Abstract
Early detection of ocular diseases is vital to preventing severe complications, yet it remains challenging due to the need for skilled specialists, complex imaging processes, and limited resources. Automated solutions are essential to enhance diagnostic precision and support clinical workflows. This study presents a deep learning-based system for automated classification of ocular diseases using the Ocular Disease Intelligent Recognition (ODIR) dataset. The dataset includes 5,000 patient fundus images labeled into eight categories of ocular diseases. Initial experiments utilized transfer learning models such as DenseNet201, EfficientNetB3, and InceptionResNetV2. To optimize computational efficiency, a novel two-level feature selection framework combining Linear Discriminant Analysis (LDA) and advanced neural network classifiers—Deep Neural Networks (DNN), Long Short-Term Memory (LSTM), and Bidirectional LSTM (BiLSTM)—was introduced. Among the tested approaches, the “Combined Data” strategy utilizing features from all three models achieved the best results, with the BiLSTM classifier attaining 100% accuracy, precision, and recall on the training set, and over 98% performance on the validation set. The LDA-based framework significantly reduced computational complexity while enhancing classification accuracy. The proposed system demonstrates a scalable, efficient solution for ocular disease detection, offering robust support for clinical decision-making. By bridging the gap between clinical demands and technological capabilities, it has the potential to alleviate the workload of ophthalmologists, particularly in resource-constrained settings, and improve patient outcomes globally.
Similar content being viewed by others
Introduction
The field of ocular disease detection has evolved significantly over the past few decades. Initially, traditional manual methods, such as fundus examination, were widely used, but these were labour-intensive and prone to diagnostic errors. The integration of image processing techniques in the early 2000s offered some improvements in identifying patterns in retinal images. However, it wasn’t until the advent of deep learning in the 2010s that significant advancements were made. Convolutional Neural Networks (CNNs) and ensemble methods, such as those combining random forests with CNNs, started to outperform earlier techniques by improving accuracy and robustness1. In recent years, unsupervised methods and more sophisticated models like VGG, ResNet, and DenseNet have pushed the boundaries of automated diagnosis, making it possible to classify a wide range of ocular diseases with remarkable precision. As of the early 2020s, the combination of deep learning, transfer learning, and feature selection techniques2,3 have paved the way for highly accurate, efficient systems capable of handling large datasets, such as the ODIR dataset used in this study. The complexity of eye’s anatomy, which has a multiplicity of the structures, as well as the number of ocular diseases as shown in Fig. 1, which are not the same and could change over time increases the difficulty in diagnosing eye disorders. However, this seldom happens in ophthalmology as the ophthalmologists may use history taking, clinical examination and imaging modalities simultaneously to diagnose a disease such as visual acuity, tonometry, optical coherence tomography, fluorescein angiography ophthalmologists may use. While such findings are of great significance, assigning accurate interpretations to them may be tricky, and sometimes they may even demand for a highly skilled and subjective specialist’s assessment. In addition, certain eye diseases like the early series diabetic retinopathy are seldom manifested in symptoms; therefore the patients do not realize there is a problem in time4. Furthermore, the availability of sophisticated tools and knowledge, especially in places with high number of people with difficulties to access care, could be the key factor to the provision of quality and full-scope eye care and its correct diagnosis. To sum up, technological progress in eye disease diagnosis is a complex and always changing process, which requires continuous team effort rooted in research, understanding and development to increase diagnosis efficiency and provide quality care for patients from all over the globe on a long term basis.
Deep learning-based ocular diagnosis is paving the way for the ophthalmology of the future, becoming a new tool supporting and enhancing the precision of diagnosing and treatment options5,6. With regards to deep learning methods, notably convolutional neural networks, CNNs, have a lot come to the use of algorithms that are able to understand medical images including retinal scans and fundus photographs and identify signs of different types of ocular diseases. The process, as a rule, comprises the initial stage of assembling large data sets which are labelled and were created basing on some kind of ocular pathology. These data provide the starting point for training some deep learning models that will then learn to recognize apart of these details and to distinguish between the different disease states. Transfer learning, a type of machine learning where pre-trained models are put into practice based on medical imaging data and used to process and narrow down the determination of barely labelled data, has demonstrated practicality for these scenarios the most. With training, these models accumulate insights and with time they get better in handling the images, therefore supporting ophthalmologists in identifying early diseases and consequently making the right diagnosis.
Despite the fact that, still, there are several barriers to completely benefit from the possibility of deep learning in retinal diseases diagnosis7. Probably the first problem is the availability quality datasets as well as diversity. Making sure that datasets in adequate quantity, variety and accurately labelled is of paramount importance to train models which not only produce outputs generalize well among real-life scenarios but also diverse patients. Moreover, the challenge of deep learning models interpretability is yet another issue in concern of these models. Understanding how a model makes its decisions is the crucial requirement for clinical adherence and trust. Nevertheless, there are initiatives to form new methods of explainable AI in order to deal with the problem. That being said, the development of deep learning systems for medical purposes also needs well-validated and governmentally-approved clinical trials to ensure that patients are safe and get the desired results8.
The deep learning integration9 into the ophthalmic care does not resolve the challenges, however, the application focuses on transforming diagnostic procedures as well as enhancing treatment outcomes for patients. As electronic methods of camera observation are combined with the expertise of ophthalmologists through deep learning technologies, this has the potential to automate image analysis, resulting in earlier diagnosis, lighter workloads and prompt intervention. Research and screening must continue, and there have to be collaborations everywhere to get over the challenges, and then deep learning technologies will be better utilized in diagnosing ocular diseases.
Eye diseases like diabetic retinopathy, glaucoma, cataract, and age-related macular degeneration are major causes of blindness globally, affecting millions, yet there is a significant shortage of ophthalmologists to diagnose and treat these conditions through traditional fundus screening methods10. The prevalence of eye diseases varies worldwide due to factors like age, gender, lifestyle, economic status, and geographic region. Deep learning techniques have shown promise in medical image analysis tasks, but few studies have provided a comprehensive diagnosis of multiple eye diseases11. The study proposes a deep learning system to classify various ocular diseases by addressing the issue of highly imbalanced datasets through binary classification using balanced data and a pretrained VGG-19 model.
In summary, the work done in this manuscript emphasizes the need for automated ocular disease detection systems using deep learning techniques to address the global burden of eye diseases and the challenges associated with manual diagnosis. Given the limitations of manual diagnosis, the growing burden of ocular diseases, and the need for efficient diagnostic tools, this research aims to develop an automated framework to enhance the precision of ocular disease classification. Leveraging deep learning models and optimized feature selection, the proposed system seeks to bridge the gap between clinical demands and current technological capabilities. The primary motivation of this work lies in addressing three key challenges:
-
1.
The need for early and accurate detection of multiple ocular diseases to prevent severe complications.
-
2.
Reducing the computational cost and complexity associated with transfer learning models, which can hinder their practical deployment.
-
3.
Providing a robust and scalable solution to support ophthalmologists, especially in settings with limited resources.
To address the challenges listed above, the main contributions of the proposed work are as follows:
-
1.
Development of a novel two-level feature selection framework: This approach combines feature extraction from pre-trained models with Linear Discriminant Analysis (LDA) to retain only the most discriminative features, ensuring improved performance with reduced computational overhead.
-
2.
Integration of advanced neural architectures for classification: Models such as DNN, LSTM, and BiLSTM were employed, with the combined feature approach achieving 100% accuracy on the training set and 98.04% on the validation set.
-
3.
Introduction of a “Combined Data” strategy: This strategy leverages features from DenseNet201, EfficientNetB3, and InceptionResNetV2 models, leading to superior classification performance compared to individual models.
-
4.
Demonstration of practical significance in ophthalmology: The system shows potential to automate diagnosis, support clinical decision-making, and improve patient outcomes, particularly in underserved areas.
This research offers a comprehensive, scalable framework for automated ocular disease classification, positioning it as a valuable tool in computer-aided diagnosis and healthcare innovation.
