Abstract
Advancements in large-scale analysis of metabolites in human peripheral blood samples revealed the links between metabolite concentrations and genetic variations. This field is known as metabolome-genome-wide association study (MGWAS). Although MGWAS is a powerful tool, it has some limitations, particularly in terms of the number of metabolites that can be measured. Whether the observed associations are directly due to genetic variation or indirectly due to changes in unmeasured metabolites is unclear. To address this, we used simulations of metabolic pathway models to investigate the influence of genetic variants on metabolite concentrations and enhance the interpretation of MGWAS results. By systematically adjusting the enzyme reaction rates to simulate genetic variants, we observed changes in the metabolite levels. Our simulations accurately represented most of the variant-metabolite pairs identified by MGWAS with significant p-values, thereby demonstrating the potential of our approach. Furthermore, our simulations revealed additional marked fluctuations in metabolite levels that the MGWAS did not detect, suggesting that some variant-metabolite pairs might become more significant with larger sample sizes. We also categorized the enzymes into three types based on their impact on metabolite concentrations, highlighting enzymes with minimal impact. This indicated that genetic variations in these enzymes may have limited biological significance. Our study not only validates key MGWAS findings, but also provides a systematic framework for understanding enzyme-metabolite relationships. This approach offers valuable insights for future experimental studies and potential therapeutic interventions.
Similar content being viewed by others
Introduction
With the advent of high-throughput technologies and bioinformatics, researchers have integrated metabolomics with genome-wide association studies (GWAS), resulting in the MGWAS field1,2,3,4,5,6,7,8. The synthesis of data from MGWAS enables a multi-layered analysis of the genotype–phenotype relationship, revealing how single nucleotide variants throughout the genome can influence metabolic traits. Understanding the complex genetic networks that govern metabolic processes is essential to interpret how genetic predispositions can lead to fluctuations in metabolite levels that may indicate health or disease states. MGWAS represents the confluence of genetics and metabolomics, offering a powerful approach to reveal the genetic factors that shape the metabolome and their broader implications for human health and disease.
MGWAS became the standard for exploring the relationship between genetic variants and metabolite levels in biological samples, such as blood plasma. Despite their usefulness, these studies have inherent limitations9. The correlations they yielded were mostly statistical, indicating that they did not provide experimental biological validation. Consequently, these associations often raise questions about causality. This can lead to false-positive findings, where an association appears significant by chance and is not due to an objective biological relationship. Another limitation is that the small sample sizes used in these studies may have missed rare genetic variants, leading to false negatives and missing true associations. Experimental confirmation of the vast array of variant-metabolite combinations identified through MGWAS is daunting, presenting considerable challenges in interpreting and validating the results.
To overcome these limitations, we proposed the application of metabolic pathway model simulations for the analysis of MGWAS results10,11. These in silico experiments offer a comprehensive approach to investigate all possible variant-metabolite combinations, probing deeper into the metabolic network than is typically feasible in MGWAS. The essential advantage of this comprehensive approach is its ability to discern true associations from false positives by validating each variant-metabolite pair using simulated perturbations. By adjusting the enzyme reaction rates within the model to reflect specific genetic variations, the simulations could predict the resulting changes in metabolite concentrations. This thorough analysis not only supports the identification of true positives but also aids in confirming true negatives—cases where no actual association exists between a variant and metabolite, which MGWAS may incorrectly suggest as a potential association. By examining the entire landscape of possibilities, simulations can help clarify whether an observed association is a genuine biological phenomenon or an artifact of a statistical process. Furthermore, our simulation approach can reveal considerable fluctuations in metabolite concentrations that suggest biological relevance for variant-metabolite pairs that fail to show significance in MGWAS due to limited sample sizes. This comprehensive capability allows researchers to prioritize genetic variants that may warrant further experimental investigation, optimize resource allocation, and direct attention to the most promising candidates for experimental validation.
With these advantages in mind, we conducted in silico experiments using a metabolic pathway model to explore the effects of altered enzyme reaction rates on metabolite concentrations. By comparing these results with those from MGWAS, we sought to enhance the accuracy of metabolomic studies, reduce the likelihood of false positives, confirm true negatives, and provide a robust foundation for interpreting the complex interplay between genetics and metabolism.
