Abstract
Lung adenocarcinoma (LUAD) is the most popular lung cancer type with highly mortality. We performed a single cell RNA-seq analysis to explore characteristic of cancer stem cells in LUAD. We downloaded the single cell RNA-seq data (GSE149655) from the GEO database, the scRNA-seq analysis was performed by using the “Seurat” and “harmony” R package. The FindMarkers function and “ClusterProlifer” package was used for differentially expressed genes (DEGs) and function enrichment analysis. The protein-protein interaction and transcriptional regulatory network were performed by STRING and ChIPBase database. Immunohistochemistry tests to be used to observe differences in the expression of specific genes in LUAD and paracancerous tissue samples. BEAS-2B and A549 cells was used for vitro assay and the qRT-PCR, western blotting, wound healing, trans-well assays, EdU tests, and flow cytometry were performed. A total of 9 cell clusters were obtained after scRNA-seq analysis, in which the cancer stem cells had higher proportion in LUAD samples. Subsequently, function enrichment analysis revealed that the amino sugar and nucleotide sugar metabolism and DNA replication pathways were activated in cancer stem cells (CSCs), which were further sub-divided into 3 subtypes, the LGR5 + stem cell is a major contributor to cancer progression, its hub genes, such as HLA-DPB1, CD74, CTSH and HLA-DRB5 mediated the unique transcriptional state. In addition, the marker genes of three CSCs were also overexpressed in LUAD cells and the CXCL3 played an important role in mediating cell proliferation, apoptosis, migration and invasion of tumor. We performed a scRNA-seq analysis and identified the LGR5 + stem cell as a major contributor in LUAD progression, our findings are expected to provide new insights into the pathogenesis of LUAD.
Similar content being viewed by others
Introduction
Lung cancer represent a malignant tumor with the highest cancer-related deaths worldwide, accounting for approximately 18% of all cases of cancer-related death1,2. Among major lung cancer subtypes, the lung adenocarcinoma (LUAD) is the most popular histological type (50%) with gradually increased incidence, and characterized by the high invasiveness, aggressiveness and resistance to therapy3,4, it was estimated that there are 234,580 new cases and 125,070 deaths in 20245. During the past decades, the intensive efforts, such as non-invasive surgical resection, chemoradiotherapy, tyrosine kinase inhibitors (TKIs) of gefitinib, anaplastic lymphoma kinase inhibitors and immunotherapy (PD-1 or PD-L1 targeting blockade)6, have only benefitted a small proportion of LUAD patients, so the outcome of LUAD patients still remains bleak, with an approximately 17.4% of 5-year overall survival (OS)7. The poor survival rate of patients may be caused by the considerable heterogeneity (drug resistance, immune response efficiency, postoperative high recurrence and metastatic propensity) that are genomic drivers8, the 75% of patients are diagnosed at late stage and immunocompromised9 due to the mutation of cancer cells that increased the invasion degree of cancer10. The tumor cells separate from the primary tissue and transfer through the blood vessels to other location, the brain metastasis represent a major cause of poor prognosis, recurrence and death for LUAD patients, of whom about 20–40% are brain metastasis eventually11. Advanced cancer and metastasis can shorten survival time of patients drastically and decrease life quality, but the specific details of LUAD progression at different disease stages remain poorly understood. Therefore, it is an urgent task to explore the underlying pathogenesis or molecular characteristics and search new therapeutic targets for advancing more precise and effective diagnostic and therapeutic strategies for LUAD patients.
Genetic, epigenetic modifications, and tumor microenvironmental characteristics may affect the intrinsic cellular programs and mediate diverse disease pathogeneses of LUAD, these underlying heterogeneities determined the poor efficiency of uniform therapeutic strategies. Such as the clinical therapeutic response between the LUAD and lung squamous carcinoma (LUSC) exist distinct difference12, of which include type II alveolar cells (AT2) and basal cells with multiple subclones that present different response to targeting treatment13. Cancer appear a multicellular community that composed of various malignant epithelial cells and cancer stem cells, and non-malignant immune and stromal cells, they exhibit dynamic interaction14. Endothelial cells expressed the vascular endothelial growth factor (VEGF) that is a key anti-angiogenic target, Zhang el at, revealed a specific tip-like endothelial cells mediated pro-angiogenesis and anti-host immune responses and its PSMA marker in a pan-cancer study15. In addition, mounting studies revealed that the significant immunogenic and cellular compositions heterogeneity between the primary and metastatic tumors16, the advanced tumors usually consist of a number of subclones with different genetic and functional alterations to mediate the immunotherapy resistance for the survival of tumor17. Tumor heterogeneity with the complex cellular subclones play an important role in cancer progression and treatment response18, but the potential molecular features that characterize contribution of cancer heterogeneity to metastasis and malignant progression are largely unknown. We all known that there are many different cell types express unique transcriptomes during the tumorigenesis, the traditional bulk sequencing only can provide an average expression value for a batch of cells, whereas the single-cell RNA sequencing (scRNA-seq) can reveal the comprehensive transcriptome profiling of small cell samples with non-biased high-throughput studies at single-cell resolution, and have larger sensitivity in quantifying rare transcripts and mutations19, and become a promising tool to characterize the complicated and rare cells, gene regulatory networks and cell differentiation trajectories20.