Literature survey
The studies primarily focus on the detection and classification of eye diseases such as diabetic retinopathy, glaucoma, and cataract, as well as brain MRI analysis for Alzheimer’s disease. Table 1 lists the reference numbers, model names or approaches used, the achieved accuracy rates, and the datasets employed for training and evaluation. Several deep learning models, including convolutional neural networks (CNNs) like VGG-19, ResNet, AlexNet, and DenseNet, as well as other techniques like ensemble random forests, fully-connected conditional random fields (CRFs), and unsupervised methods like trainable COSFIRE filters and active contour models, have been employed across these studies12. The datasets used range from public datasets like DRIVE, STARE, OIA-ODIR, ORIGA, SCES, DIARETDB0, DIARETDB1, and Eye-PACS, to institutional datasets collected from various hospitals and research centers in different countries. The accuracy rates reported in the table vary depending on the specific task, model, and dataset, with some studies achieving impressive results, such as 99% accuracy for glaucoma detection using a ResNet CNN, 98.4% accuracy for diabetic retinopathy classification on the Eye-PACS dataset, and 96.8% accuracy for non-proliferative diabetic retinopathy detection using image processing and classification techniques. Overall, this table provides a comprehensive overview of recent research efforts in ophthalmic and medical image analysis, highlighting the diverse range of models, datasets, and tasks being addressed in this field.
The key highlights of this work are as following:
-
Initially transfer learning models like DenseNet201, EfficientNetB3, and InceptionResNetV2, were applied with EfficientNetB3.
-
Applying LDA-based feature selection on the features extracted from DenseNet201, EfficientNetB3, and InceptionResNetV2, and then using DNN, LSTM, and Bi-LSTM classifiers.
The work is organized as follows: It begins by introducing the importance of Ocular Disease Classification systems using advanced technologies. It then presents a literature survey reviewing recent deep learning-based techniques for oncluar disease classification, summarizing the accuracy levels achieved by various existing methods. The “Methodology” Section describes the datasets used, the transfer learning models applied, and the proposed feature selection technique using Linear Discriminant Analysis (LDA) combined with deep learning classifiers. The experimental results evaluate the performance of the transfer learning models as well as the LDA-based feature selection approach, demonstrating the superior accuracy and computational efficiency of the latter. Finally, the conclusion summarizes the key findings and emphasizes the potential of the developed system for sustainable crop management and protection in agriculture.
Methodology
The classification of ocular disease demands a series of approaches which incorporate the combination of methods to produce an accurate and deep understanding of different eye conditions. The implementation of this methodology will enable healthcare providers to make ocular diagnosis easily with accurate identification who would be sure of getting timely diagnosis, the right treatment and enhancement in their condition.
Research design
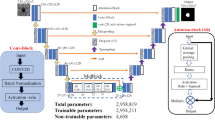
An artificial intelligence learning-based algorithm for the classification of eye diseases needs collection of ocular images in great number, as well as images with different diseases and normal ones. These images too have to go through a pre-processing phase such as normalization and augmentation so that the model makes use of gainful opportunities to prove its worth. The result of investigation will have an impact to the improvement and verification of the detection techniques of eye ailments in the future and, therefore, eye care management and diagnosis can be achieved easily using automated technologies. The key steps of the methodology are as follows and are shown in Fig. 2.
-
1.
Ocular Disease Intelligent Recognition (ODIR) dataset is considered having 8 classes including images of Normal (N), Diabetes (D), Glaucoma (G), Cataract (C), Age-related Macular Degeneration (A), Hypertension (H), Pathological Myopia (M) and other diseases/abnormalities (O).
-
2.
The dataset contains Number of images as: Cataract-293, Diabetes-1608, Glaucoma-284, Hypertension-128, Macular Degeneration-266, Normal-2873, Other Diseases-708, Pathogical Myopia-708, with 80% used for training and 20% for testing.
-
3.
Initially, three pre-trained transfer learning models: DenseNet201, EfficientNetB3, and InceptionResNetV2 were applied for classification.
-
4.
To reduce the computational cost of the transfer learning models, a novel feature selection technique using Linear Discriminant Analysis (LDA) is proposed. LDA is applied to the features extracted from the pre-trained models (DenseNet201, EfficientNetB3, and InceptionResNetV2) to identify the most relevant features for classification.
-
5.
DenseNet201 extracted 1920 features. Then LDA feature selection was further applied. It then obtained 7 features out of 1920 features. On the selected features DNN, LSTM and Bi-LSTM were applied.
-
6.
EfficientNetB3 extracted 1536 features. Then LDA feature selection was further applied. It then obtained 7 features out of 1536 features. On the selected features DNN, LSTM and Bi-LSTM were applied.
-
7.
InceptionResNetV2 extracted 4608 features. Then LDA feature selection was further applied. It then obtained 7 features out of 4608 features. On the selected features DNN, LSTM and Bi-LSTM were applied.
-
8.
After applying individually, the features extracted using DenseNet20 (i.e. 1920), EfficientNetB3 (i.e. 1536) and InceptionResNetV2 (4608) were then combined. Total 8064 features were then considered. On these features, again LDA is applied and 7 features were extracted. On the selected features DNN, LSTM and Bi-LSTM were applied.
-
9.
Then the performance of the deep learning classifiers (DNN, LSTM, and BiLSTM) is evaluated on the LDA-selected features.
-
10.
Then the performance of the deep learning classifiers (DNN, LSTM, and BiLSTM) is evaluated on the LDA-selected combined features.
Dataset
The Ocular Disease Intelligent Recognition (ODIR) dataset32 is a structured ophthalmic database consisting of fundus photographs of 5,000 patients’ eyes, along with their age, sex, and doctors’ diagnostic keywords. The dataset aims to represent real-life patient information collected from various hospitals and medical centres in China. The dataset contains colour fundus photographs of both the left and right eyes for each patient, captured using different camera models from manufacturers like Canon, Zeiss, and Kowa, resulting in varied image resolutions. The annotations were labelled by trained human readers with quality control measures in place. The dataset classifies patients into eight labels including Normal (N), Diabetes (D), Glaucoma (G), Cataract (C), Age-related Macular Degeneration (A), Hypertension (H), Pathological Myopia (M) and other diseases/abnormalities (O). The dataset is well-suited for tasks such as ocular disease recognition, classification, and computer-aided diagnosis systems. It provides a diverse set of fundus images representing various eye conditions, along with patient metadata and diagnostic keywords. The dataset’s strengths include its large size (5,000 patients), the inclusion of both left and right eye images for each patient, and the annotation of diagnostic keywords by medical professionals. However, it’s important to note that the dataset’s license and expected update frequency are not specified. Overall, the ODIR dataset offers a valuable resource for researchers and practitioners in the field of ophthalmology and computer vision, enabling the development and evaluation of intelligent systems for ocular disease recognition and diagnosis. The overall description of the dataset is shown in Tables 2 and 3.
Implementation flow of the proposed work
Implementing a Ocular Disease Classification system using feature selection involves several steps. Here’s a generalized flow of the implementation as shown in Fig. 3 also.
Step 1: Load the Ocular Disease Intelligent Recognition (ODIR) dataset containing 8 classes of images (Normal, Diabetes, Glaucoma, Cataract, Age-related Macular Degeneration, Hypertension, Pathological Myopia, and Other diseases/abnormalities).
Step 2: Split the dataset into training (80%) and testing (20%) sets.
Step 3: Apply pre-trained transfer learning models (DenseNet201, EfficientNetB3, and InceptionResNetV2) for initial classification.
Step 4: Perform feature extraction using the pre-trained models.
Step 4.1: Extract 1920 features from DenseNet201.
Step 4.2: Extract 1536 features from EfficientNetB3. Substep 4.3: Extract 4608 features from InceptionResNetV2.
Step 5: Apply Linear Discriminant Analysis (LDA) for feature selection on the extracted features.
Step 5.1: Apply LDA on the 1920 features from DenseNet201 and select 7 most relevant features.
Step 5.2: Apply LDA on the 1536 features from EfficientNetB3 and select 7 most relevant features.
Step 5.3: Apply LDA on the 4608 features from InceptionResNetV2 and select 7 most relevant features.
Step 6: Apply deep learning classifiers (DNN, LSTM, and BiLSTM) on the selected features from each pre-trained model.
Step 6.1: Apply DNN, LSTM, and BiLSTM on the 7 selected features from DenseNet201.
Step 6.2: Apply DNN, LSTM, and BiLSTM on the 7 selected features from EfficientNetB3.
Step 6.3: Apply DNN, LSTM, and BiLSTM on the 7 selected features from InceptionResNetV2.
Step 7: Combine the features from all pre-trained models (1920 + 1536 + 4608 = 8064 features).
Step 8: Apply LDA on the combined 8064 features and select 7 most relevant features.
Step 9: Apply deep learning classifiers (DNN, LSTM, and BiLSTM) on the selected 7 combined features.
