Abstract
This study aims to construct and validate noninvasive diagnosis models for evaluating significant liver fibrosis in patients with chronic hepatitis B (CHB). A cohort of 259 CHB patients were selected as research subjects. Through random grouping, 182 cases were included in the training set and 77 cases in the validation set. The nomogram was developed based on univariate analysis and multivariate regression analysis. Various machine learning models were employed to construct prediction models for significant liver fibrosis. The area under the ROC curve (AUC), sensitivity, specificity, NPV, PPV, and F1 score were used to evaluate the diagnostic performance. The new nomogram had excellent diagnostic efficiency (AUC 0.806, 95% CI: 0.740–0.872). Compared with other traditional noninvasive diagnostic models, the nomogram demonstrated higher AUC values and better prediction performance. Among six machine learning models, the random forest (RF) model achieved the highest AUC (AUC 0.819, 95% CI: 0.720–0.906). Finally, the importance of all variables in the RF model was ordered to illustrate the contribution of different variables, providing the clinical factors associated with the risk of significant liver fibrosis. This new nomogram may more reliably than other traditional models and the RF model demonstrated superior accuracy among six machine learning models.
Similar content being viewed by others
Introduction
Hepatitis B virus (HBV) infection is a major global public health problem. About 296 million people worldwide are chronic carriers of HBsAg1. Chronic HBV infection can lead to liver fibrosis, cirrhosis, and even liver cancer2,3. In 2022, about 1.1 million people died from hepatitis B1. If left untreated, up to 40% of patients with chronic hepatitis B(CHB) will develop cirrhosis4. The primary mechanism involves fibrin deposition. Hepatic inflammatory injury activates hepatic stellate cells, leading to deposition of extracellular matrix proteins in liver parenchyma. Subsequently, fibrous scar replaces normal liver tissue, resulting in impaired liver function. Recurrent liver inflammation will gradually lead to fibrosis and progression to cirrhosis and even liver cancer2,5,6,7.
Liver fibrosis is an intermediate step in the progression of hepatitis to cirrhosis, and several studies have shown that liver fibrosis can be reversed by removing the cause and controlling liver inflammation. The potential mechanisms include alteration of the inflammatory environment, elimination of activated hepatic stellate cells, and degradation of the extracellular matrix8,9,10. For patients with CHB, antiviral therapy can not only address the root cause of the disease, but also control liver inflammation, which is an important means to reverse liver fibrosis and reduce the mortality related to cirrhosis and liver cancer. Major domestic and international guidelines also consider the degree of liver fibrosis as one of the evaluation indicators for initiating antiviral therapy in CHB patients1,3,4,11,12.
Fibrosis assessment during antiviral treatment is a key step in antiviral therapy evaluation. As we all know, the gold standard for evaluating liver fibrosis is liver biopsy. However, liver biopsy is an invasive procedure with several limitations, such as complications and sampling errors13,14,15,16. Currently, there is increasing emphasis on the noninvasive diagnosis of liver fibrosis. Noninvasive methods for assessing liver fibrosis include various biochemical serum markers, imaging examinations that measure liver stiffness by physical methods, and combining multiple clinical indicators to build noninvasive diagnostic models17. To date, several serological diagnostic models have been reported, such as aspartate aminotransferase to platelet ratio index (APRI)18, fibrosis-4 score (FIB-4)19 and γ-glutamyl transpeptidase to platelet ratio (GPR)20. However, there is no unified standard for these models, and the diagnostic accuracy is still under investigation. The aim of this study is to construct accurate and convenient diagnostic models with clinical serological indicators for clinicians to evaluate the liver fibrosis in CHB patients.
Materials and methods
Materials
The study conformed to the Declaration of Helsinki. It was a single-center, retrospective study approved by the institutional ethics committee of Xiangya Hospital of Central South University (No. 202205120), and waived the requirement for the written informed consent of the patients because the selected clinical data in this study would not affect the prognosis and privacy of the patients. The trial was registered with https://register.clinicaltrials.gov (NCT05560503).
