Abstract
Given the morphological similarity and medicinal efficacy differences between Acorus tatarinowii Rhizoma and Acorus calamus Rhizoma, both belonging to the Acorus rhizome slices, as well as the phenomenon of their mixed use in the market, this study aims to achieve high-precision classification and rapid object detection of these two Acorus Species Slices using deep learning technology, thus enhancing the accuracy and efficiency of Traditional Chinese Medicine (TCM) identification. The study constructed a high-quality dataset consisting of 1,928 rigorously preprocessed and annotated images of Acorus tatarinowii Rhizoma and Acorus calamus Rhizoma specimens. The ResNet50 model was employed for classification to improve classification accuracy. Furthermore, the YOLOv8 algorithm was utilized for object detection. Experimental results indicate that the ResNet50 model can accurately distinguish between Acorus tatarinowii Rhizoma and Acorus calamus Rhizoma decoction pieces, achieving a test set accuracy of 92.8%, thereby realizing precise classification. Meanwhile, the YOLOv8 algorithm achieved rapid object detection in mixed states of the two, with a detection accuracy of 98.6% and a detection frame rate of 22fps. Meanwhile, we innovatively integrate both channel attention (SE modules) and spatial attention into ResNet50 and YOLOv8 architectures, respectively, to enhance the model’s ability to capture discriminative features of Acorus slices and provide a novel solution for real-time mixed-state detection.Compared to the baseline models, the SE module enhanced the classification accuracy of ResNet50 by 1.7%, while the spatial attention module improved the mAP50 of YOLOv8 by 1.2%, demonstrating the effectiveness of attention mechanisms in fine-grained identification of Chinese herbal materials.This study successfully applied deep learning technology to the classification and object detection of TCM decoction pieces, providing an effective means for intelligent identification and management of Chinese medicinal materials.
Similar content being viewed by others
Introduction
Motivation
Acorus tatarinowii rhizoma (hereafter referred to as Shi Chang Pu) was derived from the dried rhizomes of the Acorus tatarinowii, which belongs to the Acorus genus (Araceae), and was first recorded under the name “Changben” in the Western Zhou Dynasty’s “Rites of Zhou"1. Its inclusion as a traditional Chinese medicine (TCM) under the name “Changpu” was first documented in the “Shen Nong’s Materia Medica Classic,” where it was classified as a superior grade herb2. Due to the numerous species within the Acorus genus, their wide distribution, and incomplete early records, historical confusion regarding the names and original sources of Shi Chang Pu3, leading to frequent misuse in practical applications, particularly with the closely related species Acorus calamus L. (commonly known as Zang Chang Pu or Shui Chang Pu in the market, hereafter referred to as Shui Chang Pu). Shui Chang Pu is the dried rhizome of the Acorus calamusL., which is officially listed in the Chinese Pharmacopoeia under the name Zang Chang Pu4,5. According to the 2020 edition of the “Chinese Pharmacopoeia,” Shi Chang Pu possesses the functions of opening the senses and dispelling phlegm, refreshing the mind and enhancing intelligence, and transforming dampness to stimulate appetite4. In contrast, Shui Chang Pu is known for warming the stomach and relieving inflammation and pain. Despite their character similarities, their therapeutic effects differ significantly, posing significant challenges to the safety and efficacy of clinical medication. Hence, accurately distinguishing between these two herbs is paramount for elucidating the provenance of Shi Chang Pu decoction pieces and ensuring their consistent quality control.
Related work
In the Chinese Pharmacopoeia, 2020 edition, the identification of both Shi Chang Pu and Shui Chang Pu employs a combination of empirical identification, microscopic identification, and physicochemical identification. Currently, the main methods for distinguishing between them are as follows:
Empirical Identification6,7,8: This method relies on the visual characteristics of the calamus-type medicinal materials, such as surface color, internode length, cross-sectional features, and odor, to differentiate Shi Chang Pu and Shui Chang Pu. However, as the saying goes, “Sliced calamus, a challenge even for celestial beings to discern”, while empirical Identification is straightforward, it heavily depends on the expertise and accumulated experience of industry professionals. This inevitably introduces a degree of subjectivity, and slight differences in judgment may exist among experts, making it challenging to ensure the accuracy and consistency of identification. Furthermore, the cultivation of experts requires significant human and material resources, as well as long-term practical experience. In the context of the increasingly vast traditional Chinese medicine (TCM) market, relying solely on human labor for individual identification is undoubtedly overwhelming.