Step 10: Evaluate the performance of the deep learning classifiers (DNN, LSTM, and BiLSTM) on both the individual LDA-selected features and the combined LDA-selected features.
Pseudocode of the proposed work
This section describes the psueudocode of the proposed work. To write the psueudocode some of the notations are required which are listed in Table 4.

Technologies applied
The key technologies employed in this work include a combination of transfer learning, feature selection, and deep learning techniques. This work proposes a hybrid approach, leveraging the strengths of feature selection and advanced deep learning architectures, showcases an efficient and highly accurate solution for automated Ocular Disease Classification, addressing the trade-off between model complexity and classification performance. Various technologies used in this work as explained below:
Transfer learning
Transfer Learning33 is a machine learning technique where a model trained on one task is repurposed, or transferred, to another related task. Instead of training a model from scratch, transfer learning leverages knowledge gained from solving one problem and applies it to a different but related problem. The process of transfer learning typically involves choosing a pretrained model that was trained on a large dataset for a specific task, such as image classification (e.g., VGG, ResNet, Inception, etc.), natural language processing (e.g., BERT, GPT, etc.), or other tasks like object detection or segmentation. After that feature extraction is performed. In this approach, one freeze the parameters of the pretrained model and use it to extract relevant features from your dataset. These features are then fed into a new classifier (or other downstream task-specific layers) that is trained from scratch. This method is useful when you have a small dataset or when computational resources are limited. Practice another, one may have learned to use the pre-trained model not not only for extracting feature, but also to correct the flaw of old data model. Here, the components to be removed from the pre-trained model are selected and typically, the whole model is used for training using our data. Fine-tuning eliminates the weaknesses of the model in terms of adapting to a structure of the dataset and usually results in more efficient operation in larger datasets. Ultimately, task orienting training invites a data specialist (either building a new model or completely tuning it) for a particular post. Typically, this involves adding the delete mark or training the model and turning the back propagation of patches to typical propagation patterns. Transfer-learning is greatly employed in several areas including computer vision, natural language processing and audio processing which is among other. Active learning has now turned to be the standard practice of doing deep learning as it has proven to not only achieve better results with just fewer labelled data but also with minimal computational resources than training models from the scratch. Transfer Learning Models used in the study are discussed as follows:
-
DenseNet20134 is a convolutional network model assigned to the family of these models called Dense Net. It is exactly the 201-layer increase of the base DenseNet architecture. The Densenet is devised around the idea of dense feature reuse, which means that each layer is not only connected to its preceding layer, but also to the subsequent one. Such type of coincidence allows for the functional reuse, supports the extension, and reduces the number of arguments. This is how the applications acquire more capability and become faster. DenseNet201 is especially being used to facilitate tasks like image classification, object detection and segmentation, especially when it involves dealing with massive datasets or more complex models.
-
EfficientNetB335 of the convolutional neural networks known as EfficientNet family bags the possibility to attain the state-of-the-art accuracy and provides reduced errors and computation more compared to these predecessors network models. Specially, EfficientNetB3 denotes a neural network that has a greater size and power than the networks of EfficientNetB0 and B1 but comparable to the networks of EfficientNetB4, B5, and B7 in terms of size. The EfficientNet model was developed by exposing the basic model in three dimensions: today’s TV sets provide a wide range of offerings that include screen size, screen resolution, and viewing angle. The extension enables them to reach the next level of performance for many tasks without requiring substantial power. In larger models, the features and patterns of the object are more in depth, wide and resolved, which are allowing often more models details to be captured. It is an excellent tool which can be applied to various functions including image classification, object detection, and segmentation whenever more precision is aspired but resources to be used in computation are limited.
-
InceptionResNetV236 is a deep convolutional neural network model that incorporates the beneficial aspects of the inception and residual networks. It is similarly a counterpart to InceptionV3 that adds on and refines the performance through the interlinking of its sections. Residual connections, pioneered in ResNet, provide the ability for layers to leapfrog over some of the training, helping to mitigate the vanishing gradient problem and making the training of networks surprisingly simpler. However, the remaining connections between InceptionResNetV2 are all located underneath the middle layer of the Inception module, i.e., in the latter half. On the opposite side, the most feature of the Inception architecture is utilizing hypothetically multiple network connections, which provides the network to detect features at distinct scales. This method is capable of not just providing a better understanding of the disease but also predicts the probability of a scientist to advance the study of mathematics. InceptionResNetV2 excels in fixing up the robustness of the networks towards the distracting situations making the network highly effective in tasks demanding high precision, for instance, image classification, object detection, and image segmentation. It is well-known for its ability to build complex visualization tools and manipulate a large dataset volume.
Feature selection will occur by using pre-trained convolutional neural networks (CNNs) such as the DenseNet201, EfficientNet, and InceptionResNetV2, all of which were trained on the ImageNet dataset. These models based on the Internet with their weights that are actually rich structure-directing features of the input image. Every feature from the CNN conveys different levels of visual data ranging from the low-frequency features to the high-frequency ones which include the basic level details and high-level semantic concepts respectively. Primal component analysis or LDA are the methods of Dimensionality reduction. Usually after the selection of very informative features interpretability is improved and the performance is enhanced. Using this method, researchers exploit strong image classification features of pre-trained CNNs and ImageNet weights as shown in Table 5 is thus achieved for object detection or classification tasks.
Linear discriminant analysis
Linear Discriminant Analysis (LDA)37 is a technique used in statistics and machine learning for dimensionality reduction and classification. Its primary goal is to find a linear combination of features that characterizes or separates two or more classes of objects or events. LDA extracts the most relevant features by estimating the weight vector w that defines the linear combination of predictors that best separates the classes. This allows LDA to both identify important predictors and reduce the dimensionality of the data to a single discriminant score for classification. LDA performs feature extraction and dimensionality reduction in the following way:
-
LDA finds the linear combination function f(x) using (1).
that maximizes the ratio between the separation of the class means (µ0 - µ1) and the within-class covariance Σ. This is done by choosing the weight vector as (2)
where c is an arbitrary constant.
-
The absolute values of the weights w correspond to the variables’ importance, implying an order of feature relevance. This allows LDA to perform feature extraction by identifying the most important predictor variables for discriminating between the classes.
-
LDA assumes that the data in each class follows a multivariate normal distribution with a common covariance matrix Σ. This allows it to estimate the class means µi and the pooled covariance matrix Σ from the training data.
-
The estimated weight vector w* defines a linear projection that maps the original p-dimensional data to a 1-dimensional discriminant score. This effectively reduces the dimensionality of the data from p to 1, performing dimensionality reduction.
-
The discriminant scores can then be used to classify new observations into one of the two classes based on a decision rule that compares the discriminant scores to a threshold.
Overall, LDA extracts a number of features equal to the number of classes minus one. If there are K classes, LDA will extract K − 1 features. These features are linear combinations of the original input variables that maximize the separation between the classes while minimizing the variance within each class.
Deep learning techniques
The key advantage of using the deep learning techniques is their ability to automatically learn hierarchical features from the input data, without the need for manual feature engineering. By leveraging the feature extraction capabilities of CNNs and the sequence modeling capabilities of LSTM and BiLSTM, the authors were able to develop a highly accurate and robust classification system38.
-
Dense neural network (DNN): DNN stands for “Deep Neural Network,” which is also a form of an Artificial Neural Network (ANN), although they are separated in few layouts between the input and the output layer. Deep neural networks being the next generation and, perhaps, new potential technology beyond traditional genetic engineering and cloning see wider applications and, thus, they have been adopted in computer vision, natural language processing, speech recognition, and other fields. They can consider and analyze complicated fixed-patterns in data, as they find their way in machine learning and artificial intelligence applications algorithms. DNN is in fact a fully connected neural network. Moreover, DNN can be regarded as an additional classifier applied to the LDA selected feature sets. DNNs with a high learning ability are able to understand the hidden relationships between the feature sets for the ocular diseases and the target types.