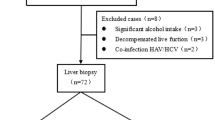
From January 2017 to May 2022, a total of 259 patients diagnosed with CHB at our hospital were retrospectively reviewed. Inclusion criteria: (1) met the diagnostic standards outlined in the Asian-Pacific clinical practice guidelines on the management of hepatitis B4; (2) have not received regular antiviral therapy; (3) underwent liver biopsy to assess the degree of liver pathological changes before antiviral treatment. Exclusion criteria: (1) co-infection with human immunodeficiency virus (HIV); (2) other causes of liver diseases, such as co-infection with other hepatitis viruses, autoimmune hepatitis (AIH), primary biliary cholangitis (PBC), primary sclerosing cholangitis (PSC), inherited metabolic liver disease, drug-induced liver injury (DILI), alcoholic liver disease, decompensated cirrhosis, liver transplantation, and hepatocellular carcinoma (HCC); (3) with other malignant tumor and major systemic diseases; and (4) incomplete clinical data.
Liver fibrosis was staged according to the Scheuer scoring system: S0 (no fibrosis), S1 (portal expansion), S2 (portal fibrosis with limited septa), S3 (numerous septa with structural disorder of the lobule), and S4 (cirrhosis)21. The degree of fibrosis S0-S1 was defined as non-significant liver fibrosis, while S2-S4 as significant liver fibrosis. The flowchart of patient enrollment is shown in Fig. 1.
Data collection
The patient data were extracted from the electronic medical record system included demographics such as age and gender, as well as examination results within 3 days before liver biopsy, including routine blood count, coagulation function test, liver and kidney function, alpha fetoprotein, and viral serology test. These variables were collected for subsequent analysis as they are indicators of routine clinical testing, easy to obtain, relatively objective, and closely related to the severity of hepatitis B.
The APRI was calculated by the formula: [(AST/ULN)×100]/PLT18. The FIB-4 was calculated by the formula: (Age×AST)/(PLT×\(\:\sqrt{\text{A}\text{L}\text{T}\:}\))19. The GPR was calculated by the formula: (GGT/ULN)/(PLT×100)20.
Statistical analysis
All statistical analyses were carried out using IBM SPSS Statistics 25.0 (IBM Corporation, Armonk, NY) and R 4.3.2 (The R Foundation for Statistical Computing, Vienna, Austria). Continuous variables were expressed as median (interquartile range (IQR)). Differences between groups were compared using the nonparametric Mann-Whitney U test. Categorical variables were described by frequency, and differences between groups were compared using the χ2 test or Fisher’s exact test. Variables with statistically significant differences between two groups in univariate analysis were used to screen possible risk factors using multivariate logistic regression analysis with forward stepwise selection and the likelihood ratio test. Forward stepwise selection added variables incrementally to avoid overfitting, while the likelihood ratio test evaluated whether each addition significantly improved model fit. The training dataset was imported into R software to establish the diagnostic nomogram with the screened variables. The scores of different variables were summed to yield a total score for each patient, which could be converted into the predicted probability of significant liver fibrosis. The performance of the nomogram was assessed by the area under the ROC curve (AUC), sensitivity, specificity, NPV, and PPV in the training set and validation set. The optimal cut-off was derived using the Youden index.
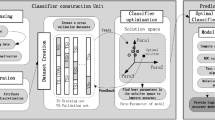
Additionally, in this study, we selected several machine learning algorithms to predict significant liver fibrosis and analyze the importance of factors, each chosen for its distinct strengths in healthcare-related applications. Random Forest (RF) is a robust ensemble learning method that combines multiple decision trees to improve classification accuracy. It excels in handling high-dimensional data and is resilient to outliers and noise, making it particularly suitable for complex clinical datasets. Logistic Regression (LR), a widely used linear model for binary classification, is advantageous for its simplicity, rapid training speed, and interpretability, allowing clinicians to understand feature contributions to predictions. This makes LR especially suitable for clinical contexts where model transparency is crucial. eXtreme Gradient Boosting (XGBoost) is a powerful gradient-boosting algorithm known for its high accuracy in classification tasks. It is capable of handling missing values and noisy data effectively, which is valuable when working with large, complex datasets that are common in healthcare research. Adaptive Boosting (AdaBoost) is effective in generating strong classifiers by assigning greater weight to misclassified samples in successive iterations. It performs particularly well on imbalanced datasets, which are often encountered in medical diagnoses where certain conditions may be rare. Support Vector Machine (SVM) is highly suitable for high-dimensional data and performs well with small sample sizes. Its use of kernel functions allows it to model non-linear relationships, a valuable feature when attempting to capture complex patterns within clinical data. Light Gradient Boosting Machine (LightGBM), a gradient-boosting framework optimized for speed and efficiency, is well-suited for large-scale datasets with numerous features. LightGBM is highly efficient, with fast training times and strong predictive performance, making it suitable for clinical datasets with diverse features.
