al. Quantitative structure–property relationship studies for direct photolysis rate constants and quantum yields of polybrominated diphenyl ethers in hexane and methanol. Ecotoxicol. Environ. Saf. 72(5), 1587–1593 (2009).
Chen, J. et al. Quantitative structure–property relationships for direct photolysis of polybrominated diphenyl ethers. Ecotoxicol. Environ. Saf. 66(3), 348–352 (2007).
Eriksson, J. et al. Photochemical Decomposition of 15 Polybrominated Diphenyl Ether Congeners in Methanol/Water pp. 3119–3125 (Environmental Science & Technology, 2004). 11.
Yap, C. W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32(7), 1466–1474 (2011).
Rogers, D. & Hopfinger, A. J. Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J. Chem. Inf. Comput. Sci. 34(4), 854–866 (1994).
Wold, S., Sjöström, M. & Eriksson, L. PLS-regression: a basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 58(2), 109–130 (2001).
Banerjee, A. & Roy, K. First report of q-RASAR modeling toward an approach of easy interpretability and efficient transferability. Mol. Diversity 26(5), 2847–2862 (2022).
Chatterjee, M. et al. A novel quantitative read-across tool designed purposefully to fill the existing gaps in nanosafety data. Environ. Science: Nano. 9(1), 189–203 (2022).
Roy, K., Kar, S. & Das, R. N. Chap. 7 - validation of QSAR models, in Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment, K. Roy, S. Kar, and R.N. Das, Editors. Academic: Boston. 231–289. (2015).
Acknowledgements
We gratefully acknowledge the support and contributions from the following entities and individuals: CMC thanks Environmental Science Technology Consultants Corporation (Internal Reference No. 113D592) for providing financial support. AB thanks LSRB, DRDO, New Delhi for a senior research fellowship. VK thanks ICMR, New Delhi for a research associateship.
Funding
This research was funded by the Provincial Secretariat for Higher Education and Scientific Research, grant number 142-451-3098.
Author information
Authors and Affiliations
Contributions
C.M.C contributed to writing, review, editing, methodology, A.B. contributed to software and methodology, V. K. contributed to software and methodology, K. R. contributed to conceptualization, writing, review, editing and methodology, E.B. contributed to writing, review, editing, and supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chang, C.M., Banerjee, A., Kumar, V. et al. The q-RASPR approach for predicting the property and fate of persistent organic pollutants. Sci Rep 15, 1344 (2025). https://doi.org/10.1038/s41598-024-84778-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-84778-2