-
Long short-term memory (LSTM): LSTM means Long Short-Term Memory, a type of RNN special neural network architecture which is designed in a way to make RNNs able to identify long dependences and be less impacted by errors in sequential data. LSTMs are used frequently in machine learning tasks like natural language processing, time series prediction, speech and identification, among others. What makes LSTMs stand out is the property of the memory cell, which is of updating its value and being able to selectively remember some input information rather than memorise everything at a given time difference. The LSTM not only shorter term but long range dependencies carry because of the architecture that is uncertainty that in some instances the error flow is constant, with preserving the information over lengthy time sequence. This results in LSTMs to be most viable for jobs where context overlong sequence is vital, for instance, language modelling, translation, and speech recognition. LSTM, which is one type of RNN, is used to classify ocular disease, relying on the feature using Latent Dirichlet Allocation methodology. LSTM -based models are built to emphasize on the long-term dependencies in the sequential data thus, spell out unique skills for learning the periodic patterns in the ocular disease data.
-
Bidirectional LSTM (BiLSTM): A BiLSTM (Bidirectional Long Short-Term Memory) is a particular type of RNN (Recurrent Neural Network) architecture which is designed specially by the deep learning, mostly for sorting out sequence modelling tasks. The second development is that this network enlarges the function capacity of the conventional LSTM (Long Short-Term Memory) by processing the input sequence in a way which is similar to two-way communication. BiLSTMs are mainly used in situations where context from two directions (both past and future) has to be taken into account, which some issues of natural language process do. For example, named entity recognition, part-of-speech tagging, sentiment analysis, and machine translation do so. On the other hand, they have been fundamental for applications in speech recognition, time series prediction, and more. BiLSTM processes its input-sequence directionally in both forward and backward directions. It achieves this by using two separate LSTM cells that process the input sequence and simultaneously reversing this sequence to form a reversed copy. BiLSTM allows to capture more data context and rules specific to the data sets and that may boost the classifier performance ratio vs. unidirectional approach.
Ethical considerations
As per our conducted research, we have utilized the already available open datasets and also have not applied our research on any human or living being. At present stage, the ethical considerations have not required. In case of real time implementation with involvement of human sources, key steps to achieve HIPAA (Health Insurance Portability and Accountability Act.) or even the GDPR (General Data Protection Regulation) will be conducted. However, respecting patient’s privacy and featuring in rules and guidelines of use of health information as provided by the HIPAA or even the GDPR is important to avoid the collapse of the public’s trust. Further, AI models have been observed to reproduce or even amplify the bias in the data given to them during training and therefore clinical diagnosis will be uniformly biased by the A1 models depending on the population sampled. If these models are trained using the data from certain populations or geographical regions, regardless of marginalized representation, the models will be less accurate when operating in different contexts and increase rather than reduce healthcare inequality. Furthermore, when it comes to the use of AI in healthcare, it is an equally big issue that requires a solution because while AI-assisted diagnostic platforms may translate into less expensive diagnostics and easy access to doctors in developed countries, these advantages may not impact developing or the deprived in developed countries’ hinterlands. Hence, ownership of AI should be revamped to be clear, objective, and safe as well as to enforce proper distribution of the systems so that they do not predispose any specific or multiple sets of people to wrong healthcare determinations.
Experimental results and evaluation
In this section, using feature selection based deep learning models in the classification problems provide a loss reduction and performance improvement. The performance of the proposed model is evaluated in terms of the improvement of model accuracy and the other evaluation parameters. For taking the results, system configurations are present in Table 6.
Analysis of baseline transfer learning
To validate the results of the proposed model, initially, Ocular Disease Classification is performed by applying Transfer Learning models including DenseNet201, EfficientNetB3, InceptionResNetV2. Figure 4 contains the training and validation data for three different deep learning models: EfficientNetB3, InceptionResNetV2, and DenseNet201. These models are trained using transfer learning techniques for oncluar data classification. Transfer learning involves using pre-trained models, trained on large datasets like ImageNet, as a starting point and fine-tuning them on a new task with a smaller dataset. The Figure shows the training and validation accuracy, loss, precision, and recall values for each epoch of the training process. For the EfficientNetB3 model, it can be observed that the training accuracy increases steadily indicating that the model is learning from the training data. However, the validation accuracy plateaus around 60%, suggesting that the model may be overfitting to the training data or that more training is required to further improve its performance on the validation data. The InceptionResNetV2 model follows a similar trend, with training accuracy increasing by epochs. However, its validation accuracy stagnates at around 50%, lower than that of EfficientNetB3, potentially indicating a less effective transfer learning process or a more challenging task. The DenseNet201 model has the lowest performance among the three, with a training and validation accuracy of around 50% by the last epoch. This could be due to various factors, such as the model architecture being less suitable for the task, the transfer learning process being less effective, or the task itself being particularly challenging for this model. Overall, the provided Figure gives an overview of the training progress and performance of three different deep learning models using transfer learning techniques. While all three models show improvement in training accuracy over the epochs, their validation accuracy differs, indicating varying degrees of success in transferring knowledge from the pre-trained models to the new task.
The evaluation metrics presented in the Table 7 file shows insights into the performance of three Transfer learning models: DenseNet201, EfficientNetB3, and InceptionResNetV2. Among these models, EfficientNetB3 emerges as the top performer across multiple metrics. It achieved the highest accuracy of 62.234% on the training set and 62.128% on the validation set, indicating its ability to generalize well to unseen data. Furthermore, EfficientNetB3 exhibited the highest precision of 71.906% on the training set and 66.279% on the validation set, demonstrating its capability to minimize false positives effectively. Regarding recall, which measures the model’s ability to minimize false negatives, EfficientNetB3 once again outperformed the other two models, attaining a recall of 44.201% on the training set and an impressive 55.243% on the validation set. This high recall value suggests that EfficientNetB3 excelled in correctly identifying positive instances, which can be crucial in scenarios where missing positive cases is undesirable. Additionally, EfficientNetB3 exhibited the lowest loss of 1.215 on the training set and 1.489 on the validation set, indicating better convergence and optimization during the training process. InceptionResNetV2, the second-best performing model, achieved an accuracy of 59.241% on the training set and 58.059% on the validation set, slightly lower than EfficientNetB3. Its precision and recall values were 69.799% and 40.681% on the training set, and 64.894% and 51.8% on the validation set, respectively. While these metrics are commendable, they fall short of EfficientNetB3’s performance. The loss values for InceptionResNetV2 were 1.3 on the training set and 1.43 on the validation set, indicating higher loss compared to EfficientNetB3. DenseNet201, the third model, exhibited the lowest performance among the three. Its accuracy was 51.946% on the training set and 54.617% on the validation set, while its precision and recall values were 61.909% and 25.621% on the training set, and 63.717% and 43.349% on the validation set, respectively. DenseNet201 also had the highest loss of 1.422 on the training set and 1.313 on the validation set, suggesting potential issues with model convergence or optimization during training.
Analysis of pre-trained algorithms based feature selection and DenseNet classification
The selection of features with a two-level architecture, where Level 1 contains three models: DenseNet for extraction of complex features, EfficientNetB3 for extraction of efficient features and InceptionResNetV2 for extraction of inception features. In the case of deep learning models, the data set is used to instruct the model to extract patterns and features that it considers important. The features from each model are extracted and put together to form a vector of enriched features. At Level 2, Linear Discriminant Analysis (LDA) is taken as a dimensionality reduction method to narrow on the most discriminating features in the combines feature set. LDA is an algorithm that seeks to project the original high-dimensional domain to a lower dimensional axis while concentrating on the largest class of separation, such class separation being the most significant factor in category recognition whereas others are considered nominal. This two-level approach tries to utilize the strength of the deep learning (NL) features and LDA for the NL selection. Finally, this mixture should improve the model interpretability and the prediction performance.
Analysis of DNN (dense neural network) on extracted features
As described in above sections, feature extraction is created by means of models that are called DenseNet 201, EfficientNetB3, and InceptionResNetV2 respectively. This served as the input for Deep Neural Network (DNN) that was used for further processing and prediction. The DNN, which is basically constructed as several consecutive layers of interlinked neurons, may recognize the complicated patterns in the features that were extracted manually. Trees of non-linear transformations with increasing resolutions are consecutively fed into the DNN to reflect the image into a structure which has better representations of the inherent data. Training the DNN is mainly applied to change the parameters of the network, like weights and biases, so as to obtain a global minimum loss function, which is typically done by using a method such as stochastic gradient descent. Once the DNN is trained, its metrics can be relied on to make predictions on new data with the help of the learned representations for classification or regression purposes. By doing so, it enables the user to utilize the hierarchical representations that were previously learned by the pre-trained model and alter the model appropriately for the particular issue at hand, thus leading to enhanced predictive capabilities.