Microscopic Identification9,10: This method involves using optical or electron microscopes to conduct microscopic analysis of the tissues and powders of Shi Chang Pu medicinal materials or decoctions. It is highly specific but necessitates the support of specialized personnel and equipment.
Physicochemical Identification11,12,13: This approach differentiates Shi Chang Pu and Shui Chang Pu based on their physicochemical properties. For instance, fluorescence identification is employed to distinguish Shi Chang Pu, Shui Chang Pu, and Anemone altaica Fisch. (Jiujie Chang Pu). Additionally, ultra-performance liquid chromatography coupled with orbitrap mass spectrometry (UPLC-Orbitrap-MS/MS) and gas chromatography-mass spectrometry (GC-MS), in conjunction with chemometric methods, are used to characterize and identify Shi Chang Pu and its adulterants. However, these methods tend to be inefficient, time-consuming, and require expensive instrumentation and specialized operation.
Biological Methods14,15,16,17: These include intronic transcription sequencing, DNA barcode sequence analysis, PCR-RFLP, and other techniques to differentiate Shi Chang Pu and Shui Chang Pu. Nevertheless, due to their high degree of specialization and high costs, these methods are challenging to popularize and apply at the grassroots level.
Due to the numerous limitations in distinguishing between Shi Chang Pu and Shui Chang Pu using the aforementioned identification methods, such as the subjectivity of empirical identification and the requirement for personnel professionalism, as well as the professional thresholds and equipment dependencies of microscopic, physicochemical, and biological identification, there is a pressing need to explore more precise and efficient identification methods. With the rapid development of artificial intelligence technology, particularly the outstanding performance of deep learning in image recognition, new possibilities have emerged for the rapid identification of traditional Chinese medicinal materials. Therefore, the introduction of deep learning image recognition technology into the field of traditional Chinese medicinal material identification has been proven to enable precise and efficient identification of these materials, as evidenced by relevant studies18,19,20,21.
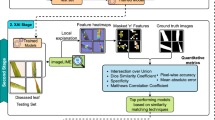
Therefore, this research focuses on two traditional Chinese herbal slices with similar appearances but different efficacies that are easily confused in the market-Acorus tatarinowii rhizoma (Shi Chang Pu) and Acorus calamus rhizoma (Shui Chang Pu), delving into the innovative application of deep learning technology in the classification and object recognition of these Chinese herbal slices. By converting traditional expert identification experience into “machine experience,” we aim to construct an efficient and accurate intelligent recognition system. In terms of specific technical implementation, this paper adopts advanced deep learning models: For the task of classification, we select the ResNet (Residual Network) model to effectively distinguish the characteristics of Shi Chang Pu and Shui Chang Pu in subtle differences in morphology, texture, and other features. For the object detection task, we employ the You Only Look Once version 8 (YOLOv8) model, meeting the needs for real-time, fast, and high-precision recognition, laying a foundation for further research on locating and identifying decoction targets in complex backgrounds (Fig. 1). Meanwhile, this study pioneers the integration of Squeeze-and-Excitation (SE) modules with YOLOv8, proposing a lightweight spatial attention mechanism tailored for detecting mixed states of Chinese herbal slices. This innovation offers a novel strategy for real-time recognition in complex backgrounds. By translating expert knowledge into machine-learnable features, we achieved highly efficient and precise intelligent identification.Taking this research as a strong starting point, we aim to extend the same technical framework and strategies to the identification of other traditional Chinese medicinal materials. This not only serves as a beneficial supplement to traditional identification methods of traditional Chinese medicinal materials but also represents an important step forward in pushing the identification technology towards intelligence and automation. Its application prospects are vast and worthy of further research and promotion.