Materials and methods
Metabolic pathway model
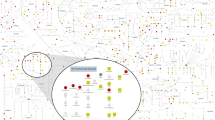
The human liver cell folate cycle model developed by Nijhout et al.12, acquired from BioModels13,14, was used. The model was structured using differential equations, with initial metabolite concentrations and enzyme reaction rates derived from experimental data to accurately replicate the normal in vivo environment. Notably, this model did not explicitly incorporate the genetic variants investigated in our study. The model comprised two compartments: cytosol and mitochondria. While simulations were performed considering both compartments, our analysis predominantly focused on metabolites and enzymes within the cytosol (Fig. 1A). Although certain enzymes and metabolites were shared between cytosolic and mitochondrial compartments, they were treated as different entities in each compartment to reflect their separate environments and dynamics. Nevertheless, for molecules, such as sarcosine and dimethylglycine, which are presumed to diffuse freely across compartmental boundaries, a single concentration represents their levels in the cytosol and mitochondria. This model assumes that the total concentration of folate derivatives remains constant. Therefore, the model retained the total amounts of THF, DHF, 10-formyl-THF, 5-methyl-THF, 5,10-methenyl-THF, and 5,10-methylene-THF.
Overview of the model and MGWAS results. (A) Schematic representation of cytosolic part of the folate cycle model (modified from12). Black rectangles represent variable metabolites, and enzymes were labeled to the corresponding reaction. Non-enzymatic reactions were omitted. The red arrow indicated glycine cleavage system, which sub-localized in mitochondria. Enzymes mentioned in MGWAS part were showed with the corresponding gene within parentheses. Based on the enzyme classification in the simulation part, names of enzymes impacting a wide array of metabolites were colored in red, and those of enzymes with a targeted impact were colored in blue. Metabolites whose total amount is kept constant as a conserved quantity are underlined. (B) Manhattan plots of MGWAS results. The genome-wide significant level and the suggestive level were shown by red and blue horizontal lines. The green dots indicate the variants associated with the genes in the folate cycle pathways. Gene assignment for each peak was judged based on the biology of genes included in that peak. ADGRV1, TRIB1, JMJD1C, SFXN2, and LMO1 were the genes with the highest p-value in the peak because the relationship between the biological function of genes in the peaks and metabolites was uncertain. Genes assigned to non-validated peaks were shown by asterisk.
MGWAS
Metabolite measurement
From the Tohoku Medical Megabank Community-Based Cohort Study (TMM CommCohort Study)15, we chose metabolites implicated in the folate cycle model, including formate, serine, glycine, methionine, and N; N-dimethylglycine, as measured by Nuclear Magnetic Resonance(NMR) spectrometry; and homocysteine and sarcosine, as measured by targeted-MS . A complete list of all the measured metabolites is available on our website (https://jmorp.megabank.tohoku.ac.jp/)16. The participants used for the MS measurements were a subset of the participants used for the NMR measurements. Details of both measurements have been previously described (NMR:15,17, MS: 18,19). Metabolite extraction was followed by NMR spectroscopy using a Bruker 600 MHz spectrometer at 298 K. Standard NOESY and CPMG spectra were obtained for each sample, and the data were processed using the Chenomx NMR Suite. Our internally developed software enables the automated quantification of metabolites (Aoki et al., in preparation). MS measurements were performed using an MxP Quant 500 Kit (Biocrates Life Sciences AG, Innsbruck, Austria) with a Xevo TQ-XS MS/MS. The LC and FIA modes followed the kit guidelines, with the relevant parameters adjusted accordingly. Subsequently, the concentration was standardized using the MetIDQ Oxygen software.
This study was a part of the Tohoku Medical Megabank Organization Omics Study approved by the Ethics Committee of Tohoku University. Our study was carried out in accordance with the approved guidelines. All the participants provided written informed consent.