While each algorithm presents unique strengths, they also exhibit certain limitations. For instance, RF can be computationally intensive and lacks interpretability, making it challenging to analyze the importance of individual features. LR struggles to capture non-linear relationships, which may reduce its effectiveness on complex datasets. XGBoost and LightGBM, despite their accuracy, can require intensive computation and careful parameter tuning, increasing model complexity and resource demands. AdaBoost, while effective on imbalanced data, is sensitive to noise and outliers, which can lead to overfitting. SVM, although powerful for small and high-dimensional data, is computationally inefficient on large datasets and can be challenging to tune. To address these limitations, we conducted comparative testing to balance predictive accuracy with interpretability, aiming to select the optimal algorithm suited to the characteristics of the clinical dataset in this study.
Additionally, given the imbalance between the number of significant and non-significant patients, a range of resampling techniques were applied, including over-sampling (SMOTE, ROSE), under-sampling (Tomek Links), and mixed-sampling (SMOTE + Tomek Links). Five-fold cross-validation was used to obtain the optimal parameters and the most appropriate imbalance treatment for each machine learning algorithm in the training set. The selected parameters and the performance of the models were then evaluated using the validation set. The validation set, consisting of data not seen during the training phase, provided an unbiased measure of the model diagnostic capability. This evaluation included metrics such as AUC, accuracy, precision, recall, and F1 score. The use of the validation set ensured that the model’s effectiveness was assessed on data simulating real-world scenarios. P < 0.05 was considered statistically significant.
Results
General characteristics
A total of 259 patients with CHB, comprising 178 males and 81 females, aged 18–65 years, were included in this study. By random grouping, 182 patients were assigned to the training set, and 77 patients were assigned to the validation set. The results of statistical analysis showed no significant difference in the demographic data, clinical indicators, and pathological results between two groups (P > 0.05) (Table 1).
Screening for predictive factors
According to the Scheuer scoring system21, the patients in the training set were divided into two groups: the non-significant liver fibrosis group (S0-S1, n = 113) and the significant liver fibrosis group (S2-S4, n = 69). The factors screened by the univariate analysis were entered into the multivariable logistic regression analysis, using forward stepwise selection with LR. When P > 0.05, the variables were eliminated. According to the final result of regression analysis [reported as odds ratios (ORs) and 95% confidence intervals (CI), the ALB [0.837(0.750–0.934), P = 0.002], ALP [1.023(1.009–1.037), P = 0.001], HBV DNA [0.746(0.641–0.869), P < 0.001], and PTA [0.944(0.913–0.977), P = 0.001] were independently associated with significant liver fibrosis (Table 2).
Base on the logistic regression analysis, the convenient nomogram (R2 = 0.355, C-index = 0.806) was constructed using the four variables (Fig. 2). The value of each variable was assigned a score from 0 to 100. A score was obtained by projecting the values of each factor onto the first scale axis. By adding the scores of each factor to obtain the final score and projecting the final score to the total point score axis, we could predict the probability of significant liver fibrosis. For each patient, a higher final score indicated a higher risk of significant liver fibrosis.
Then, the data in the training set and validation set were imported into the nomogram model, APRI, FIB-4, and GPR. In the training set, the results showed that the AUC of the nomogram (0.806, 95%CI 0.740–0.872) was significantly higher than APRI (0.573, 95%CI 0.488–0.658), FIB-4 (0.639, 95%CI 0.554–0.724), and GPR (0.649, 95%CI 0.565–0.734) (Fig. 3A). The same conclusion was obtained in the validation set (Fig. 3B; Table 3).