Materials and methods
Sample collection
To establish a high-quality dataset of Shi Chang Pu and Shui Chang Pu, this study adopted a diversified sample collection strategy, gathering samples of both types through multiple channels. The majority of the samples were purchased from traditional Chinese medicine markets, traditional Chinese medicine materials companies, while a small portion was collected directly from the Chinese medicinal materials planting base. This base is located in Lisong Village, Lixian County, Hunan Province. All samples were authenticated by Liu Haitao (Deputy Chief Pharmacist) from the Xiangya Hospital of Central South University. All samples were carefully stored in the Traditional Chinese Medicine Identification Laboratory of Hunan Food and Drug Vocational College, with voucher specimens deposited under voucher ID number SHI20230807, SHI20230608, SHI20230901, SHI20230715, SHI20230908, SHUI20230908, SHUI20230618, and SHUI20230705, respectively. No specific permissions were required for the collection of these specimens, as they were obtained from publicly accessible areas (as shown in Table 1) and did not involve endangered or protected species.
Dataset construction and preprocessing
In previous studies on the identification of Chinese herbal medicines (CHMs), the image datasets used for establishing recognition models varied. Some consisted of images of single CHM slices against a clear background22, while others featured multiple CHM slices stacked or mixed together against a complex background. The latter often contained irrelevant and redundant information, which could introduce biases and affect the training process. The quality of the dataset is crucial to the creation of the model, and thus a rich and representative dataset is of paramount importance23. The construction process of the dataset in this study is as follows:
Sample coverage
To establish a high-quality dataset, we first collected samples from multiple diverse locations such as herbal medicine markets, herbal planting bases, traditional Chinese medicine pharmacies, and so on (as shown in Table 1), conducting image data acquisition and striving to broaden the sample coverage as much as possible.
Shooting tools and parameters
The shooting tool used was a mobile phone camera, which features high resolution and excellent imaging quality, ensuring the clarity and accuracy of the image data. The camera parameter settings are detailed in Table 2, ensuring consistency in subsequent data processing and analysis.
Image data acquisition and preprocessing
In this study, all images were captured under uniform lighting conditions and against a clear, uncluttered background(as shown in Table 2), ensuring image quality and consistency that clearly showcase the morphological characteristics of the slices. During photography, Both of the Shi Chang Pu and Shui Chang Pu were laid flat on white paper, and individual or non-overlapping multiple slices were photographed to avoid mixing or stacking (Fig. 2). Simultaneously, we also excluded the images that did not meet the required standards.This approach allowed us to leverage professional knowledge and expert experience to guide the algorithm in capturing details and identifying more relevant features from the CHM images, making its learning process more akin to that of practitioners in traditional Chinese medicine appraisal.
To improve the model performance, we conducted rigorous preprocessing work on the original image data. Firstly, we cropped and resized the images to ensure that all images had a uniform resolution before being input into the model (224 × 224 pixels for ResNet50 and 640 × 640 pixels for YOLOv8). Secondly, we performed color correction and normalization to eliminate the influence of external factors such as lighting and shooting equipment on image quality. Finally, L.L., an herbal medicine identification expert from our research group annotated the CHS images, where the X-AnyLabeling toolbox was used for sample annotation during the YOLOv8 training process (Fig. 3). This annotation work converted “expert experience” into “machine experience,” laying a foundation for subsequent image classification and object recognition tasks, thereby enhancing the generalization ability and robustness of the model.
Dataset partitioning
To evaluate the generalization ability of the model, this study divided the annotated image dataset of Shi Chang Pu and Shui Chang Pu into a training set, a validation set, and a test set in a ratio of 8:1:1 (Table 3). Among them, the training set is primarily used for model training, enabling the model to learn and extract patterns and features from the data. The validation set is utilized for model tuning, assessing model performance, and preventing the occurrence of overfitting. The test set is mainly employed to evaluate the generalization ability of the model after training, that is, to assess the accuracy of the model’s predictions on new data that have not appeared during training and validation.