In the present study, we downloaded the GSE149655 dataset from the Gene Expression Omnibus (GEO) database, in which the scRNA-seq data including 2 LUAD and 2 healthy samples were retained. After quality control and cell clustering, we found that the Cancer stem cells, Mast cells, Myeloid cells and Fibroblast cells had higher proportion in LUAD samples. Especially, the Cancer stem cells is a major culprit of metastases and relapse of cancer, we calculated the DEGs of this cell among LUAD and healthy samples and analyzed their function enrichment, the Amino sugar and nucleotide sugar metabolism and DNA replication pathways were significantly activated in the cancer stem cells. Further, these cancer stem cells were sub-divided into various cell sub-population and the LGR5 + stem cell is a specific cell cluster that contributed to progression, and its hub genes were identified by using the protein-protein interaction (PPI) network and transcription regulatory network analysis. Delving into the intricate molecular mechanism of LUAD pathogenesis are imperative for the development of more effective therapeutic strategies, our finding may provide several novel insights into the target identification and medicinal development for LUAD intervention.
Materials and methods
ScRNA-seq data acquisition of lung cancer samples
We downloaded the single-cell RNA sequencing (scRNA-seq) data of GSE149655 from GEO database (https://www.ncbi.nlm.nih.gov/geo/), including 2 LUAD and 2 healthy control samples based on an 10x Genomics DNA library and NovaSeq 6000 high-throughput sequencing platform21,22.
ScRNA-seq data preprocessing
The scRNA-seq analysis was performed by using R packages, including the “Seurat” and “harmony”23. First, the Read10X function was used to read the raw matrix of scRNA-seq data with three filtering measures for high-quality data, cells that expressed 200 ~ 6000 genes, and < 10% mitochondrial genes, and > 1000 Unique Molecular Identifiers (UMI) were retained, and these genes were contained in at least three cells. Second, we used the SCTransform function to normalize the filtered data, the RunPCA function was used to perform the Principal Component Analysis (PCA, setting dims = 1:20) to reduce the dimension based on top 2000 genes. Then, the “harmony” R package was used to remove the batch effect between samples, and the top 20 principal components was used for Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction. The FindNeighbors and FindClusters functions was used to perform the cell clustering analysis (distance = Euclidean, resolution = 0.1)23,24,25. Finally, the gene markers from the CellMarker2.0 database was used for the cell type annotation, and the same single cell clustering was performed for the subtype of cancer stem cell (resolution = 0.3).
Gene set enrichment analysis (GSEA)
We used the FindAllMarkers function to calculate the differentially expressed genes (DEGs) of each cell clusters among various samples by using the Wilcoxon-Mann-Whitney tests26. Then, the gseGO and gseKEGG function in the package of “ClusterProlifer” was used to visualize the enriched pathways of these genes in biological process (BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG)26,27,28.
Protein-protein interaction (PPI) network for hub genes screening
STRING database (https://cn.string-db.org/) is a useful tool to explore the functional interaction relationship between all known and predicted proteins29. In this study, we constructed a LGR5 + stem cell associated PPI network based on the DEGs in LGR5 + stem cell and the criterion of minimum interaction confidence > 0.150. Several hub genes with highly interaction intensity in local PPI may represent molecular complex and play specific biological function, these genes were identified for further analysis. The CytoHubba plug-in the Cytoscape software was used to calculate the score of DEGs in LGR5 + stem cell and obtained the top10 identified DEGs in the PPI network as hub genes by using the Maximal Clique Centrality (MCC) algorithm. Further, the GeneMANIA online website was used to screen the similar functional protein that associated with the hub genes and constructed a PPI network.
Association of the hub genes with prognosis and drug sensitivity in patients with LUAD
The RNA-seq expression data in transcripts per million (TPM) format from GDC TCGA-LUAD and patients’ clinical follow-up information were downloaded via UCSC Xena (https://xenabrowser.net/datapages/), and samples with complete overall survival (OS) were retained, and a total of 500 samples were finally included. Subsequently, 10 hub genes were cox analyzed using the survival package30, and Kaplan-Meier (KM) curves were plotted using the survminer package31 to compare survival differences between different gene expression groups. In addition, we used the DepMap (https://depmap.org/portal/) website and set the filtering criteria to Lineage=’lung’, Lineage Sub-subtype=’Lung Adenocarcinoma’ to download the expression matrix of LUAD cell line samples and IC50 values of drugs. Next, we screened for drugs that had IC50 values in at least 75% of the samples in the LUAD cell line and used the spearman method to calculate the correlation between the expression of the 10 hub genes and the IC50 of the drugs.
Construction of transcription regulatory network
We uploaded these hub genes into the miRDB database to obtain the microRNA (miRNA) of hub genes and visualized the mRNA-miRNA regulatory network by using the Cytoscape software. Then, we used the ChIPBase database to search the transcription factor (TFs) of hub genes and analyzed its potential regulating effect, the mRNA-TF regulatory network was visualized by using the Cytoscape software32.
Human tissue sample collection, cell culture, and qPCR test
Tissues from human specimens of LUAD and adjacent non-tumor tissues (n = 6 for each) were sourced from individuals diagnosed with LUAD who were admitted to our hospital. Every enrolled patient satisfied these criteria: (i) no prior chemotherapy or radiotherapy treatment; (ii) no other cancer diagnoses, autoimmune conditions, or infectious diseases, among others. All samples were acquired through biopsy or surgical removal and were washed in saline prior to storage in liquid nitrogen for subsequent expression analysis. The ethic committee of the Third Affiliated Hospital of Guangzhou Medical University has reviewed and approved the conduction of our current study (ethical approval code: S2025-068), and every volunteer has provided the written informed consent to participate in this study. The study strictly followed the ethical guidelines outlined in the Declaration of Helsinki.