Selection of participants for MGWAS
The participants included in this study were selected from the TMM CommCohort Study based on the following criteria: (1) non-pregnant individuals with plasma metabolite concentrations measured using NMR spectroscopy up to the year 2021 (\(N\) = 28,975); (2) individuals whose samples were stored in the Biobank on the day of blood sampling or the following day (\(N\) = 27,865); (3) individuals with measured genotype data from a previous study (\(N\) = 22,916); and (4) individuals who passed both sex and ethnicity checks (\(N\) = 22,561). Ethnicity was visually estimated by performing principal component analysis with the 1000 Genomes Project data20 for LD-pruned variants using a 500 kb window size and an \({r}^{2}=0.2\) threshold with PLINK 2.0 software21,22. After removing distantly related ancestries, \(F\)-statistics of chromosome X without a pseudoautosomal region and the presence of the Y genotype count were calculated using PLINK 1.9 software21,23 for the pruned variant. A linear regression model was used for the MGWAS owing to the small number of participants on the MS platform, related participants were removed by applying a threshold of 0.0884 for kinship coefficients using PLINK 2.021,24. Finally, the following numbers of participants were used for each MGWAS: NMR: formate (\(N\) = 22,465), serine (\(N\) = 22,454), glycine (\(N\) = 22,486), methionine (\(N\) = 22,462), and N, N-dimethylglycine (\(N\) = 22,447); MS: homocysteine (\(N\) = 5020) and sarcosine (\(N\) = 5127). To ensure reproducibility, we included additional participants from the same group, ensuring that there was no overlap with the following participant numbers: NMR: formate (n = 5377), serine (n = 5376), glycine (n = 5386), methionine (n = 5379), N, N-dimethylglycine (n = 5378), MS: homocysteine (n = 1299), and sarcosine (n = 1036).
MGWAS calculation
We performed MGWAS for genotyped and imputed single-nucleotide variations (SNVs) with the following filters: minor allele frequency \(<0.01\), \(p\)-value of the Hardy–Weinberg equilibrium test \(<0.00001\), and missing genotype rate \(>0.05\). In addition, SNVs with INFO scores of < 0.9 in each MGWAS dataset were removed. Finally, approximately 90 million SNVs were used for the MGWAS. MGWAS for metabolites measured using NMR was conducted with BOLT-LMM v2.3.625 and linear regression implemented in GCTA v1.94.026 was used for MGWAS for metabolites measured using MS. All metabolite concentrations were log-transformed. Outliers with a \(p\)-value \(<0.001\) in the Grubbs test27 implemented in the outlier package28 of R 4.1.229 were removed for each metabolite. We calculated the residuals for the log-transformed plasma metabolite concentration with linear regression analysis using age, BMI, sex (male = 1, female = 2), the date interval for storage in the biobank (0 or 1), and the top 10 principal components as covariates to use GWAS phenotype. In the replication study, \(p\)-value was adjusted using the Bonferroni correction based on the number of genome-wide significant peaks detected in the discovery GWAS for each metabolite. Annotation of genes and exonic functions for each detected variant was performed using ANNOVAR30.
Gene assignments to each protein
Genes for each enzyme in the original simulation were selected from the corresponding reactions of the KEGG pathways31,32, and subcellular localization was confirmed by UniProt33. The gene assignments for each enzyme are provided in Supplementary Table 1.
Simulation protocols
We used PySCeS version 1.1.0 to conduct our simulations34. The model was initially converted from the SBML format available in BioModels to the PySCeS Model Description Language (https://www.ebi.ac.uk/biomodels/MODEL1007200000). We made minor modifications to simplify the process. Specifically, we set the initial THF concentration in both the cytosolic and mitochondrial compartments. The model file is located in the GitHub repository with a detailed description (https://github.com/kodates/pathway_sim). With this modification, we successfully simulated the steady state by replicating the results described in the original paper12. The steady-state concentration of each metabolite in the model was recorded and served as a baseline for subsequent comparisons with the modified model results.
We evaluated the correlation between patterns of metabolite concentration changes in the simulation and those observed in the GWAS. Specifically, correlations were calculated in binary signs, with + 1 representing an increase and − 1 a decrease. For this analysis, we employed Kendall’s correlation35, which usage was collectively supported for sign-based data in previous studies36,37,38. The data and code used in this analysis are available on GitHub (https://github.com/kodates/pathway_sim).
In our simulations, we introduced a rate-adjusting coefficient into the model for each enzymatic reaction, which enabled us to simulate the effects of genetic variants on the model. In the initial simulation, we reduced the reaction rate of methylenetetrahydrofolate reductase (MTHFR) by 70% (coefficient = 0.3). This adjustment was followed by a simulation run at a steady state, with the resultant steady-state concentration of each metabolite compared to the baseline. In subsequent simulations, we systematically decreased the reaction rate of each enzyme in the model individually in 10% steps (i.e., coefficients = 1.0, 0.9, 0.8, …, 0.1). This was followed by simulation runs to a steady state for each case, with differences in the steady-state metabolite concentrations calculated relative to the baseline.