Experimental environment configuration
The experimental environment configuration for this study is presented in Table 4.
All of these activities were strictly in accordance with the guidelines and regulations of relevant institutions, countries, and international organizations.
Image classification method based on ResNet50
Model architecture
This study employs the classical deep learning model ResNet50 for image classification24. Proposed by He Kaiming and his colleagues in 2015, this model is renowned for its robust feature extraction capabilities and residual learning mechanism, which have proven exceptional performance in various image classification tasks. By introducing residual connections, ResNet50 effectively addresses the issue of gradient vanishing/explosion during the training of deep CNNs, enabling the construction of deeper network structures capable of extracting richer image features. Image data traverses through multiple convolutional layers and residual blocks within ResNet50, progressively abstracting low-level to high-level image features. These features encapsulate crucial information such as texture and shape patterns in the decoction piece images, playing a vital role in subsequent classification tasks. At the terminal of ResNet50, a fully connected layer is appended, which maps the extracted feature vectors onto predefined class labels (i.e., Shi Chang Pu and Shui Chang Pu).
Training process
Loss Function Design: The Cross-Entropy Loss function is employed in this study for multi-class classification due to its intuitive ability to measure the discrepancy between predictions and true labels, its seamless integration with softmax outputs, and its clear optimization objective of driving predicted probabilities closer to the true labels, effectively accelerating model training and enhancing classification accuracy25. Consequently, this function is adopted to evaluate the difference between the model’s predictions and the actual labels. The formula is as follows25:
where L represents the cross-entropy loss value; n denotes the total number of the samples; \({{y}}_{i}\) is a binary indicator (0 or 1) that equals 1 if the sample belongs to the positive class and 0 otherwise; \(\hat{{y}}_{i}\) is the probability predicted by the model that sample belongs to the positive class.
When \(y_{i} = 1\), indicating that the sample belongs to the positive class (Acorus tatarinowii), the loss function simplifies to: \(l_{i} = \log (\hat{y}_{i} )\). In this case, the closer the predicted probability \(\hat{y}_{i}\) is to 1, the smaller the loss function value L becomes, indicating a more accurate model prediction.
When \(y_{i} = 0\), indicating that the sample belongs to the negative class (Acorus calamus), the loss function simplifies to: \(l_{i} = \log (1 - \hat{y}_{i} )\). Similarly, the closer the predicted probability \(\hat{y}_{i}\) is to 0, the smaller the loss function value L becomes, also indicating a more accurate model prediction.
Optimization Algorithm: The Adam algorithm, a variant of Stochastic Gradient Descent (SGD), is utilized within the back-propagation algorithm to optimize the model parameters26. By minimizing the loss function, Adam aims to improve the classification accuracy. It incorporates adaptive learning rates for individual parameters, which helps in faster convergence and potentially better optimization.
Hyperparameter Tuning: The process of adjusting hyperparameters such as the learning rate, batch size, and number of epochs to achieve optimal model performance is conducted through the Random Search method. This method randomly samples values from predefined distributions for each hyperparameter, allowing for an efficient exploration of the hyperparameter space. The initial values set for these hyperparameters during the training process are provided in Table 5.
The visualization results of the training process, as shown in Fig. 4, indicate that the loss function on the training set stabilizes after approximately 50 iterations (Fig. 4a), while the accuracy on the validation set stabilizes after approximately 60 iterations (Fig. 4b).
Model architecture with attention mechanism
To enhance the model’s focus on discriminative features of Acorus species slices, we integrated Squeeze-and-Excitation (SE) blocks into the ResNet50 architecture. Specifically, SE modules were inserted after each residual block to adaptively recalibrate channel-wise feature responses. The SE block consists of two steps: (1) Squeeze: Global average pooling aggregates spatial information into channel descriptors.(2) Excitation: Two fully connected layers learn channel-wise attention weights, amplifying critical features while suppressing irrelevant ones.This modification introduces minimal computational overhead but significantly improves feature representation for subtle morphological differences between Shi Chang Pu and Shui Chang Pu.