Human normal lung epithelial cell line BEAS-2B and lung adenocarcinoma cell line A549 were obtained from the American Type Culture Collection (ATCC, USA) and cultured in Dulbecco’s Modified Eagle’s Medium (Gibco) supplemented with 10% fetal bovine serum, 2 mM L-glutamine and 0.1% (v/v) penicillin-streptomycin (Gibco) in in a humidified environment with 5% CO2 at 37 °C33. After 24 h, the cells were harvested for total RNA extraction by using the TRizol Reagent (Invitrogen), and the The ReverTra Qpcr RT Master Mix kit (TOYOBO) was used for the cDNA synthesis according to its instructions. Then, we performed the qPCR for target genes detection by using the SYBR Green PCR Master Mix (Applied Biosystems) on LightCycler 96 (Roche) based on manufacturer’s instructions. The specific primers are listed in (Table 1).
Western blot analysis
To assess the expression of the CXCL3 protein, a western blot analysis was conducted. In brief, BEAS-2B and A549 cell lines were lysed using RIPA buffer that contained protease inhibitors. Following this, proteins were separated on 15% SDS-PAGE gels, then transferred to PVDF membranes (Beyotime, Shanghai, China) and subsequently blocked. The membranes were incubated overnight at 4 °C with primary antibodies: anti-CXCL3 (1:100, Abcam, ab10064) and anti-GAPDH (1:5000, Millipore, Billerica, MA). Afterward, HRP-conjugated secondary antibodies were applied for 1 h at room temperature, and the bands were detected using the ECL system (Amersham Biosciences, Inc). ImageJ software was employed to evaluate the grayscale values of the developed strips and to perform the calculations.
Immunohistochemistry (IHC) assay
The staining of LUAD tissue samples using immunohistochemistry was performed in accordance with established protocols. Initially, the LUAD specimens were fixed in 4% paraformaldehyde (Servicebio, Wuhan, China), after which they were embedded in paraffin and sectioned to a thickness of 4 μm. Tissue sections underwent deparaffinization and rehydration, followed by a treatment with 4% hydrogen peroxide for 10 min in a dark environment at room temperature. Subsequently, they were autoclaved at 95 °C for 10 min in a 0.05 mol/L Tris-EDTA solution (pH 9.0). After this, the sections were incubated at 37 °C for 2 h with a rabbit polyclonal antibody targeting human CXCL3 (1:100 dilution, Bioss, China). Following three washes with PBS, the sections received a treatment with a secondary antibody (goat anti-rabbit at a dilution of 1:1,000; Beijing ZhongShan Biotechnology) for 45 min at 37 °C. Finally, they were counterstained using hematoxylin and covered with a glass cover slip. The processed samples were examined under an Olympus microscope.
Wound healing and trans-well assay
We purchased the si-CXCL3 regent (Thermo Fisher Scientific) to obtain the silenced A549 cells, the Lipo-fectamine 3000 (Invitrogen) was used for cell transfection. The wound healing was performed for the cell migration, a density of 4 × 106 cells were seeded into 6-well plates (Corning) until confluency, then a scratch was created by a pipette tip, after overnight, the 4% paraformaldehyde (Servicebio) was used for cell fixation (15 min) and 0.1% crystal violet (Servicebio) was used for staining (15 min), the wound closure was detected by an inverted microscope (Leica)33. For invasion assays, the trans-well assay was performed, a total of 4 × 104 cells were incubated into the upper chamber well of 24-well plates (Corning, 8-mmpore) with 200 µLserum-free DMEM and the lower chamber was added into 800 µL of DMEM with 20% FBS (Thermo Fisher Scientific), after 48 h, the migrating cells on lower chamber were fixed with 4% paraformaldehyde (15 min) and stained with 0.1% crystal violet (15 min) for imaging by inverted microscope (Leica)33,34,35.
Cell proliferation assay
A549 cells that had been transfected were grown for 48 h until they reached the logarithmic growth phase before being moved to 96-well plates. Cell proliferation was assessed using the EdU Cell Proliferation Assay Kit (RiboBio, Guangzhou, China). In accordance with the provided protocol, the cells underwent staining, were examined, and captured images under a fluorescence microscope (Nikon). The quantification of EdU-positive cells was conducted using ImageJ software.
Flow cytometry
A549 cells that were transfected with either CXCL3-specific siRNA or si-NC were gathered, washed with PBS, and subsequently resuspended in 195 µL of annexin-V FITC (BD Biosciences, Franklin Lakes, NJ, USA) mixed with 5 µL of propidium iodide (PI), following the protocol provided. After a 10-minute incubation of the cells in darkness at room temperature, flow cytometry was employed for the analysis. Each experiment was conducted with three technical replicates.
Statistical analysis
All statistical analysis was performed by using R software (version 4.3.2). The data are presented as mean ± standard deviation (SD). The experimental results were assessed utilizing GraphPad Prism 8.0 software. Comparisons among multiple groups were conducted via a Student’s t-test or One-way analysis of variance (ANOVA). p values < 0.05 were considered significant.