Figure 5a–h shows the results of training multiple deep learning models having Features Selection methods as (Multi-Features, DenseNet201, EfficientNetB3, and InceptionResNetV2) and DNN as a Classifier on some dataset. The accuracy values are reported for three different feature selection techniques: DenseNet201, EfficientNetB3, and InceptionResNetV2, across 200 epochs classified by DNN. Additionally, the accuracy for a “Combined Data” approach is also provided. DenseNet201 achieved an accuracy of around 76–77% in the later epochs. EfficientNetB3 performed better than DenseNet201, reaching an accuracy of around 72% in the later epochs. InceptionResNetV2 had a slightly lower accuracy compared to EfficientNetB3, hovering around 71–72% in the later epochs. The “Combined Data” approach, which combines the features from all three techniques, achieved significantly higher accuracy, ranging from 99 to 99.3% in the later epochs. This suggests that combining different feature selection techniques can significantly improve the overall performance of the Deep Neural Network (DNN) classifier. In case of validation accuracy, DenseNet201 achieved a validation accuracy of around 74–75% in the later epochs. EfficientNetB3 performed better than DenseNet201 on the validation set, reaching a validation accuracy of around 71–72% in the later epochs. InceptionResNetV2 had a similar validation accuracy to EfficientNetB3. The “Combined Data” approach, achieved significantly higher validation accuracy in the later epochs, This suggests that combining different feature selection techniques can significantly improve the overall performance of the Deep Neural Network (DNN) classifier on the validation set. For DenseNet201, the loss starts around 1.3 and decreases steadily, in the later epochs. EfficientNetB3 has a higher starting loss but it decreases more rapidly, which is higher than DenseNet201. InceptionResNetV2 also starts with a higher loss and decreases in the later epochs, performing similarly to EfficientNetB3. The “Combined Data” approach loss values decreases significantly in the later epochs. This suggests that combining different feature selection techniques can significantly reduce the overall loss of the Deep Neural Network (DNN) classifier. Validation Loss, for DenseNet201, decreases gradually in the later epochs. EfficientNetB3 has a higher starting validation loss but it decreases more rapidly compared to DenseNet201 in the later epochs. InceptionResNetV2 validation loss decreases in the later epochs, performing better than EfficientNetB3 on the validation set. The “Combined Data” approach significantly lower validation loss compared to the individual approaches. Lower validation loss generally indicates better model performance on the validation set. The “Combined Data” approach clearly outperforms the individual feature selection techniques in terms of validation loss, suggesting that combining different feature extraction methods can lead to improved model performance on unseen data. For DenseNet201, the precision indicates that the model correctly classified around 80% of the instances on average. The values do not show a clear increasing or decreasing trend, suggesting that the model’s performance did not significantly improve or degrade over the training epochs. EfficientNetB3 exhibits lower precision values compared to DenseNet201. This indicates that the model correctly classified approximately 75–77% of the instances on average. Similar to DenseNet201, the values do not show a clear trend, implying that the model’s performance remained relatively stable throughout the training process. InceptionResNetV2 has precision values slightly lower than DenseNet201. This suggests that the model correctly classified approximately 75–78% of the instances on average. Again, the values do not exhibit a clear increasing or decreasing trend, indicating that the model’s performance did not significantly improve or degrade over the training epochs. The Combined data, shows significantly higher precision values. This suggests that the combined model correctly classified approximately 99-99.3% of the instances on average, indicating excellent performance. Overall, the Combined Data approach achieved the highest precision, outperforming the individual feature selection techniques, while EfficientNetB3 exhibited the lowest precision among the techniques evaluated. For DenseNet201, the validation precision indicates that the model correctly classified approximately 79% of the validation instances on average. The values do not show a clear increasing or decreasing trend, suggesting that the model’s performance on unseen data did not significantly improve or degrade over the training epochs. EfficientNetB3 exhibits lower validation precision values compared to DenseNet201. The values do not display a clear trend, implying that the model’s performance on data remained relatively stable throughout the training process. InceptionResNetV2 has validation precision values slightly higher than EfficientNetB3. The Combined Data approach, shows significantly higher validation precision values. This means that the combined model correctly classified approximately 97-98.5% of the validation instances on average, indicating excellent performance on unseen data. The values do not display a clear trend, implying that the model’s performance on current data remained consistently high throughout the training process. Overall, the Combined Data approach achieved the highest validation precision, outperforming the individual feature selection techniques, while EfficientNetB3 exhibited the lowest validation precision among the techniques evaluated. For DenseNet201, the recall values indicate that the model correctly identified approximately 71% of the positive instances in the validation set on average. The values do not show a clear increasing or decreasing trend, suggesting that the model’s ability to identify positive instances did not significantly improve or degrade over the training epochs. EfficientNetB3 exhibits lower recall values compared to DenseNet201. This means that the model correctly identified approximately 64–66% of the positive instances in the validation set on average. Similar to DenseNet201, the values do not display a clear trend, implying that the model’s ability to identify positive instances remained relatively stable throughout the training process. InceptionResNetV2 has recall values slightly lower than DenseNet201. The Combined Data approach, shows significantly higher recall values. This means that the combined model correctly identified approximately 99-99.3% of the positive instances in the validation set on average, indicating excellent performance in identifying positive instances. For DenseNet201, the validation recall values indicate that the model correctly identified approximately 70% of the actual positive instances in the validation set on average. EfficientNetB3 exhibits lower validation recall values compared to DenseNet201 means that the model correctly identified approximately 63–65% of the actual positive instances in the validation set on average. Similar to DenseNet201, the values do not display a clear trend, implying that the model’s ability to identify positive instances on unseen data remained relatively stable throughout the training process. InceptionResNetV2 has validation recall values slightly higher than EfficientNetB3. The Combined Data approach, shows significantly higher validation recall values, means that the combined model correctly identified approximately 97-97.7% of the actual positive instances in the validation set on average, indicating excellent performance in identifying positive instances on given data.
Analysis of LSTM on extracted features
On the contrary, the computational model can be fed with features, invariably generated using models such as DenseNet 201, EfficientNetB3, and InceptionResNetV2. The subsequent feature input can be timed using Long Short-Term Memory networks (LSTM). LSTMs are an RNN-type temporal network (RNN) that allows capturing the correlations of temporal dependencies within sequential data. The features are detached and in turn are subsequently read by the LSTM model the way they are supposed to be scheduled which means that for each feature sequence every time step gets processed as soon as it is due. What LSTM’s architecture allows it to retain is long-term links in a selective way to revision of the information and its consequent rejection through the memory cells. While in training, this LSTM attempts to capture the patterns commonly associated with each data, known as sequences, by adjusting its internal parameters using the back propagation method to minimize the prediction errors. After the training, a LSTM can be either used for forecasting or classification of time series data as it has already identified the patterns in features that it extracted from the data. This makes it a very useful and powerful tool in tasks that requires evidence of temporal data.
Figure 6a–h shows the training and validation metrics for four different deep learning models having Features Selection methods as (Multi-Features, DenseNet201, EfficientNetB3, and InceptionResNetV2) and LSTM as a Classifier. All models exhibit an increasing trend in accuracy and a decreasing trend in loss as the number of epochs increases, which is expected during the training process. The Combined Data (Multi-Features Data) model starts with a significantly higher initial accuracy compared to the other models indicating better performance from the beginning. The Combined Data model achieves very high accuracy relatively quickly, around epoch 100–110, while the other models take longer to reach similar accuracy levels. Precision and recall values generally increase for all models as training progresses, indicating improved model performance in correctly classifying positive and negative instances. The Combined Data model starts with a higher initial precision compared to the other models, suggesting better initial performance. The Combined Data model reaches very high precision and recall values relatively quickly, around epoch 100–110, while the other models take longer to achieve similar levels. Validation accuracy and validation loss follow similar trends as their training counterparts, with validation accuracy increasing and validation loss decreasing as training progresses. The Combined Data model shows consistently higher validation accuracy and lower validation loss compared to the other models throughout the training process. Validation precision and recall also improve for all models, with the Combined Data model exhibiting higher values throughout the training process. Overall, the Combined Data model demonstrates superior performance compared to the other individual models, achieving higher accuracy, precision, and recall, as well as lower loss values, both during training and validation.