Developing of prediction model using machine learning
Potential predictors for model development were consistent with all variables subjected to the univariate analyses described above. Prediction algorithms used for model development included Random Forest, Logistic Regression, SVM, XGBoost, AdaBoost, and LightGBM. We comprehensively evaluated the performance of the models in terms of accuracy, sensitivity, specificity, NPV, PPV, F1 score, and AUC for the six machine learning models (Table 4). The ROC curves for the six models in the training set (Fig. 4A) and validation set (Fig. 4B) are illustrated. The estimated AUC values for RF and Logistic Regression were over 0.8 in the training set and validation set, while the AUC value for RF was the highest in the validation set (AUC = 0.819) and training set (AUC = 0.816).
The importance variable of RF model
As the AUC value of RF was higher than other machine learning models, indicating good predictive power. The importance of different variables were showed as which indicated the degree of contribution to the RF prediction model. All variables were ranked from the RF analysis. The top 10 variables were PLT, GGT, HBV DNA, ALB, ALP, ALT, HBsAg, PTA, PT, and AFP (Fig. 5).
Discussion
HBV infection leads to complex and dynamic pathological changes at various stages1,3,22, activating the immune response and damaging hepatocytes. This results in recurrent inflammatory necrosis, fibrous tissue deposition, and ultimately liver fibrosis, which can progress to cirrhosis or hepatocellular carcinoma6. The extent of these pathological alterations is a key indicator for guiding antiviral therapy in CHB patients. Early recognition of liver changes and prompt initiation of antiviral treatment can mitigate the risk of poor outcomes. Major international guidelines recommend starting antiviral therapy when significant hepatic pathological changes are observed (inflammation grade G ≥ 2 and/or fibrosis stage S ≥ 2)1.
Liver biopsy remains the gold standard for assessing fibrosis, but its invasiveness presents limitations such as surgical risks, sampling errors, reading inaccuracies, and high costs13,15. These challenges restrict its use in clinical practice and make it unsuitable for routine monitoring of liver status in CHB patients. Consequently, there is a growing focus on noninvasive methods to evaluate liver fibrosis, including the development of serum biomarkers, imaging techniques, and diagnostic models integrating clinical parameters. However, progress in serum biomarker research has been slow, and no definitive liver-specific marker exists to accurately reflect fibrosis severity. Novel imaging technologies, particularly those based on ultrasound and magnetic resonance elastography, are gaining traction17,23. Transient elastography (TE), a widely used tool, assesses liver stiffness via ultrasound but relies on the examiner’s skill. Additionally, factors like acute hepatitis, cholestasis, respiratory conditions, congestive heart failure, alcohol intake, and body mass index can influence test results24,25,26. The implementation of advanced imaging techniques requires specialized equipment and expertise, which may hinder their use in non-tertiary hospitals.
CHB patients face prolonged disease progression and treatment, resulting in a significant disease burden. Developing a diagnostic model that incorporates standard clinical parameters offers a cost-effective and practical solution. This approach enables continuous monitoring of liver fibrosis without adding financial strain on patients, making it an optimal noninvasive diagnostic approach.
The first part of this study employed traditional logistic regression analysis to screen indicators and develop a diagnostic model, complemented by a nomogram for visual representation to enhance clinical applicability. The nomogram model exhibited substantial diagnostic efficacy for significant liver fibrosis, outperforming the APRI and FIB-4 models recommended in current guidelines. A cutoff probability of 41.0%, determined by the Youden Index, provides an optimal balance of sensitivity and specificity for identifying patients at risk of significant liver fibrosis. Clinically, this threshold aids in risk stratification: patients with probabilities above 41.0% are at higher risk and may require further invasive testing or closer monitoring, while those below this level may only need routine follow-up. Although statistically derived, this cutoff offers practical value in guiding initial screening and prioritizing patients for further evaluation. Notably, the nomogram model excludes ALT to mitigate the impact of short-term liver protective treatments on results, facilitating preliminary assessment of liver fibrosis severity.
On this basis, to obtain more efficient diagnostic models, we designed prediction models based on machine learning algorithm within our cohort. However, some research suggests these methods may exhibit optimism and overfitting tendencies27. To mitigate overfitting and enhance model stability, this study combined multiple resampling techniques and 5-fold cross-validation. After multiple attempts, we identified models with relatively high diagnostic efficacy for significant liver fibrosis, which demonstrated robust diagnostic performance, providing a reliable theoretical basis for the future development of simple, user-friendly, and accurate tools for liver fibrosis evaluation. This advancement is particularly significant for extensive and standardized prospective clinical applications.