Results and discussion
MGWAS for seven metabolites in folate cycles
Novel associations
We performed large-scale MGWAS calculations for metabolites in the human liver cell folate cycle model and metabolome analyses of the Tohoku Medical Megabank Cohort Study. More specifically, the following seven metabolites were analyzed: formate, glycine, serine, N, N-dimethylglycine, and methionine, as measured by NMR spectrometry; and sarcosine and homocysteine, as measured by targeted-MS. The results of the GWAS calculations for these metabolites are shown in Fig. 1B.
Thirty-five genome-wide significant peaks were identified, 27 of which were replicated in the reproduction dataset. Furthermore, 115 genes were identified in the peaks, as designated in Fig. 1A, by adding parentheses to the gene names if they were included in the folate pathway. Among these, 11 metabolite-gene pairs were not found in the GWAS catalog as at August 30, 2024: formate–MTHFR, formate–carbamoyl-phosphate synthase 1 (CPS1), formate–MTRR, formate–SHMT1, serine–alanine–glyoxylate aminotransferase (AGXT), serine–aldehyde dehydrogenase 2 (ALDH2), glycine–glucokinase regulator (GCKR), glycine–tribbles pseudokinase 1 (TRIB1), glycine–ALDH2, methionine–LIM domain only 1 (LMO1), and N,N-dimethylglycine–asparaginase (ASPG).
Formate
In a previous study6, we identified an association between formate and MTHFR. This study revealed that CPS1, MTRR, and serine hydroxymethyltransferase 1 (SHMT1) were associated with blood formate levels. MTRR influences MTR, which encodes the MS involved in the simulation, while SHMT1 also encodes SHMT. Therefore, three of the four genes associated with formate were included in the folate cycles. This suggests that the folate cycle is the major biological pathway affecting blood formate levels.
Glycine and serine
Glycine and serine are neighboring metabolites in the metabolic pathway, and are converted by SHMT and AGXT. Serine is associated with GCKR, CPS1, AGXT, phosphoserine phosphatase (PSPH), and ALDH2. These were subsets of genes associated with glycine, except AGXT, whereas glycine was further associated with five other genes (ALDH1L1, TRIB1, glycine decarboxylase [GLDC], solute carrier family 38 member 4 [SLC38A4], and glycine cleavage system protein H [GCSH]). In previous studies, GCKR and CPS1 were identified as pleiotropic genes for amino acids39,40. This is likely due to their role in encoding proteins that affect the rate-limiting steps of glycolysis and urea cycle. ALDH2 is associated with alcoholic consumption41 and involved in various biological pathways, potentially affecting a wide range of metabolic processes. PSPH and AGXT were the genes encoding the proteins closely related to both glycine and serine in the biological pathway. Therefore, the similarity in the MGWAS results between serine and glycine was likely due to their close relationships within the pathway.
In contrast, glycine is associated with genes related to the glycine cleavage system. Although the association study is unable to demonstrate the absence of associations, the result suggested that serine concentration is not affected from the glycine cleavage system. This may exemplify sublocalization within biological pathways, providing robustness to the entire pathway, as discussed in both experimental42 and theoretical contexts43.
Metabolites involved in methionine metabolism
N, N-dimethylglycine, methionine, sarcosine, and homocysteine are metabolites involved in methionine metabolism, which is related to folate metabolism via MTHFR and MS (Fig. 1A). Among these metabolites, homocysteine was associated with MTHFR, and N, N-dimethylglycine was associated with BHMT. Our results suggest that MTHFR affects not only the folate cycle, but also methionine metabolism. Notably, the results related to homocysteine and sarcosine were limited compared with those of other metabolites, owing to the small sample sizes for homocysteine and sarcosine.
Modifying single enzyme reaction rate
MGWAS analyses are a powerful method for identifying associations between metabolite concentrations and genes. However, there are limitations in understanding how genes affect concentrations. To overcome this difficulty, we used a human liver cell folate cycle model to perform simulations and investigate the effects of changes in enzyme reaction rates, which reflect genetic variants, on the steady-state concentrations of metabolites. Initially, focused on the MTHFR enzyme, which converts 5,10- MTHFR into 5- MTHFR. Previous studies have shown that individuals with the rs1801133 (MTHFR A222V) variant exhibit an approximately 70% reduction in MTHFR enzyme activity, which is associated with a 15% decrease in formate concentration in the blood plasma44. To replicate these findings via simulations, we reduced the MTHFR reaction rate in our model by 70% to mimic the effects of genetic variation. The simulation revealed a 14.7% decrease in the steady-state formate concentration in the modified model, which is closely aligned with previously reported results.