Analysis of BiLSTM on extracted features
With the utilization of these models namely DenseNet 201, EfficientNetB3, and InceptionResNetV2, we can now capture the bidiuretic nature of the sequential data using the bidirectional recurrent networks i.e. Bi-LSTM. Bi-LSTMs consist of two LSTM layers: One neural network working in forward fashion and the other one working in backward manner on the next step. Further, this sort of mechanism gives the network an edge over its ability to track past and also future by capturing all information for each time step which is a development in terms of temporal dependency representation within the extracted features. Bi-LSTM unravels crucial properties from the input sequence’s opposite directions whilst encoding information. Hence, it conveys the complex patterns and relationships of the sequence. After having been retrained, the Bi-LSTM will be able to be used for tasks as e.g., sequence classification, sequence labeling or sequence-to-sequence prediction, benefitting in each case from the bidirectional architecture in order to achieve improved performance in tasks where sequence data is involved.
Figure 7a–h shows the results of a Deep learning model having Features Selection methods as (Multi-Features, DenseNet201, EfficientNetB3, and InceptionResNetV2) and BiLSTM as a Classifier. All models exhibit an increasing trend in accuracy and a decreasing trend in loss as the number of epochs increases, which is expected during the training process. The Combined Data (Multi-Features) model starts with a significantly higher initial accuracy compared to the other models, indicating better initial performance. The Combined Data model achieves very high accuracy (above 0.99) relatively quickly, around epoch 100–110, while the other models take longer to reach similar accuracy levels. Precision and recall values generally increase for all models as training progresses, indicating improved model performance in correctly classifying positive and negative instances. The Combined Data model has a higher initial accuracy than other models which might indicate its better initial performance, or it might indicate higher initial accuracy. The Combined Data model attains much higher accuracy and recall fast, near 100–110 epochs, than the other ones that do it slower, around 150–200 epochs later. Validation accuracy and validation loss show similar patterns as the training parameters, where they both converge to optimum level, giving more accuracy and producing more loss while training progresses. The validation accuracy and likelihood show continuously higher values with validation loss being lower for the Combined Data model compared to the others models throughout the training. Not only is the validation of precision and recall increased over time for all models, the Combined Data model takes the lead on these two metrics. Altogether, Combined Data model provides the best results compared to the other models. It has a smaller loss value both during the training and validation sides, besides achieving an increased accuracy, precision, and recall. This suggests that combining data from different sources or using an ensemble approach can lead to better model performance. Additionally, the trends indicate that all models improve their performance as training progresses, with the Combined Data model converging to very high accuracy and precision/recall values more rapidly than the other models.
The provided Table 8 presents evaluation metrics for various deep learning models combined with different classifiers, including BiLSTM, Dense, and LSTM. Among these models, MultiData with BiLSTM classifier stands out with an impressive accuracy of 100% on both the training and validation sets (98.04%), along with 100% precision and recall on the training set, and 98.04% precision and 97.91% recall on the validation set. Moreover, it achieved the lowest loss of 0.00 on the training set and 0.06 on the validation set, indicating excellent model convergence and generalization. Other models, such as DenseNet201 with BiLSTM classifier, EfficientNetB3 with BiLSTM classifier, and InceptionResNetV2 with BiLSTM classifier, also exhibited high accuracy, precision, and recall on the training set but struggled to maintain similar performance on the validation set, suggesting potential overfitting or generalization issues. The Dense and LSTM architectures generally underperformed compared to the BiLSTM models, with lower accuracy, precision, recall, and higher loss values across all model combinations.
Comparison with state of the art techniques
The proposed work distinguishes itself from recent State-of-the-Art (SOTA) approaches by offering a more comprehensive, efficient, and scalable solution for multi-disease classification from ocular fundus images. While many existing studies focus on individual diseases such as glaucoma, cataract, or diabetic retinopathy, the proposed system addresses the classification of eight different ocular diseases using the ODIR dataset. In contrast to methods like ResNet50, VGG-19, and DenseNet201, which demonstrate high accuracy for specific diseases but struggle with generalizability across multiple conditions, the proposed approach combines three pre-trained models—DenseNet201, EfficientNetB3, and InceptionResNetV2—into a powerful ensemble. A key innovation lies in the two-level feature selection process, where Linear Discriminant Analysis (LDA) is applied to extract only the most discriminative features, significantly reducing computational overhead while retaining high predictive power. Additionally, while SOTA techniques like ResNet-based models can encounter challenges with imbalanced datasets and computationally intensive operations, the proposed work mitigates these issues by using the LDA-selected features in combination with advanced classifiers such as Deep Neural Networks, Long Short-Term Memory, and Bidirectional LSTM. The “Combined Data” approach—integrating features from all three transfer learning models—further enhances the model’s accuracy, achieving 100% on the training set and 98.04% on the validation set, outperforming existing methods in both precision and recall. This hybrid framework not only advances diagnostic accuracy but also provides a practical solution for reducing the workload on healthcare professionals, making it well-suited for real-world clinical applications where early detection and efficient diagnosis are essential. By addressing the limitations of existing models, such as limited disease coverage, imbalanced data handling, and computational complexity, the proposed work sets a new benchmark for automated ocular disease classification.
Table 9 presents a detailed comparison of various methods for ocular disease detection, showcasing accuracy rates and datasets used across different studies. Earlier approaches like the fully convolutional neural network and extreme learning machine achieved accuracy rates of 95.33% and 96.7% respectively on the DRIVE dataset. VGG-19 implementations demonstrated strong performance across different studies, achieving 98.10% accuracy on the ODIR dataset with 5000 fundus images and 97.94% on the OIA-ODIR dataset. When focusing on specific conditions like glaucoma, CNN achieved 80% accuracy using ORIGA and SCES datasets, while an ANN classifier reached 93.3% accuracy for cataract detection using images from Kasturba Medical College Hospital. Some approaches showed more modest results, such as CRNN achieving 70.7% on the ACHIKO-NC dataset and a custom CNN reaching 68.36% accuracy for cataract grading using Korean hospital data. More recent approaches using advanced architectures showed promising results, with ResNet-50 achieving 94.8% accuracy for multi-classification of cataract, diabetic retinopathy, and maculopathy on Singapore eye disease study data, and various combinations of modern architectures like ResNet50, InceptionResNetV2, and VGG variants achieving 95.7% accuracy for glaucoma detection. The proposed work, which combines DenseNet201, EfficientNetB3, and InceptionResNetV2 with LDA and BiLSTM, demonstrates superior performance compared to all previous approaches, achieving 100% accuracy on the training set and 98.04% on the validation set using the ODIR dataset of 5000 images. This indicates a significant advancement in the field of automated ocular disease detection, particularly noteworthy given that it handles multiple disease classifications simultaneously rather than focusing on a single condition like many of the compared approaches.
The proposed method stands out for its comprehensive approach to multi-disease classification in ocular fundus images, achieving superior performance compared to existing methods. While previous studies have achieved high accuracy on specific diseases such as glaucoma or cataracts, the proposed model addresses eight different ocular conditions simultaneously by leveraging the ODIR dataset. For instance, models such as VGG-19 and ResNet50 have achieved 98.10% and 94.8% accuracy, respectively, but they were typically optimized for single-disease classification, lacking the versatility required for multi-disease diagnostics. The integration of three pre-trained models—DenseNet201, EfficientNetB3, and InceptionResNetV2—within the proposed system allows it to capture diverse feature representations across the full range of conditions. This multi-model strategy enables a more generalized solution applicable across various ocular diseases, an advancement over models that focus on specific conditions. The proposed system has resulted an innovative two-level feature selection process further distinguishes it from state-of-the-art techniques. By applying LDA to reduce dimensionality, the proposed method efficiently selects the most discriminative features from each transfer learning model, significantly minimizing computational overhead without compromising predictive power. This feature selection approach allows the model to maintain high accuracy, achieving 100% on the training set and 98.04% on the validation set. This is a notable improvement over other advanced methods, such as ResNet50 and DenseNet201, which can struggle with imbalanced data and computational intensity. Additionally, the “Combined Data” approach, which aggregates features from all three pre-trained models, further optimizes performance by enhancing the model’s robustness and precision. Advanced classifiers such as BiLSTM were used to capitalize on the extracted features’ temporal dependencies, leading to further improvements in accuracy, precision, and recall on the validation set. Together, these features make the proposed model a robust and practical solution that not only outperforms other models in diagnostic accuracy but also offers a scalable and efficient system suitable for real-world clinical applications.
Computational efficiency
The section analyzes the time and resource demands of training and validating each transfer learning (TL) model used in the study, including EfficientNetB3, DenseNet201, and InceptionResNetV2, as well as the subsequent Deep Neural Network (DNN) model that classifies the extracted features. To measure efficiency, the study quantifies the time needed per epoch and the total time required to complete training and validation across 10 epochs for each model.