In comparison to conventional liver fibrosis assessment models like APRI, FIB-4, and GPR model, the nomogram model established in this study through logistic regression analysis and the machine learning-based RF model demonstrate a substantial improvement in diagnostic efficacy for significant liver fibrosis. These models exhibit significant potential in non-invasive liver fibrosis assessment. They solely rely on outpatient follow-up in CHB patients routine serological test indicators for, unlike models explored previously incorporating unconventional test indicators such as alpha-2-macroglobulin, apolipoprotein A1, haptoglobin, hyaluronic acid, TIMP-1, etc28,29. Additionally, they eliminate the need for extra specialized examinations or technical personnel, offering a clear cost and convenience advantage. Particularly advantageous for non-tertiary hospitals conducting regular significant liver fibrosis screenings, these models are appealing due to the higher patient volume and limited technical expertise available. Both the nomogram and RF models developed in this study demonstrated slightly higher specificity than sensitivity, indicating their effectiveness in excluding patients without significant liver fibrosis and reducing the risk of misdiagnosis. However, these models may have limitations in accurately identifying positive cases. Additionally, the positive and negative predictive values of the models were comparable, suggesting a balanced approach to case identification. While the high specificity is advantageous, there is potential for improving sensitivity through the introduction of novel biomarkers or the optimization of algorithms in future research. Overall, these models hold promise for clinical application in the non-invasive diagnosis of HBV-associated significant liver fibrosis, particularly in screening high-risk populations.
Interestingly, in this study, the nomogram model demonstrated superior diagnostic performance for significant liver fibrosis compared to most of these machine learning models. This result may be due to the simplicity and interpretability of traditional statistical methods, which focus on key clinical variables directly associated with liver fibrosis. Nomograms are well-suited to smaller datasets and avoid the potential overfitting seen in complex machine learning models when applied to limited sample sizes. Additionally, the nomogram’s intuitive design allows clinicians to easily interpret and apply the results in practice, a critical advantage over the “black-box” nature of many machine learning algorithms. Certainly, machine learning models offer distinct practical advantages. As clinical data continues to accumulate, these models can iteratively update and optimize to enhance predictive accuracy, making them highly promising for personalized diagnosis and treatment. These findings underscore the importance of selecting models that align closely with the specific characteristics of the data and the practical demands of clinical application. Overall, the nomogram model established through traditional indicator screening methods offers a convenient and relatively accurate approach for evaluating liver fibrosis severity in CHB patients, while machine learning models demonstrate considerable potential in data-rich, complex environments. Together, these approaches provide valuable insights for future research and development in noninvasive diagnostic tools.
We conducted an additional analysis to determine the relative importance of all indicators in the RF models. Through integration with clinical characteristics analysis, we observed that patients with significant liver fibrosis exhibited lower HBV DNA levels, consistent with previous research findings30,31. Recent studies have also revealed that patients with low HBV DNA levels may still manifest significant liver fibrosis, implying potential ongoing viral replication and progression of liver disease32. This highlights the need for clinicians to proactively address and potentially modify antiviral therapy regimens in order to achieve complete virological responses.
Notable indicators include ALP and GGT, as they play a crucial role in the RF outcomes. The nomogram model incorporates ALP. For CHB patients, the association between ALP, GGT, and liver fibrosis may be more significant than the traditional transaminase levels reflecting liver cell damage. ALP and GGT predominantly indicate biliary system health. In CHB patients, persistent inflammation and extracellular matrix deposition can result in biliary tract damage, leading to elevated ALP and GGT levels33. Previous studies have also shown that GGT can serve as a biomarker for advanced fibrosis in non-alcoholic fatty liver disease34. In general, clinicians prioritize the assessment of transaminase levels in CHB patients, as these levels are widely recognized to correlate with liver inflammation. The findings of this study indicate that, in addition to transaminase levels, clinicians should also consider the ALP and GGT levels in their patients, as these may be indicative of the extent of liver fibrosis.
Additionally, this study has inherent limitations. As a single-center retrospective analysis with a small sample size and lacking external validation, the study was limited by the available data, which restricted the collection of sufficient imaging and transient elastography data for comprehensive statistical analysis. Furthermore, the applicability of these models to antiviral therapy—an essential factor for clinical application—has not been evaluated. Therefore, future research should build on these findings through prospective, multicenter studies with larger sample sizes to improve the generalizability and reliability of the results. These studies should also comprehensively validate the models’ effectiveness across diverse clinical scenarios by incorporating antiviral treatment-related factors.