Results
Single-cell transcriptome atlas in LUAD samples
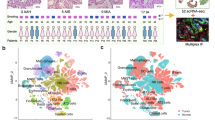
To elucidate the cellular diversity in lung cancer samples, the scRNA-seq analysis was performed on tumor and normal samples from 4 patients. After the initial quality control and cell clustering, a total of 9 mainly cell clusters from 41,322 cells were obtained (Fig. 1A, B), including the cancer stem cell, ciliated cells, dendritic cells, endothelial cells, epithelial cells, fibroblast cells, mast cells, myeloid cells and T cells. Results of initial quality control assessment as described previously showed that the UMI in each cell are more than 200 (Fig. 1C), the gene numbers in each cell are 200 ~ 6000 (Fig. 1D), and the proportion of mitochondrial genes are < 10% (Fig. 1E). The definition of these cell lineages was as follows: (1) the endothelial cells characterized with CLDN5 and FCN3, (2) the epithelial cells expressing SCEL and TNNC1, (3) fibroblast cells highly expressing DCN and LUM, (4) dendritic cells with high expression of LAMP3 and MFSD2A, (5) ciliated cells specifically express the markers of MORN5 and ENKUR, (6) T cells expressing CD3E and CD3D, (7) cancer stem cell expressing CEACAM5 and ANPEP, (8) mast cells expressing MS4A2 and TPSAB1, (9) myeloid cells expressing MS4A4A and SPI1 (Fig. 1F). The expression of marker genes in each cell are showed in (Fig. 2A), the proportions of cellular clusters existed huge diverse among cancer and normal samples, in which the Cancer stem cells, T cells, Myeloid cells, Ciliated cell, Fibroblast cells and Mast cells had higher proportion in cancer samples (Fig. 2B, C), suggesting these cells may be a major contributor of tumor heterogeneity.
Single cell atlas in lung cancer and healthy control tissue. (A) UMAP plot of cell dimension reduction clustering in different samples. (B) UMAP plot of 9 mainly cell clusters. (C) UMI distribution of each cell in various samples. (D) The distribution of the number of genes in each cell. (E) The distribution of the proportion of mitochondrial genes in each cell. (F) Bubble plot of marker gene expression levels for each cell type.
The pathway difference of cancer stem cell in LUAD and healthy samples
Cancer stem cells (CSCs) represent a major culprit of metastases and relapse of cancer, we explored the function difference of CSC in cancer and healthy samples for tumorigenicity and progression. The function enrichment analysis of DEGs showed that the Amino sugar and nucleotide sugar metabolism, DNA replication, primary immunodeficiency and biosynthesis of nucleotide sugars pathways were significantly enriched in the cancer samples of CSC, while the CSC in the healthy samples showed heightened activities of NF-kappa B signaling pathway and IL-17 signaling pathway (Fig. 3A), indicating that the cancer samples of CSC were associated to tumor growth and proliferation. The genes of CSF3, IL6 and CCl2 in the IL-17 signaling pathway were significantly overexpressed in the healthy samples (Fig. 3B). In addition, due to the DNA replication and Amino sugar and nucleotide sugar metabolism play an important role in the cancer progression, these genes include the MCM3, RNASEH1, RFC2, PRIM2, RFC5, RPA2 (Fig. 3C), and the CHIT1, AMDHD2, FPGT, CHIA, NANS, CYB5R1, HEXB and UXS1 (Fig. 3D) were significantly upregulated in the cancer samples, their difference of expression levels are showed in (Fig. 3E,F), implying that the DNA replication and Amino sugar and nucleotide sugar metabolism are closely associated with the carcinogenesis of LUAD.
Functional enrichment of Cancer stem cell in lung cancer and healthy control samples. (A) The enrichment of Cancer stem cell in lung cancer and healthy control group. (B) Gene set enriched in IL-17 signaling pathway. (C) Gene set enriched in DNA replication pathway. (D) Gene set enriched in Amino sugar and nucleotides sugar metabolism pathway. (E) The bubble plot of gene expression of DNA replication pathway among various samples. (F) The bubble plot of gene expression of Amino sugar and nucleotides sugar metabolism pathway among various samples.
Highly infiltration of LGR5 + stem cell is a key type of CSC contributing to LUAD progression
The identified CSCs had huge carcinogenic potential, we further investigated the heterogeneity of CSC in the carcinogenesis of LUAD by using the cell clustering analysis. The CSCs were sub-divided into 3 mainly cell clusters (Fig. 4A,B), including the Basal cell, LGR5 + stem cell and Paneth cells. Their marker genes are showed in the (Fig. 4C), in which the Basal cell was characterized with the DSC3, PKP1 and CXCL3 markers, the LGR5 + stem cell with the highly expression of ARSE and GJB1 and the Paneth cells expressing CXCL3 and PLCG2 (Fig. 4D). In addition, we found that the LGR5 + stem cells had higher proportion in the cancer samples (Fig. 4E), implying this type of CSC involved in the LUAD progression.
Cancer stem cell landscape of lung cancer and healthy control tissues. (A) UMAP plot of subdivision of Cancer stem cell cells in lung cancer and healthy control tissue. (B) UMAP plot of Cancer stem cell subtype. (C) The violin plot of expression level of marker gene in Cancer stem cell. (D) Bubble plot of marker gene expression levels in each Cancer stem cell subpopulation. (E) Proportion of each Cancer stem cell subpopulation in lung cancer and healthy controls, respectively.