Results and discussion
This study conducted subsequent testing using an independent test set (images that were not exposed to the model during the training process) to validate the effectiveness of the ResNet50 model in this classification task. The experimental results (Table 6) indicate that the accuracy and recall rates for Shi Chang Pu are slightly higher than those for Shui Chang Pu. This may be related to the morphological characteristics of the two species, where the former has a stronger fibrous cross-section, while the latter has a spongy fracture. According to expert identification experience, the strong fibrous nature of Acorus tatarinowii’s cross-section gives it a more distinct layered appearance visually, making it easier to accurately distinguish in image recognition. Furthermore, in terms of dataset distribution, there are more samples of Shi Chang Pu than Shui Chang Pu, which may have allowed the model to gain a deeper understanding and mastery of the characteristics of Shi Chang Pu during the learning process, resulting in higher accuracy and recall rates during classification.
To further validate the effectiveness of the SE module, we compared the original ResNet50 with the SE-enhanced ResNet50 (ResNet50-SE). As shown in Table 7, ResNet50-SE achieved a 94.5% accuracy on the test set, outperforming the baseline ResNet50 with a 92.8% accuracy. This demonstrates that the attention mechanisms introduced by the SE module can better capture discriminative features in TCM slice images, enhancing the model’s classification performance.
In summary, this method achieves high classification accuracy for both species and demonstrates good generalization ability.
Target recognition method based on YOLOv8
YOLOv8 model
In response to the issue of mixed usage of Shi Chang Pu and Shui Chang Pu in the market, this study further adopts the advanced target detection model, YOLOv8, to achieve more precise and accurate recognition of these two decoctions pieces when they appear in the same field of view27. While maintaining the efficiency of the YOLO series models, YOLOv8 enhances detection accuracy and speed through the introduction of a series of optimization strategies28. Preprocessed images are input into the YOLOv8 model, which automatically identifies the decoction targets in the images and provides their categories and bounding box information. During the training process, we utilize the built-in loss function of YOLOv8 to simultaneously optimize both classification and localization tasks, and further enhance model performance by adjusting hyperparameters and training strategies.
Training process
Loss function design: A comprehensive loss function inherent to YOLOv8 is employed, which aims to simultaneously evaluate the model’s errors in both classification and localization. Specifically, this function integrates a classification loss and a localization loss. The classification loss, typically adopting a cross-entropy loss function or binary cross-entropy loss function, gauges the accuracy of the model’s predictions for target categories. Meanwhile, the localization loss, which may employ mean squared error or its variants along with IoU-related loss functions, assesses the precision of the model’s predictions for target locations. By comprehensively considering these two aspects of loss, the YOLOv8 loss function can effectively guide the model to optimize both classification and localization performance during training.
Training process optimization: Preprocessed image data is fed into the model, and parameters are adjusted through multiple iterations of training to minimize the loss function until the model achieves satisfactory performance on the training set.
Hyperparameter tuning: The selection of hyperparameters during training plays a pivotal role in determining the model’s performance. Therefore, based on training feedback, continuous adjustments are made to the hyperparameters to find the optimal configuration. The initial hyperparameter settings are outlined (Table 8).
Spatial attention for detection
To improve the localization accuracy of mixed Acorus slices, we incorporated a lightweight spatial attention module into the YOLOv8 neck network. This module generates a spatial attention map by combining max-pooling and average-pooling operations across channels, followed by a sigmoid activation. The attention map highlights regions containing target slices while suppressing background noise. This adaptation requires only < 1% additional parameters but enhances detection robustness in cluttered environments.
Results and discussion
Recognition Process: During the recognition phase, the images to be identified are input into the pre-trained YOLOv8 model. The model performs forward propagation on the input images, extracting image features through convolutional layers, pooling layers, and other structures, and ultimately outputs target category and bounding box information. This information includes the category label of each target in the image and its position and size within the image, typically represented in the form of bounding boxes. In this manner, the YOLOv8 model is capable of achieving accurate recognition and localization of targets within images (Fig. 5).