Data availability
The data supporting the main findings of this study are available from the corresponding authors upon reasonable request.
Abbreviations
- CHB:
-
Chronic hepatitis B
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the ROC curve
- DCA:
-
Decision curve analysis
- APRI:
-
Aspartate aminotransferase to platelet ratio index
- FIB-4:
-
Fibrosis-4 score
- GPR:
-
γ-glutamyl transpeptidase to platelet ratio
- OR:
-
Odds ratio
- CI:
-
Confidence intervals
- IQR:
-
Interquartile range
- LR:
-
Likelihood ratio test
- HIV:
-
Human immunodeficiency virus
- AIH:
-
Autoimmune hepatitis
- PBC:
-
Primary biliary cholangitis
- PSC:
-
Primary sclerosing cholangitis
- DILI:
-
Drug-induced liver injury
- HCC:
-
Hepatocellular carcinoma
- ALB:
-
Albumin
- Tbil:
-
Total bilirubin
- ALT:
-
Alanine aminotransferase
- AST:
-
Aspartate aminotransferase
- GGT:
-
γ-glutamyl transferase
- ALP:
-
Alkaline phosphatase
- WBC:
-
White blood cell count
- NEUT:
-
Neutrophil count
- HGB:
-
Hemoglobin
- PLT:
-
Platelet count
- PTA:
-
Prothrombin activity
- INR:
-
International normalized ratio
- APTT:
-
Activated partial thromboplastin time
- AFP:
-
Alpha fetoprotein
- NPV:
-
Negative predictive value
- PPV:
-
Positive predictive value
- RF:
-
Random forest
- LR:
-
Logistic regression
- XGBoost:
-
EXtreme Gradient Boosting
- AdaBoost:
-
Adaptive Boosting
- SVM:
-
Support vector machine
- LightGBM:
-
Light Gradient Boosting Machine
References
World Health Organization. Guidelines for the prevention, diagnosis, care and treatment for people with chronic hepatitis B infection. (World Health Organization, 2024).
Revill, P. A. et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. 4, 545–558 (2019).
You, H., Wang, F. & Li, T. Guidelines for the prevention and treatment of chronic hepatitis B (2022 version). Chin. J. Infect. Dis. 41, 3–28 (2023).
Sarin, S. K. et al. Asian-pacific clinical practice guidelines on the management of hepatitis B: A 2015 update. Hepatol. Int. 10, 1–98 (2016).
Yuen, M. F. et al. Hepatitis B virus infection. Nat. Rev. Dis. Primers. 4, 1–20 (2018).
Iannacone, M. & Guidotti, L. G. Immunobiology and pathogenesis of hepatitis B virus infection. Nat. Rev. Immunol. 22, 19–32 (2022).
Friedman, S. L. Liver fibrosis – from bench to bedside. J. Hepatol. 38, 38–53 (2003).
Ellis, E. L. & Mann, D. A. Clinical evidence for the regression of liver fibrosis. J. Hepatol. 56, 1171–1180 (2012).
Lo, R. C. & Kim, H. Histopathological evaluation of liver fibrosis and cirrhosis regression. Clin. Mol. Hepatol. 23, 302–307 (2017).
Zoubek, M. E., Trautwein, C. & Strnad, P. Reversal of liver fibrosis: From fiction to reality. Best Pract. Res. Clin. Gastroenterol. 31, 129–141 (2017).
Terrault, N. A. et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 67, 1560–1599 (2018).
European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 67, 370–398 (2017).
Seeff, L. B. et al. Complication rate of Percutaneous Liver biopsies among persons with advanced chronic liver disease in the HALT-C trial. Clin. Gastroenterol. Hepatol. 8, 877–883 (2010).
Piccinino, F., Sagnelli, E., Pasquale, G. & Giusti, G. Complications following percutaneous liver biopsy. A multicentre retrospective study on 68,276 biopsies. J. Hepatol. 2, 165–173 (1986).
Rockey, D. C. et al. Liver Biopsy. Hepatology 49, 1017–1044 (2009).
Borg, F. et al. A survey of liver pathology in needle biopsies from HBsAg and anti-HBe positive individuals. J. Clin. Pathol. 53, 541–548 (2000).