In vitro cellular assays to validate potential biological functions of key marker genes
Subsequently, we detected the expression of marker genes of three CSCs in BEAS-2B and A549 cells, including the DSC3, PKP1, ARSE, GJB1, CXCL3 and PLCG2, and found that excepting for the DSC3, others five genes were significantly up-regulated in the A549 cells (p < 0.05, Fig. 5A), suggesting these genes as indictors of cancer cells were effective. As shown in Fig. 5B, we observed the expression of CXCL3 in six LUAD and para-cancerous tissue samples, and the protein expression level of CXCL3 was also significantly elevated in LUAD cell lines (p < 0.001, Fig. 5C). After silencing of CXCL3 gene, the wound closures of A549 cells were significantly decreased in the wound healing assay (p < 0.05, Fig. 5D), meanwhile, the numbers of A549 cells were significantly declined in the trans-well assay (p < 0.05, Fig. 5E). In addition, we found, based on the results of EdU assay and flow cytometry, that CXCL3 silencing also significantly reduced the proliferative capacity of A549 cells (p < 0.001, Fig. 5F), and increased the level of apoptosis in this cell line (p < 0.001, Fig. 5G), implying that the CXCL3 played a crucial role in mediating the development of cancer cells.
qPCR, wound healing and trans-well assay. (A) qPCR for the detection of ARSE, CXCL3, DSC3, GJB1, PKP1 and PLCG2. (B) IHC assays showed CXCL3 expression in LUAD and para-cancerous tissue samples (n = 6 for each), respectively. (C) Western blotting-based assay to analyze the difference in protein level expression of CXCL3 in BEAS-2B and A549 cells. (D) Wound healing assay for cell migration. (E) Trans-well assay for cell invasion. (F) EdU-based assay to validate the effect of CXCL3 silencing on the proliferative capacity of A549 cells. (G) Flow cytometry was employed to verify the impact of CXCL3 silencing on the apoptotic ability of A549 cells. All procedures were repeated three times as independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p<0.0001.
Identifying 10 hub genes that supported the LGR5 + stem cell function
A PPI network between proteins encoded by 35 DEGs in LGR5 + stem cell was constructed through the STRING database, and comprised 35 nodes and 92 edges (Fig. 6A), in which 21 nodes exhibited obvious interaction relationship (Fig. 6B). These genes, such as CTSH, HLA-DRB1, HLA-DRA, HLA-DPB1, HLA-DQA1, CD74, HLA-DPA1, HLA-DRB5, HLA-DMA, HLA-C had higher interaction intensity in local PPI (Fig. 6A, B), indicating they may be key factors in mediating cancer stem cell activation. Then, we calculated the maximal clique centrality score of these DEGs and ranked the score, the Top10 DEGs was used to constructed a new PPI network (Fig. 6C), the color from red to yellow represent the score from the high to low and these 10 genes including the CD74, HLA-DRB1, HLA-DPB1, HLA-DRB5, HLA-DPA1, HLA-DMA, HLA-DQA1, HLA-DRA, CTSH and HLA-C were defined as hub genes. We constructed a PPI network between hub genes and its predicted target genes, the different colors lines indicated the co-expression and shared protein domains information of proteins, and 20 target genes with similar functions of hub genes were identified (Fig. 6D). Notably, we observed that nine of these ten hub genes, except for HLA-C, were protective factors (hazard ratio < 1) and all were significantly associated with the prognosis of LUAD patients (Figure S1, p < 0.05). Drug sensitivity analysis showed that HLA-DMA, HLA-DQA1, HLA-DRA, and CTSH genes in LUAD were significantly correlated with IC50 values of some drugs (e.g., AST-1306, CD532, and AR-12, etc.) (Figure S2, p < 0.05). This suggests that these genes may play a key role in the mechanism of chemosensitivity or resistance in LUAD.
Protein-protein interaction network. (A) Protein-protein interaction network of LGR5 + stem cell of DEGs. (B) Cytoscape visualization network of LGR5 + stem cell differential genes in 21 Cancer stem cells. (C) Protein-protein interaction networks of top10 hub genes. (D) Interaction networks of functionally similar genes with hub genes.
Constructing a gene regulatory network
First, we obtained the targeted miRNA of hub genes and constructed a mRNA-miRNA regulatory network (Fig. 7A), in which 331 miRNAs associated with 10 hub genes were identified, and the HLA-DPB1 play a central role that regulated by more miRNAs. In addition, the mRNA-TFs regulatory network analysis showed that the 35 TFs targeted the CD74, CTSH and HLA-DRB5 were identified (Fig. 7B). These evidences suggested that these hub genes including the HLA-DPB1, CD74, CTSH and HLA-DRB5 may be potential target of CSC in LUAD treatment.
Discussion
Worldwide, the LUAD is a major malignancy subtype (40%) of lung cancer that contributed to approximately 2.1 million annual deaths and 20% global cancer-related mortality6. Increasing evidence has demonstrated that the CSCs are a specific subpopulation of tumor cells that can drive the initiation, establishment and relapses of tumor36, and often associated with the heterogeneous, aggressive and therapy-resistant tumors37, causing the highest mortality in the malignant neoplasms, such as lung, liver, colorectal cancers and stomach cancer38. Previous studies have shown that CSCs promote tumor progression and affect patient prognosis by regulating stemness-related genes in LUAD. Based on mRNA stemness index (mRNAsi) analysis, Zhang et al. showed that the expression level of CSCs-related genes was elevated with the pathological stage in LUAD and correlated with poor prognosis39. In addition, S-palmitoylation, as a post-translational modification of proteins, plays an important role in the regulation of CSCs, and ZDHHC5 modifies INCENP through S-palmitoylation to enhance its function in LUAD stem cells and is closely associated with advanced LUAD40. These studies further revealed the important role of CSCs in the progression of LUAD.