Evaluation Metrics: For the trained YOLOv8 model, this study employs precision (P), recall (R), and mean average precision (mAP) as evaluation criteria to assess the model’s performance. The evaluation metrics and their respective results are presented in Fig. 6; Table 9.
Impact of Spatial Attention on Detection: As shown in Table 10, the YOLOv8 model with spatial attention achieved a mAP50 of 95.3% (vs. 94.1% for the baseline), with a detection frame rate maintained at 21 fps. The improved precision (98.9% vs. 98.6%) and recall (91.5% vs. 90.8%) further validate the effectiveness of attention mechanisms in real-time TCM slice detection.
Based on the aforementioned data, YOLOv8 has demonstrated exceptional performance in detecting Shi Chang Pu decoction pieces. Its high precision significantly reduces false positives, ensuring reliable results. The commendable recall ensures that the model comprehensively covers all relevant targets. Furthermore, the high mAP score underscores the model’s accuracy, robustness, and ability to generalize. Collectively, these results validate YOLOv8’s suitability for processing Traditional Chinese Medicine (TCM) and underscore its potential for broader applications.
Notably, these two methods each have their own advantages and disadvantages. The reason for adopting both ResNet50 and YOLOv8 for classification and recognition is to adapt to different application scenarios and meet various practical needs.
ResNet50 excels in image classification tasks. Its deep residual network structure enables the model to learn deeper image features, thereby improving classification accuracy. It may perform better in the more meticulous field of decoction piece identification. However, the ResNet50 model has a larger structure, requiring high computational resources and lacking in real-time performance, which is also confirmed in experiments: the detection frame rate of YOLOv8 is more than twice that of ResNet50, highlighting its real-time capabilities and rapid performance. Therefore, it may be limited in scenarios requiring rapid identification.
On the other hand, YOLOv8 adopts a single-stage object detection framework, capable of accurately detecting and recognizing targets in images of Chinese medicine decoction pieces in a short period of time. Therefore, it is highly suitable for applications requiring high real-time performance, such as decoction piece inspection and warehousing, which often need to be completed within a short period of time. While maintaining real-time performance, YOLOv8 also has high detection accuracy, capable of accurately identifying and distinguishing different targets in images, such as the identification of Shi Chang Pu and Shui Chang Pu decoction pieces in this paper.
In summary, ResNet50 and YOLOv8 each have their own advantages and disadvantages. When selecting an identification method, it is necessary to weigh and choose based on the actual application scenario and requirements.
From image to insight
Deep learning technology has not only revolutionized the precision of identifying and detecting Acorus species slices, specifically Shi Chang Pu and Shui Chang Pu, but it has also illuminated deeper insights into their distinguishing characteristics. Addressing the prevalent market issue of mixed usage, this study delves into the realm of deep learning-driven classification methodologies, harnessing the prowess of ResNet50 for image classification and the cutting-edge YOLOv8 for object detection.
In the realm of image classification, our research achieves a high-precision recognition of Shi Chang Pu and Shui Chang Pu decoction pieces, fueled by the ResNet50 framework. Experimental outcomes underscore ResNet50’s unparalleled accuracy and robust generalization capabilities, positioning it as a formidable candidate for practical deployment. This approach offers a paradigm shift in the identification of traditional Chinese medicinal materials, providing an objective, precise, and efficient alternative that mitigates the inaccuracies and inconsistencies inherent in conventional identification methods.
For object detection, we capitalized on the YOLOv8 model’s advanced capabilities, achieving remarkable precision in recognizing Shi Chang Pu and Shui Chang Pu decoction pieces within images. YOLOv8’s experimental prowess, characterized by its high detection accuracy coupled with a swift inference speed, ensures seamless identification and classification of decoction pieces. This formidable performance underscores its practical applicability in real-time scenarios, presenting a groundbreaking tool for the identification, quality control, and standardization of traditional Chinese medicinal materials.