After consistently observing similarities between the simulation and a previous report, we assessed the effect of MTHFR modification on other metabolites within the model. Our findings showed that the direction of the concentration change in the simulation was consistent with the beta direction pattern of associations in the MGWAS with small p-values (Table 1). As a notable case, the non-synonymous variant, rs1801133_A, on MTHFR was positively associated with homocysteine and glycine and negatively associated with formate and serine, exceeding the suggestive significance threshold (p < 1 × 10−4). This increasing and decreasing pattern of beta perfectly aligns with the direction of concentration changes in the corresponding metabolites in the simulated results of MTHFR activity fluctuation. In another case, rs4646700_C was positively associated with serine, formate, and N,N-dimethylglycine, whereas it was negatively associated with glycine at the suggested level. This increasing and decreasing pattern aligns with the simulation results of FTD activity fluctuations, except for N,N-dimethylglycine. To evaluate the qualitative agreement, we calculated Kendall’s tau for the correlation between the direction of change in simulation and the sign of GWAS beta (τ = 0.77, p = 0.04). The comparison showed a significant correlation, suggesting that our simulation captures the directionality of metabolic changes observed in the GWAS. Without employing the simulation approach used in this study, it would be difficult to determine whether the observed associations between MTHFR activity and metabolite concentrations are due to a direct effect of decreased enzyme activity, or if they merely represent correlations without causal links. This distinction is essential to clarify whether changes in glycine, homocysteine, serine, and formate concentrations are primarily the result of reduced MTHFR activity or associated with it due to other underlying factors.
MGWAS was used to investigate the statistical correlations between the genome and metabolite concentrations in plasma, as described previously. In addition, the simulation employs a model that represents only a segment of the metabolic pathway within a specific organ, the liver, and does not fully replicate the complex interactions within a living organism. Despite these limitations, the concordance in outcomes between the two analyses was unexpected and noteworthy. Notably, the MGWAS values pertain to plasma concentrations, whereas our simulation outputs reflected cytosolic concentrations. One potential explanation for this concordance is the dynamic equilibrium between the intracellular and plasma metabolite concentrations. Changes in liver enzyme activity could alter intracellular metabolite levels, which might then be reflected in plasma concentrations owing to the close interaction between the liver and circulatory system. In mice, hierarchical modeling analysis has shown that the gut, kidney, and liver contribute the most to the plasma metabolic pattern45. Furthermore, genetic variations, such as those in MTHFR, can have systemic effects that influence both organ-specific metabolism and overall plasma metabolite levels. This concordance not only strengthens the MGWAS findings, but also underscores the validity of our simulation approach in replicating the effects of altered enzyme reaction rates.
Systematic analysis of enzyme-metabolite relationships
We shifted our focus to examining the relationships between all enzymes and metabolites in the model. We performed steady-state simulations for each setup by systematically reducing the reaction rate of each enzyme in 10% steps to 10% (the general results remained the same even when the reaction rate was increased; see Supplementary Materials Figure S1, S2). The corresponding changes in metabolite concentration were calculated (Fig. 2A). Our simulations showed that when we reduced the reaction rates by 50% for each enzyme, the direction of the fluctuations matched the MGWAS findings for variant-metabolite pairs with small p-values (Table 1, Supplementary Materials for all genes). Notably, our simulations showed marked changes in metabolite concentrations in some gene-metabolite pairs that were considered insignificant in the MGWAS. The reasons for these inconsistencies are as follows: (1) the simulation may be incorrect; (2) the sample size (N) of the MGWAS was too small to detect the association; and (3) there were no naturally occurring variants that altered the enzyme activity of the corresponding gene. The presence of genes included in case 2 should be demonstrated in future studies. To identify the genes that were included in case 3, we counted the functional variants in the exons of all genes analyzed in this study (Supplementary Table 2). Although this was a preliminary assessment, no common non-synonymous variants were found in six genes, whereas only one variant was observed in four genes. In contrast, six common non-synonymous variants were identified in MTHFR and FTD, which were associated with multiple metabolites in the pathway. This suggests that consistency between the simulation and GWAS more likely results in genes that permit evolutionary changes, whereas more than half of the analyzed genes may not tolerate such changes.