Compared to hematological malignancies, solid tumors (such as LUAD) exhibit a more complex tumor microenvironment (TME) and spatial heterogeneity41. Single-cell sequencing in solid tumors not only enables the characterization of cancer stem cells CSCs but also reveals their interactions with immune and stromal cells within the TME. In contrast, in hematological malignancies (such as acute myeloid leukemia), CSCs are more dependent on cell cycle and mitotic pathway regulation, and mRNAsi analysis is more susceptible to variations in tumor differentiation stages42. This distinction highlights the value of single-cell sequencing in studying different tumor types and suggests that future research should incorporate spatial transcriptomics to achieve a more comprehensive understanding of CSCs in solid tumors. In this study, we downloaded the scRNA-seq data of LUAD from the GEO database and identified 9 mainly cell clusters including the CSCs that had highly proportion in the LUAD samples and supported the cancer progression with the significantly activation of amino sugar and nucleotide sugar metabolism and DNA replication pathways. Furthermore, these cells were sub-divided into various sub-types, such as the Basal cell, LGR5 + stem cells and Paneth cells, among of them the LGR5 + stem cells are obvious enriched in the LUAD samples, and several hub factors mediated the unique transcriptional state were identified.
Tumor initiation can be driven by the transformed tissue resident stem cells or transformed differentiated cells43, this transformation can occur in tissue regeneration or be initiated as a response to infections, radiation, toxins or metabolic influences causing mutations44. Further, the tumor suppressors are inactivated and oncogenes are overexpressed causing the uncontrolled growth of the cells, thus these cells de-differentiate and acquire stemness cell features43. Due to tissue resident stem cells or progeny performed a specific set of genomic procedure allowing uncontrolled, niche-independent proliferation, these transformed stem cells and their progeny only need few genomic changes45, such as more than 10% gastric cancers originate from the tissue resident stem cells. The increasing glycolysis is one of the hallmarks of cancer at oxygen presence and can provide enough energy and multiple precursors supporting rapid cell division46, Shimizu et al., revealed the IL-8 induced O-GlcNAcylation increasing is necessary for the generation and maintenance CSCs in lung cancer through upregulating the GLUT3 and GFPT1 expression47, the amino sugar and nucleotide sugar metabolism pathways were significantly activated in CSCs in our study, indicating this pathway may be also mediated some key process providing a protective effect and pluripotent state for cancer stem cells. Basal cells located in main bronchi and trachea can self-renew and form heterogeneous spheres48, and cause epithelial hypoplasia and basal cell hyperplasia, eventually resulting in dysplasia or squamous cell metaplasia, which is a precursors of squamous cell lung carcinomas49. Previous studies reported that the Lgr5 stem cells contributed to the self-renewing homeostasis of small intestinal crypts based on the Paneth cell providing an essential niche signal50, meanwhile we revealed that the Lgr5 stem cells are the mainly contributor to tumor proliferation and progression in LUAD. In addition, the markers of three CSCs were also overexpressed in the A549 cells and the silencing of CXCL3 affected the migration and invasion of tumor cells, these genes can be developed to the potential therapeutic target.
Among these identified hub genes, including the HLA-DPB1, CD74, CTSH and HLA-DRB5, in which Human leukocyte antigens (HLA)-DPB1 encode the Major Histocompatibility Complex, Class II, DP Beta 1 that are responsible for the immune presentation of extracellular antigens51, a study showed that the HLA-DPB2/HLA-DPB1 axis can promote high immune infiltration via binding with has-miR-370-3p in breast cancer, supporting better prognosis52, and the interaction of HLA-DPB1 and HLA-DRB5 can accelerate neuroinflammation, such as activation of proinflammatory intracellular signaling, release of cytokines/interleukins (TNF-α, TGF-β and IL-1β) in Alzheimer’s disease53, but the reports of HLA-DPB1 in LUAD are lacking. CD74 encode a transmembrane glycoprotein of MHC class II invariant chain, and is increased in many tissue injury disorders, including the Alzheimer disease, heart ischemia-reperfusion injury and malignant cells as therapeutic target to malignancy54. Cathepsin H (CTSH) was reported to overexpress in the human hepatoma cells and increase tumor invasion through the regulating of thyroid hormone receptors55, another study also demonstrated that the CTSH influences lung cancer risk56. These evidences suggested these hub genes mediated several crucial biological processes supporting the function of cancer stem cells and may be as potential therapeutic targets. However, there has several limitations in this study. Firstly, the retrospective data of this study from public databases, more multi-center and prospective cohorts and trails of LUAD are required to eliminate bias. Secondly, the LGR5 + stem cells acted as a specific CSC that promoted LUAD progression via some tumor-related signaling pathways, the specific mechanism and some validations on LUAD tissues needed to be performed.