Meanwhile, we have also introduced attention mechanisms into the existing framework. These mechanisms enhance detection accuracy by selectively focusing on salient features amidst complex backgrounds, enabling it to maintain robust performance even in challenging environments. This innovation will further consolidate our method’s leading position in the field of traditional Chinese medicinal material identification.
Conclusions and future prospects
While acknowledging the promising outcomes achieved in this study, we are mindful of the potential challenges that may arise during the practical deployment of our deep learning-based methods.
Limitations of the proposed method
Regional Sample Bias: Although our dataset covers several provinces in southern China where calamus is produced, samples from some special habitats have not been fully incorporated. This may limit the generalization ability of the model in extreme environments.
Computational Resource Requirements: The YOLOv8 + Attention mechanism requires a mid-range GPU, which restricts its real-time application on devices without a graphics card.
Practical recommendations for users
Periodic Model Updates and Lightweight Deployment: Retrain the model with local samples annually and compress it using knowledge distillation techniques to make it suitable for low-end devices.
Human-Machine Collaboration Optimization: Initiate manual review for low-confidence predictions (such as<90%) and record ambiguous cases to iteratively optimize the model.
Future research directions
Enhancing Sample Diversity: Actively explore cooperation with calamus origin resource centers and establish an open data platform to encourage community contributions.
Edge Computing Adaptation: Develop lightweight models (such as YOLOv8-Tiny + Adaptive Attention) that support real-time CPU inference with a target frame rate of ≥ 15fps.
Automated Human-Machine Collaboration: Explore active learning frameworks that automatically request expert annotations based on model uncertainty, reducing manual workload.
In conclusion, the deep learning-powered identification strategies presented in this study, tailored specifically for Shi Chang Pu (Acorus tatarinowii rhizoma) and Shui Chang Pu (Acorus calamus rhizoma) decoction pieces, have opened up novel and effective pathways for the precise recognition of traditional Chinese medicinal materials. We are confident that with the relentless pursuit of technological advancements and continuous refinement of our methods, this innovative approach will increasingly shape the future of quality control in the traditional Chinese medicinal materials industry, ushering in an era of heightened automation and intellectualization.
Data availability
Data availabilityThe datasets used and analysed during the current study available from the corresponding author on reasonable request.
References
Cui, G. Rites of Zhou (Liaoning Education, 1997).
Weng, Q. et al. Textual research on classical prescription of Acorus Tatarinowii. Chin. J. Mater. Med. 44, 5256–5261 (2019).
Wu, J. et al. Herbal textual research on Shichangpu (Acori Tatarinowii Rhizoma). Liaoning J. Tradit Chin. Med. 24, 148–156 (2022).
National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China: Volume I. (China Medical Science Press, 2020). (2020).
harmacopoeia Commission of the Ministry of Health of the People’s Republic of China. Standards for Pharmaceutical Preparations of Traditional Chinese Medicines:Volume 2,Issued by the Ministry of Health of the People’s Republic of China (Pharmacopoeia Commission of the Ministry of Health, 1990).
Luo, H. et al. Pharmacognostic identification on four species of Acorus. Shanxi J. Tradit Chin. Med. 23, 25–35 (2022).
Cao, H. & Jiang, Y. Identification of Acorus Tatarinowii and Acorus calamus. Strait Pharm. J. 28, 33–35 (2016).
Wen, F. Identification of traditional Chinese Medicine-Acorus Tatarinowii and its easily confused species. J. Basic. Chin. Med. Sci. 19, 947–952 (2013).
Wang, X. et al. Identification study of Acorus Tatarinowii and Acorus calamus. Chin. Ethnomed. Pharmacol. 25, 17–21 (2016).
Gao, Y. & Zhu, J. Microscopic tissue structure of three species of Acorus. J. Chin. Med. Mater. 2, 28–30 (1986).