Table 10 in the “Computational efficiency” Section provides a detailed breakdown of the training and validation times per epoch for three pre-trained transfer learning (TL) models: EfficientNetB3, DenseNet201, and InceptionResNetV2. Each of these models was assessed over 10 epochs to establish baseline computational demands. The per-epoch time varied across the models, with EfficientNetB3 completing an epoch in approximately 3.5 min, DenseNet201 in 3.8 min, and InceptionResNetV2 in 4.1 min. These differences reflect the distinct architectures and resource needs of each model, with InceptionResNetV2 being the most complex and thus the most computationally intensive. EfficientNetB3, although slightly faster, required substantial computational time due to its sophisticated architecture tailored for efficiency. The total time to complete 10 epochs varied accordingly, with EfficientNetB3 taking 350 min, DenseNet201 taking 380 min, and InceptionResNetV2 reaching 410 min. This information highlights the resource demands associated with using advanced TL models for feature extraction in medical imaging tasks, guiding users in selecting a model based on both computational constraints and desired performance outcomes. EfficientNetB3’s faster processing might be advantageous in environments with limited resources, while InceptionResNetV2 may be preferable for applications where slightly higher computational costs are acceptable for a potential increase in model accuracy.
The Table 11 focuses on the feature extraction phase and the training and validation time requirements of the Deep Neural Network (DNN) model, which relies on features extracted by each TL model. The table outlines the feature extraction time per image for EfficientNetB3, DenseNet201, and InceptionResNetV2, along with the total time required to process 6,392 images. EfficientNetB3, with its optimized structure, extracted features at a rate of 0.2 min per image, taking 21 min in total. DenseNet201 was slightly slower at 0.24 min per image, totalling 25.5 min, while InceptionResNetV2 required 2.7 min per image, culminating in 28.7 min for all images. After feature extraction, a DNN model was trained using the combined feature set from all three models, requiring an additional 0.45–0.6 min per epoch over 100 epochs. The aggregated approach enhanced classification accuracy and reduced loss values by leveraging the strengths of each TL model, but at a slightly increased computational cost. This table emphasizes that although the combined model approach entails higher processing demands, it offers a balanced solution for applications requiring high diagnostic precision without compromising significantly on computational efficiency, making it feasible for clinical settings that require high-volume, rapid diagnostics.
Bridging complexity and clarity: the role of explainability in AI decision-making
Explainable models are very prominent to visually analyze the impact of applied deep learning models. In Fig. 8, Gradient-Weighted Class Activation Mapping (Grad-CAM) has been utilized to explore the features and present the highlighted outcome in form of heatmap. Each sub image in Fig. 8a or b reflecting the applied pretrained model and Grad-CAM enhanced heatmap outcome. This Section “Bridging complexity and clarity: the role of explainability in AI decision-making” emphasizes the importance of integrating explainability into AI models used for medical diagnosis, particularly in ocular disease classification. This section discusses the use of explainable AI tools like Gradient-Weighted Class Activation Mapping (Grad-CAM) to visually interpret deep learning models’ predictions. The section underscores that while deep learning models are highly effective for classification tasks, their black-box nature can make it difficult to understand how decisions are made. Explainability helps bridge this gap by allowing humans to see which features influenced a particular decision, thereby fostering trust and transparency. It also showcases the application of Grad-CAM, a tool that visualizes the regions of an input image contributing most to the model’s output. These technique overlays a heatmap on the image, highlighting areas that were crucial for the model’s classification. Such visual tools enable researchers and practitioners to validate that the model focuses on relevant features, like specific parts of a retinal image in ocular disease diagnosis. By employing Grad-CAM, practitioners can gain insights into the model’s decision-making process. For instance, if a deep learning model misclassifies a retinal condition, analyzing the Grad-CAM heatmap can reveal whether the model focused on the wrong image areas, pointing to potential areas for improvement. Figure 8 illustrates how Grad-CAM is applied to pretrained models, showing both the original input images and the resulting heatmaps. These visualizations demonstrate the highlighted areas that the model deemed important for making its predictions. Overall, this section highlights the necessity of using explainable AI to ensure that advanced diagnostic systems are transparent, interpretable, and aligned with medical expectations. This approach enhances trust in automated systems and supports their integration into clinical practice by making complex AI decisions comprehensible to healthcare professionals.
Conclusion
This study presents ans effective and efficient framework for the automated classification of ocular diseases using fundus images. The integration of deep learning techniques with novel feature selection significantly enhanced performance and reduced computational complexity. Transfer learning models—DenseNet201, EfficientNetB3, and InceptionResNetV2—were initially employed for classification, with EfficientNetB3 achieving the highest baseline accuracy of 62.23%. However, recognizing the computational burden of these models, a two-level feature selection approach was introduced. In the first level, features were extracted from the pre-trained models. In the second level, Linear Discriminant Analysis (LDA) was applied to select the most discriminative features, reducing the dimensionality while preserving important information. These selected features were further classified using Deep Neural Network, Long Short-Term Memory, and Bidirectional LSTM architectures. The “Combined Data” approach, leveraging features from all three transfer learning models, demonstrated exceptional performance, with the BiLSTM classifier achieving 100% accuracy, precision, and recall on the training set and 98.04% accuracy on the validation set. The results emphasize the significance of combining advanced feature selection with deep learning techniques for improved accuracy and computational efficiency. The proposed system showcases the potential to support ophthalmologists by automating disease detection, enabling faster and more accurate diagnosis. This approach can significantly alleviate the challenges associated with manual diagnosis, particularly in resource-limited settings, and enhance patient outcomes by facilitating early treatment. The findings highlight the importance of continuing research into automated healthcare solutions to improve diagnostic precision and reduce the burden on healthcare professionals.
However, cross-dataset validation can provide critical insights into the model’s generalizability across different imaging conditions and population demographics. In this study, we primarily focused on achieving high classification accuracy on the ODIR dataset, which is well-suited for multi-disease classification. However, we recognize that validating our model on additional datasets, such as DRIVE, STARE, or APTOS, could further demonstrate its effectiveness across diverse datasets. In future work, we plan to implement cross-dataset validation to assess model performance under varying conditions, addressing potential biases and improving adaptability for broader clinical use. Moreover, despite its high performance, the reliance on a single dataset (ODIR) limits the model’s generalizability to diverse clinical environments. Future work could explore cross-dataset validation and incorporate explainable AI techniques to improve transparency and clinical adoption.
Data availability
Data will be available on reasonable request from corresponding author.
Change history
21 May 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-02912-0
References
Li, Y. et al. Self-supervised anomaly detection, staging and segmentation for retinal images. Med. Image Anal. 87, 102805. https://doi.org/10.1016/j.media.2023.102805 (2023).
Zhang, X., Xu, M., Zhou, X. & RealNet: A feature selection network with realistic synthetic anomaly for anomaly detection 16699–16708. Available: http://arxiv.org/abs/2403.05897 (2024).
Eljialy, A. E. M., Uddin, M. Y. & Ahmad, S. Novel Framework for an intrusion detection system using multiple feature selection methods based on deep learning. Tsinghua Sci. Technol. 29, 948–958. https://doi.org/10.26599/TST.2023.9010032 (2024).
Kaewboonma, N., Lertkrai, P., Chanakot, B. & Lertkrai, J. Thai rubber leaf disease classification using deep learning techniques. In ACM Int. Conf. Proceeding Ser. 84–91. https://doi.org/10.1145/3639592.3639605 (2023).
Sharma, G., Anand, V. & Gupta, S. Harnessing the strength of ResNet50 to improve the ocular disease recognition. In 2023 World Conf. Commun. Comput. WCONF 2023 1–7. https://doi.org/10.1109/WCONF58270.2023.10234986 (2023).
Shourie, P., Anand, V., Upadhyay, D., Singh, V. & Gupta, S. Leveraging the capabilities of EfficientNetB3 to classify eye diseases with reliability. In 4th Int. Conf. Adv. Electr. Comput. Commun. Sustain. Technol. ICAECT 2024 1–5. https://doi.org/10.1109/ICAECT60202.2024.10469017 (2024).
Balance, I. Retracted: Deep learning for ocular disease recognition: An inner-class balance. Comput. Intell. Neurosci.. https://doi.org/10.1155/2023/9838475 (2023).