The interactions between all enzymes (represented by columns) and all metabolites (represented by rows) within the cytosol compartment of the model. “Non-enzymatic” indicates the non-enzymatic reaction between THF and 5,10-Methylene-THF12. Each panel depicts the changes in metabolite concentration resulting from alterations in the corresponding enzyme’s reaction rate, which was adjusted from 100 to 10% in decrements of 10%. The color intensity at each point correlates with the magnitude of change on a yellow-blue-purple scale, where a shift toward purple indicates a larger change. Enzymes are arranged from left to right in descending order of the mean change in metabolite concentration measured at 50% enzyme activity. (A1) Enzymes impacting a wide array of metabolites. (A2) Enzymes that exhibit minimal impact on the metabolome. (A3) Enzymes with a targeted impact. (B) Schematic representation of the difference between AICART and DHFR.
Our analysis of enzymatic activities revealed three distinct types of enzyme behaviors based on their impact on metabolite concentrations, as shown in Fig. 2A. The first type of enzyme acts as a central metabolic pathway node, influencing a wide range of metabolites within the model (Fig. 2A1). Changes in the reaction rate of MTHFR can markedly affect various metabolites within the model, including 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), methionine, serine, and glycine. Enzymes involved in the methionine cycle, such as GNMT, DNMT, CBS, MAT-I, MAT-III, BHMT, MS, and SAHH belong to this category (Fig. 2A1). Altering the reaction rates of these enzymes can trigger a series of reactions throughout the network, ultimately affecting the concentrations of several metabolites.
The second type of enzyme has a minimal impact on the metabolome. This indicates that changes in their reaction rates did not lead to considerable changes in metabolite concentrations, as shown in Fig. 2A2. These enzymes may be involved in reactions where their activity does not restrict metabolic flow, or their function may be compensated for by other enzymes or pathways, thus maintaining metabolic balance. According to our hypothesis, genetic variants in the genes encoding these enzymes are unlikely to substantially affect metabolism.
The third type of enzymes, AICART and DHFR, exerted a targeted effect, specifically affecting the concentration of a single metabolite (Fig. 2A3). More precisely, changes in the reaction rate of AICART only influenced the concentration of AICAR. This behavior can be explained by the network structure. Structural sensitivity analysis allowed us to determine the qualitative effect of enzyme reaction rate fluctuations on metabolite concentrations at steady-state from the network structure alone46,47,48,49,50. It was mathematically proven that fluctuations in reaction rates within that structure only impact the metabolite concentrations inside the structure and have no effect on metabolites outside of it if a network contains a sub-network called a “buffering structure.” The buffering structure is a subnetwork that satisfies the following two conditions: (1) it is output complete, meaning that none of the reaction rates in the subnetwork depend on the concentrations of chemical metabolites outside the subnetwork, and (2) the index, defined as the number of metabolites minus the number of reactions plus the number of stoichiometric cycles, is equal to zero. This condition ensures that the steady-state responses to parameter perturbations within the buffering structure are confined to it49.
We explored buffering structures in the folate model using ibuffpy49. As a result, AICART-AICAR was identified as a buffering structure, that is, the network structure determines that fluctuations in the AICART reaction rate only affect the concentration of AICAR. Additionally, fluctuations in the reaction rates of all other cytosolic enzymes affected the concentrations of all metabolites. These findings were consistent with the simulation results (Fig. 2A). Notably, DHFR-DHF does not satisfy the conditions for a buffering structure due to a conserved quantity, but the simulation results for DHFR are quite similar to AICART, so we clarified it as a “quasi” buffering structure (Fig. 2B). Although mathematical analysis can describe the behavior of a reaction network based on its structure alone, its application is often limited to qualitative scenarios. Specifically, it is most effective in cases where there is a clear separation between the states that affect metabolite concentrations and those that do not. However, this theoretical approach, although valuable for providing insights into network behavior, is restricted in scope. Simulations remain essential for quantitative assessments, such as determining the precise impact of DHFR on metabolite concentrations. Thus, combining theoretical and simulation approaches offers a more comprehensive understanding, with each method complementing the other.