Yi, C. & Ai, W. Brief discussion on the identification of Acorus tatarinowii, Acorus calamus, and Acorus gramineus. N Pharm. 9, 104 (2012).
Ma, S. et al. Discrimination of Acori Tatarinowii rhizoma from two habitats based on GC-MS fingerprinting and LASSO- PLS-DA. J. Cent. South. Univt. 25, 1063–1075 (2018).
Zhang, X., Shi, S. & Yi, L. Chromatographic fingerprinting for volatiles of grass leaf sweet flag rhizome native to Sichuan. Chin. J. Tradit Chin. Med. Pharm. 2, 459–462 (2016).
Cai, T. et al. Identification of mutual adulteration of Acorus Tatarinowii and Acorus calamus using RNase H-dependent PCR combined with melting curve method. Herald Med. 42, 328–333 (2023).
Liu, L. et al. Identification of 5 species of medicinal plants for Acorus L. based on DNA barcoding sequences. Cent. South. Pharm. 13, 388–392 (2015).
Lam, K. et al. Authentication of acori Tatarinowii rhizoma (Shi Chang Pu) and its adulterants by morphological distinction, chemical composition and ITS sequencing. Chin. Med. 11, 1–11 (2016).
Du, X., Song, S. & Ni, L. Identification of Acorus calamus adulterated in Acorus Tatarinowii medicinal materials and Decoction pieces by PCR-RFLP. Chin. J. Info Tradit Chin. Med. 30, 121–126 (2023).
Sun, X. & Qian, H. Chinese herbal medicine image recognition and retrieval by convolutional neural network. PloS One. 11, 1–19 (2016).
Weng, J., Hu, M. & Lan, K. Recognition of easily-confused TCM herbs using deep learning. Proc. the 8th ACM on Multimedia Systems Conference 233–234ACM, (2017).
Wu, C. et al. Intelligent identification of fritillariae cirrhosae bulbus, crataegi fructus and pinelliae rhizoma based on deep learning algorithms. Chin. J. Exp. Tradit Med. Formulae. 26, 195–201 (2020).
Chen, W. et al. An easy method for identifying 315 categories of commonly-used Chinese herbal medicines based on automated image recognition using automl platforms. Inf. Med. Unlocked. 25, 1–10 (2021).
Tao, O. et al. Research on identification model of Chinese herbal medicine by texture feature parameter of transverse section image. Mod. Tradit Chin. Med. Materia Med. -World Sci. Technol. 16, 2558–2562 (2014).
Luo, C. et al. How does the data set and the number of categories affect CNN-based image classification performance?? J. Softw. 14, 168–181 (2019).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. Proc. https://doi.org/10.1109/CVPR.2016.90 (2016) (2016 IEEE Conference on Computer Vision and Pattern Recognition).
Bishop, C. M. & Nasrabadi, N. M. Pattern recognition and machine learning (springer, 2006).
Kingma, D. P., Ba, J. & Adam A Method for Stochastic Optimization. Proc. the 3rd International Conference on Learning RepresentationsICLR, (2015).
Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You only look once: unified, real-time object detection. Proc. https://doi.org/10.1109/CVPR.2016.91 (2016) (2016 IEEE Conference on Computer Vision and Pattern Recognition).
Jocher, G., Chaurasia, A., Qiu, J. & Ultralytics, Y. O. L. O. Available online: (2023). https://github.com/ultralytics/ultralytics
Acknowledgements
This work was funded by Natural Science Foundation of Hunan Province Fund (2023 JJ60123) and supported by the scientific research and innovation team construction project of Hunan Food and Drug Vocational College (No.KCTD202201).
Author information
Authors and Affiliations
Contributions
Y.L. designed the experimental scheme and wrote the main manuscript text, H.L. did experimental verification, Y.D. and L.L. constructed the image datasets. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Liu, H., Li, L. et al. From image to insight deep learning solutions for accurate identification and object detection of Acorus species slices. Sci Rep 15, 15734 (2025). https://doi.org/10.1038/s41598-025-00038-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-00038-x