Loomba, R. & Adams, L. A. Advances in non-invasive assessment of hepatic fibrosis. Gut 69, 1343–1352 (2020).
Wai, C. T. et al. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology 38, 518–526 (2003).
Sterling, R. K. et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology 43, 1317–1325 (2006).
Lemoine, M. et al. The gamma-glutamyl transpeptidase to platelet ratio (GPR) predicts significant liver fibrosis and cirrhosis in patients with chronic HBV infection in West Africa. Gut 65, 1369–1376 (2016).
Scheuer, P. J. Classification of chronic viral hepatitis: A need for reassessment. J. Hepatol. 13, 372–374 (1991).
Liaw, Y. F., Brunetto, M. R. & Hadziyannis, S. The natural history of chronic HBV infection and geographical differences. Antivir Ther. 15, 25–33 (2010).
Castera, L., Friedrich-Rust, M. & Loomba, R. Noninvasive Assessment of Liver Disease in patients with nonalcoholic fatty liver disease. Gastroenterology 156, 1264–1281e4 (2019).
Patel, K. & Sebastiani, G. Limitations of non-invasive tests for assessment of liver fibrosis. JHEPReport 2, 100067 (2020).
Wagner, M. et al. Technical failure of MR Elastography examinations of the liver: Experience from a large single-center study. Radiology 284, 401–412 (2017).
Boursier, J. et al. Precise evaluation of liver histology by computerized morphometry shows that steatosis influences liver stiffness measured by transient elastography in chronic hepatitis C. J. Gastroenterol. 49, 527–537 (2014).
Zhang, C. et al. A machine learning-based model analysis for serum markers of liver fibrosis in chronic hepatitis B patients. Sci. Rep. 14, 12081 (2024).
Poynard, T. et al. Overview of the diagnostic value of biochemical markers of liver fibrosis (FibroTest, HCV FibroSure) and necrosis (ActiTest) in patients with chronic hepatitis C. Comp. Hepatol. 3, 8 (2004).
Poordad, F. F. FIBROSpect II: A potential noninvasive test to assess hepatic fibrosis. Expert Rev. Mol. Diagn. 4, 593–597 (2004).
Northup, P. G. & Caldwell, S. H. Coagulation in Liver Disease: A guide for the Clinician. Clin. Gastroenterol. Hepatol. 11, 1064–1074 (2013).
Andreani, T., Serfaty, L., Poupon, R. & Chazouilleres, O. Need to strictly define Hepatitis B Virus Immunotolerant patients to avoid unnecessary liver biopsy. Gastroenterology 135, 2155–2156 (2008).
Sun, Y. et al. Persistent low level of Hepatitis B Virus promotes fibrosis progression during therapy. Clin. Gastroenterol. Hepatol. 18, 2582–2591e6 (2020).
Herlong, H. F. & Mitchell, M. C. Jr Laboratory Tests. in Schiff’s Diseases of the Liver 17–43 (John Wiley & Sons, Ltd, 2011).
Ha, Y., Chon, Y. E., Kim, M. N., Lee, J. H. & Hwang, S. G. Gamma-Glutamyl transpeptidase dynamics as a biomarker for advanced fibrosis in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 37, 1624–1632 (2022).
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 81974080, 82170640); Natural Science Foundation of Hunan Province (grant number: 2022JJ30954); and the Scientific Research Project of Chinese Chronic Hepatitis B Clinical Cure Project (grant number: ZFGC202211207).
Author information
Authors and Affiliations
Contributions
S.P. designed the experiments and supervised the study; C.J., J.L., R.L., K.C., Y.X., and W.P. collected the clinical data; C.J., J.L., and D.C. performed the experiments and wrote the manuscript; Z.X. revised the manuscript and L.F. guided the writing of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study obtained the ethical approval from the Institutional Ethics Committee of Xiangya Hospital of Central South University, with informed consent waived (NO.202205120). The trial was registered with https://register.clinicaltrials.gov (NCT05560503).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, C., Xu, Z., Liu, J. et al. Noninvasive diagnosis of significant liver fibrosis in patients with chronic hepatitis B using nomogram and machine learning models. Sci Rep 15, 571 (2025). https://doi.org/10.1038/s41598-024-85012-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-85012-9