Fu, H., Xu, Y., Wong, D. W. K. & Liu, J. Retinal vessel segmentation via deep learning network and fully-connected conditional random fields. In Proc. - Int. Symp. Biomed. Imaging Vol. 2016 698–701. https://doi.org/10.1109/ISBI.2016.7493362 (2016).
Wang, S. et al. Hierarchical retinal blood vessel segmentation based on feature and ensemble learning. Neurocomputing 149, 708–717. https://doi.org/10.1016/j.neucom.2014.07.059 (2015).
Liskowski, P. & Krawiec, K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans. Med. Imaging 35 (11), 2369–2380. https://doi.org/10.1109/TMI.2016.2546227 (2016).
The Journal. of Engineering – 2024 - Al-Naji - Computer vision for eye diseases detection using pre‐trained deep learning.pdf.
Wang, S. et al. Advances and prospects of multi-modal ophthalmic artificial intelligence based on deep learning: A review. Eye Vis. 11, 1–13. https://doi.org/10.1186/s40662-024-00405-1 (2024).
Li, N., Li, T., Hu, C., Wang, K. & Kang, H. A benchmark of ocular disease intelligent recognition: One shot for multi-disease detection. In Lect Notes Comput. Sci. (Including Subser. Lect Notes Artif. Intell. Lect Notes Bioinformatics) Vol. 12614 177–193 (LNCS , 2021). https://doi.org/10.1007/978-3-030-71058-3_11.
Dasgupta, A. & Singh, S. A fully convolutional neural network based structured prediction approach towards the retinal vessel segmentation. In Proc. - Int. Symp. Biomed. Imaging 248–251. https://doi.org/10.1109/ISBI.2017.7950512 (2017).
Zhu, C. et al. Retinal vessel segmentation in colour fundus images using Extreme learning machine. Comput. Med. Imaging Graph. 55, 68–77. https://doi.org/10.1016/j.compmedimag.2016.05.004 (2017).
Azzopardi, G., Strisciuglio, N., Vento, M. & Petkov, N. Trainable COSFIRE filters for vessel delineation with application to retinal images. Med. Image Anal. 19, 46–57. https://doi.org/10.1016/j.media.2014.08.002 (2015).
Khan, M. A. U. et al. Automatic retinal vessel extraction algorithm based on contrast-sensitive schemes. In Int. Conf. Image Vis. Comput. New. Zeal 1–5. https://doi.org/10.1109/IVCNZ.2016.7804441 (2016).
Zhao, Y., Rada, L., Chen, K., Harding, S. P. & Zheng, Y. Automated vessel segmentation using infinite perimeter active contour model with hybrid region information with application to retinal images. IEEE Trans. Med. Imaging 34, 1797–1807. https://doi.org/10.1109/TMI.2015.2409024 (2015).
Chen, X., Xu, Y., Kee Wong, D. W., Wong, T. Y. & Liu, J. Glaucoma detection based on deep convolutional neural network. In Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS Vol. 2015 715–718. https://doi.org/10.1109/EMBC.2015.7318462 (2015).
Tanito, M. et al. Validation of a deep learning model to screen for glaucoma using images from different Ryo Asaoka Acknowledgement 23. Available: https://www.sciencedirect.com/science/article/pii/S2589419619300614 (2019).
Maheshwari, S., Kanhangad, V. & Pachori, R. B. CNN-based approach for glaucoma diagnosis using transfer learning and LBP-based data augmentation 1–7. Available: http://arxiv.org/abs/2002.08013 (2020).
Acharya, R. U. et al. Identification of cataract and post-cataract surgery optical images using artificial intelligence techniques. J. Med. Syst. 34, 619–628. https://doi.org/10.1007/s10916-009-9275-8 (2010).
Gao, X., Lin, S. & Wong, T. Y. Automatic feature learning to grade nuclear cataracts based on deep learning. IEEE Trans. Biomed. Eng. 62, 2693–2701. https://doi.org/10.1109/TBME.2015.2444389 (2015).
Kim, D., Jun, T. J., Eom, Y., Kim, C. & Kim, D. Tournament based ranking CNN for the cataract grading. In Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS 1630–1636. https://doi.org/10.1109/EMBC.2019.8856636 (2019).
Tham, Y. C. et al. Referral for disease-related visual impairment using retinal photograph-based deep learning: A proof-of-concept, model development study. Lancet Digit. Health 3, e29–e40. https://doi.org/10.1016/S2589-7500(20)30271-5 (2021).
Guangzhou An, K. O. Glaucoma diagnosis with machine learning based on optical جديد.pdf. J. Healthc. Eng. 2019. https://www.hindawi.com/journals/jhe/2019/4061313/ (2019).
Phan, S. et al. Evaluation of deep convolutional neural networks for glaucoma detection. Jpn. J. Ophthalmol. 63, 276–283. https://doi.org/10.1007/s10384-019-00659-6 (2019).
Diaz-Pinto, A. et al. CNNs for automatic glaucoma assessment using fundus images: An extensive validation. Biomed. Eng. Online 18, 1–19. https://doi.org/10.1186/s12938-019-0649-y (2019).
Al Hazaimeh, O. M., Nahar, K. M. O., Al Naami, B. & Gharaibeh, N. An effective image processing method for detection of diabetic retinopathy diseases from retinal fundus images. Int. J. Signal. Imaging Syst. Eng. 11, 206. https://doi.org/10.1504/ijsise.2018.10015063 (2018).
Gharaibeh, N. et al. Swin Transformer-based segmentation and multi-scale feature pyramid fusion module for Alzheimer’s disease with machine learning. Int. J. Online Biomed. Eng. 19, 22–50. https://doi.org/10.3991/ijoe.v19i04.37677 (2023).
Al-hazaimeh, O. M., Abu-Ein, A. A., Tahat, N. M., Al-Smadi, M. A. & Al-Nawashi, M. M. Combining artificial intelligence and image processing for diagnosing diabetic retinopathy in retinal fundus images. Int. J. Online Biomed. Eng. 18, 131–151. https://doi.org/10.3991/ijoe.v18i13.33985 (2022).
Ocular Disease Recognition. Accessed 07 May 2024. Available: https://www.kaggle.com/datasets/andrewmvd/ocular-disease-recognition-odir5k.
Shaha, M. & Pawar, M. Transfer learning for image classification. In Proc. 2nd Int. Conf. Electron. Commun. Aerosp. Technol. ICECA, 2018 656–660. https://doi.org/10.1109/ICECA.2018.8474802 (2018).
Zhao, C. et al. Dermoscopy Image classification based on StyleGAN and DenseNet201. IEEE Access 9, 8659–8679. https://doi.org/10.1109/ACCESS.2021.3049600 (2021).
Batool, A. & Byun, Y. C. Lightweight EfficientNetB3 model based on depthwise separable convolutions for enhancing classification of leukemia white blood cell images. IEEE Access 11, 37203–37215. https://doi.org/10.1109/ACCESS.2023.3266511 (2023).
Ferreira, C. A. et al. Classification of breast cancer histology images through transfer learning using a pre-trained inception Resnet V2. In Lect Notes Comput. Sci. (Including Subser. Lect Notes Artif. Intell. Lect Notes Bioinformatics) Vol. 10882 763–770 (LNCS, 2018). https://doi.org/10.1007/978-3-319-93000-8_86.
Graf, R., Zeldovich, M. & Friedrich, S. Comparing linear discriminant analysis and supervised learning algorithms for binary classification—A method comparison study. Biometrical J. 66, 1–12. https://doi.org/10.1002/bimj.202200098 (2024).
Ozcan, A., Catal, C., Donmez, E. & Senturk, B. A hybrid DNN–LSTM model for detecting phishing URLs. Neural Comput. Appl. 35, 4957–4973. https://doi.org/10.1007/s00521-021-06401-z (2023).
Author information
Authors and Affiliations
Contributions
Conceptualization, I.K. and V.K; methodology, P.S. and S.S.; software, P.S.; writing---original draft preparation, S.S and A.J.S; writing---review and editing, J.A.H and A.J.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained a repeated error where the term ‘ocular’ was incorrectly given as ‘oncular’. Full information regarding the corrections made can be found in the correction for this Article.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kansal, I., Khullar, V., Sharma, P. et al. Multiple model visual feature embedding and selection method for an efficient ocular disease classification. Sci Rep 15, 5157 (2025). https://doi.org/10.1038/s41598-024-84922-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-84922-y