Conclusion
In summary, we performed a scRNA-seq analysis and characterized the cancer stem cell in LUAD. The activated Amino sugar and nucleotide sugar metabolism and DNA replication pathways are crucial to maintain the pluripotent state of cancer stem cell, the LGR5 + stem cells may be interacted with the Paneth cells supporting the self-renewal of cancer stem cell and some hub genes that play an important role for the function of cancer stem cells were identified in our study. Our findings are expected to provide novel insight to develop the LUAD treatment strategies.
Data availability
The datasets generated and/or analyzed during the current study are available in the [GSE149655] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149655].
Abbreviations
- LUAD:
-
Lung adenocarcinoma
- DEGs:
-
Differentially expressed genes
- CSC:
-
Cancer stem cells
- TKIs:
-
Tyrosine kinase inhibitors
- LUSC:
-
Lung squamous carcinoma
- VEGF:
-
Vascular endothelial growth factor
- scRNA-seq:
-
Single-cell RNA sequencing
- GEO:
-
Gene Expression Omnibus
- PPI:
-
Protein-protein interaction
- UMI:
-
Unique Molecular Identifiers
- PCA:
-
Principal Component Analysis
- UMAP:
-
Uniform Manifold Approximation and Projection
- BP:
-
Biological process
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- TFs:
-
Tanscription factor
References
Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Ullah, A. et al., Sanguinarine attenuates lung cancer progression via oxidative stress-induced cell apoptosis. Curr. Mol. Pharmacol. 17 (2024).
Nooreldeen, R. & Bach, H. Current and future development in lung Cancer diagnosis. Int. J. Mol. Sci. 22 (2021).
Qian, X-J., Wang, J-W., Liu, J-B. & Yu, X. The mediating role of miR-451/ETV4/MMP13 signaling Axis on epithelialmesenchymal transition in promoting Non-small cell lung Cancer progression. Curr. Mol. Pharmacol. 17 (2023).
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74, 12–49 (2024).
Xu, L. et al., MiR-21/Sonic Hedgehog (SHH)/PI3K/AKT pathway is associated with NSCLC of primary EGFR-TKI resistance. 24, 579–590 (2022).
Denisenko, T. V., Budkevich, I. N. & Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell. Death Dis. 9, 117 (2018).
Lin, J. J., Riely, G. J., Shaw, A. T. & Targeting, A. L. K. Precision medicine takes on drug resistance. Cancer Discov. 7, 137–155 (2017).
Kong, Q. et al. Analysis of the susceptibility of lung cancer patients to SARS-CoV-2 infection. Mol. Cancer. 19, 80 (2020).
Muliaditan, T. et al. Macrophages are exploited from an innate wound healing response to facilitate cancer metastasis. Nat. Commun. 9, 2951 (2018).
Bhatt, V. R., Kedia, S., Kessinger, A. & Ganti, A. K. Brain metastasis in patients with non-small-cell lung cancer and epidermal growth factor receptor mutations. J. Clin. Oncol. 31, 3162–3164 (2013).
Lei, Y. et al. Applications of single-cell sequencing in cancer research: progress and perspectives. J. Hematol. Oncol. 14, 91 (2021).
Zhang, L. et al. Integrated single-cell RNA sequencing analysis reveals distinct cellular and transcriptional modules associated with survival in lung cancer. Signal. Transduct. Target. Ther. 7, 9 (2022).
Bischoff, P. et al. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene 40, 6748–6758 (2021).
Zhang, J. et al. Single-cell analysis of multiple cancer types reveals differences in endothelial cells between tumors and normal tissues. Comput. Struct. Biotechnol. J. 21, 665–676 (2023).
Liu, Y. et al. Single-cell transcriptome analysis demonstrates inter-patient and intra-tumor heterogeneity in primary and metastatic lung adenocarcinoma. Aging (Albany NY). 12, 21559–21581 (2020).
Fan, M., Xia, P., Clarke, R., Wang, Y. & Li, L. Radiogenomic signatures reveal multiscale intratumour heterogeneity associated with biological functions and survival in breast cancer. Nat. Commun. 11, 4861 (2020).
Cook, D. J., Kallus, J., Jörnsten, R. & Nielsen, J. Molecular natural history of breast cancer: leveraging transcriptomics to predict breast cancer progression and aggressiveness. Cancer Med. 9, 3551–3562 (2020).
Kim, N. et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11, 2285 (2020).
Duma, N., Santana-Davila, R. & Molina. Non-Small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc. 94, 1623–1640 (2019).
Dost, A. F. M. et al. Organoids model transcriptional hallmarks of oncogenic KRAS activation in lung epithelial progenitor cells. Cell. Stem Cell. 27, 663–678e668 (2020).
Xu, X., Huang, Y., Han, X. & Single-Nucleus, R. N. A. Sequencing reveals cardiac macrophage landscape in hypoplastic left heart syndrome. Congenit. Heart. Dis. 19, 233–246 (2024).
Zulibiya, A. et al. Single-Cell RNA Sequencing reveals potential for endothelial-to-mesenchymal transition in tetralogy of fallot. 18, 611–625 (2023).
Zulibiya, A. et al. Single-Cell RNA sequencing reveals potential for Endothelial-to-Mesenchymal transition in tetralogy of fallot. Congenit. Heart. Dis. 18, 611–625 (2023).
Cao, H. et al. Identification of prognostic molecular subtypes and model based on CD8 + T cells for lung adenocarcinoma. Biocell 48, 473–490 (2024).
Tan, Z. et al. Comprehensive analysis of scRNA-Seq and bulk RNA-Seq reveals dynamic changes in the tumor immune microenvironment of bladder cancer and establishes a prognostic model. J. Transl Med. 21, 223 (2023).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017).
Therneau, T. M. & Lumley, T. Package ‘survival’ R Top. Doc. 128, 28–33 (2015).
Kassambara, A., Kosinski, M., Biecek, P. & Fabian, S. Package ‘survminer’. Drawing Survival Curves using ‘ggplot2’(R package version 03 1) (2017).
Huang, J. et al. ChIPBase v3.0: the encyclopedia of transcriptional regulations of non-coding RNAs and protein-coding genes. Nucleic Acids Res. 51, D46–d56 (2023).
Liang, W. et al. KRT17 promotes T-lymphocyte infiltration through the YTHDF2-CXCL10 Axis in colorectal Cancer. Cancer Immunol. Res. 11, 875–894 (2023).
Wang, J., Cai, Y., Sheng, Z. & Dong, Z. EGFR inhibitor CL-387785 suppresses the progression of lung adenocarcinoma. Curr. Mol. Pharmacol. 16, 211–216 (2023).
Zhang, L. et al. miR-30a-5p/PHTF2 axis regulates the tumorigenesis and metastasis of lung adenocarcinoma. Biocell 48, 581–590 (2024).
Capp, J. P. Cancer stem cells: From historical roots to a new perspective. J. Oncol. 2019, 5189232 (2019).
Kuşoğlu, A. & Biray Avcı, Ç. Cancer stem cells: A brief review of the current status. Gene 681, 80–85 (2019).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Zhang, Y. et al. mRNAsi index: machine learning in mining lung adenocarcinoma stem cell biomarkers. Genes (Basel) 11 (2020).
Zhang, Y. et al. Potential role of S-Palmitoylation in Cancer stem cells of lung adenocarcinoma. Front. Cell. Dev. Biol. 9, 734897 (2021).
Wang, C. et al. The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing. Signal. Transduct. Target. Ther. 7, 289 (2022).
Zhang, Y. et al. Identification of biomarkers for acute leukemia via machine learning-based stemness index. Gene 804, 145903 (2021).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Basu, A. K. DNA damage, mutagenesis and Cancer. Int. J. Mol. Sci. 19 (2018).
Batlle, E. & Clevers, H. Cancer stem cells revisited. Nat. Med. 23, 1124–1134 (2017).
Abbaszadeh, Z., Çeşmeli, S. & Biray Avcı, Ç. Crucial players in glycolysis: Cancer progress. Gene 726, 144158 (2020).
Shimizu, M. & Tanaka, N. IL-8-induced O-GlcNAc modification via GLUT3 and GFAT regulates cancer stem cell-like properties in colon and lung cancer cells. Oncogene 38, 1520–1533 (2019).
Hardavella, G., George, R. & Sethi, T. Lung cancer stem cells-characteristics, phenotype. Transl Lung Cancer Res. 5, 272–279 (2016).
Fukui, T. et al. Lung adenocarcinoma subtypes based on expression of human airway basal cell genes. Eur. Respir J. 42, 1332–1344 (2013).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Cheng, L., Guo, Y., Zhan, S. & Xia, P. Association between HLA-DP gene polymorphisms and cervical Cancer risk: A Meta-Analysis. Biomed. Res. Int. 2018, 7301595 (2018).
Lyu, L. et al. Overexpressed pseudogene HLA-DPB2 promotes tumor immune infiltrates by regulating HLA-DPB1 and indicates a better prognosis in breast Cancer. Front. Oncol. 10, 1245 (2020).
Hampel, H. et al. A path toward precision medicine for neuroinflammatory mechanisms in Alzheimer’s disease. Front. Immunol. 11, 456 (2020).
Otterstrom, C. et al. CD74: a new prognostic factor for patients with malignant pleural mesothelioma. Br. J. Cancer. 110, 2040–2046 (2014).
Wu, S. M. et al. Cathepsin H regulated by the thyroid hormone receptors associate with tumor invasion in human hepatoma cells. Oncogene 30, 2057–2069 (2011).
Luyapan, J. et al. Candidate pathway analysis of surfactant proteins identifies CTSH and SFTA2 that influences lung cancer risk. Hum. Mol. Genet. 32, 2842–2855 (2023).
Acknowledgements
None.
Funding
This study was supported by grants from Natural Science Foundation of Guangdong Province, China (2023A1515011600).
Author information
Authors and Affiliations
Contributions
All authors contributed to this present work: [GW] & [SMW] designed the study, [SQN] & [SQM] acquired the data. [GW] & [SMW] drafted the manuscript, [GW], [SQN], [SMW] and [SQM]revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The ethic committee of the Third Affiliated Hospital of Guangzhou Medical University has reviewed and approved the conduction of our current study (ethical approval code: S2025-068), and every volunteer has provided the written informed consent to participate in this study. The study strictly followed the ethical guidelines outlined in the Declaration of Helsinki.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wen, G., Niu, S., Mei, S. et al. The cancer stem cells characteristics analysis of LGR5 + cells that influence lung cancer risk by using single cell RNA-seq analysis. Sci Rep 15, 16085 (2025). https://doi.org/10.1038/s41598-025-00585-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-00585-3