By systematically categorizing enzymes, we can effectively prioritize our research efforts, focusing on those that noticeably affect the metabolome. This classification not only guides us in identifying potential therapeutic targets but also offers insights into the metabolic basis of specific phenotypes. Moreover, this approach provides a framework for interpreting our simulation and MGWAS findings, suggesting potential pathways for further exploration of experimental designs.
When shifting our focus from enzymes to metabolites, it becomes evident that while various enzymes influence metabolite concentrations, certain metabolites, such as AICAR, serine, and glycine, are predominantly affected by a limited number of enzymes, as demonstrated by the corresponding metabolite rows in Fig. 2A. This suggests that these metabolites exhibit robustness or resilience to changes in enzyme activity, which may have implications for their roles in metabolic regulation.
However, it is essential to recognize the limitations of our model, which only represents a subset of the entire metabolic pathway. The minimal changes observed in metabolites, such as serine and glycine, could be attributed to the incomplete representation of their pathways49,50. Additionally, our study primarily focused on qualitative analysis, examining the directionality of metabolite concentration changes, rather than quantifying the magnitude of these concentration changes. Future research should address these limitations by expanding the scope of the model and incorporating quantitative assessment.
The relationship between genes and metabolites has previously been indicated with limited resolution based on biological links. By contrast, our study demonstrates that the simulation approach provides deeper insights. The observed beta directions in associations with small p-values in the MGWAS results were consistent with the direction of the concentration changes in the simulation (Table 1). This consistency provides evidence that these genes are likely causal for these associations. However, this consistency can only be confirmed in variants significantly associated with multiple metabolites, as it is often unclear whether a given variant increases enzyme activity. Future MGWAS that identify additional genes associated with these metabolites will further validate the utility of this simulation approach.
Our findings underscore the importance of a dual approach that combines theoretical analysis with simulations. Although theoretical models provide valuable qualitative insights, simulations are indispensable to understand the quantitative nuances of metabolic networks. This comprehensive approach enhances our ability to interpret complex biological systems and paves the way for more targeted and effective research.
Data availability
In response to reasonable requests for these data (contact us at dist@megabank.tohoku.ac.jp), we will share the stored data after assembling the dataset and obtaining approval from the Ethics Committee and the Materials and Information Distribution Review Committee of the Tohoku Medical Megabank Organization. The model and simulation codes are available in the GitHub repository (https://github.com/kodates/pathway_sim).
References
Gieger, C. et al. Genetics meets metabolomics: A genome-wide association study of metabolite profiles in human serum. PLoS Genet. 4, e1000282 (2008).
Adamski, J. & Suhre, K. Metabolomics platforms for genome wide association studies—linking the genome to the metabolome. Curr. Opin. Biotechnol. 24, 39–47 (2013).
Rhee, E. P. et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 18, 130–143 (2013).
Shin, S.-Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014).
Long, T. et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 49, 568–578 (2017).
Koshiba, S. et al. Identification of critical genetic variants associated with metabolic phenotypes of the Japanese population. Commun. Biol. 3, 662 (2020).
Tadaka, S. et al. jMorp updates in 2020: Large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res. 49, D536–D544 (2021).
Kojouri, M. et al. Metabolome-wide association study on physical activity. Sci. Rep. 13, 2374 (2023).
Vallarino, J. G. et al. Limitations and advantages of using metabolite-based genome-wide association studies: Focus on fruit quality traits. Plant Sci. 333, 111748 (2023).
Gombert, A. K. & Nielsen, J. Mathematical modelling of metabolism. Curr. Opin. Biotechnol. 11, 180–186 (2000).
Kitano, H. Computational systems biology. Nature 420, 206–210 (2002).
Nijhout, H. F. et al. In silico experimentation with a model of hepatic mitochondrial folate metabolism. Theor. Biol. Med. Model. 3, 1–11 (2006).
Glont, M. et al. BioModels: Expanding horizons to include more modelling approaches and formats. Nucleic Acids Res. 46, D1248–D1253 (2018).
Malik-Sheriff, R. S. et al. BioModels—15 years of sharing computational models in life science. Nucleic Acids Res. 48, D407–D415 (2020).
Koshiba, S. et al. Omics research project on prospective cohort studies from the Tohoku Medical Megabank Project. Genes Cells 23, 406–417 (2018).
Tadaka, S. et al. jMorp: Japanese multi-omics reference panel update report 2023. Nucleic Acids Res. 52, gkad978 (2023).
Koshiba, S. et al. The structural origin of metabolic quantitative diversity. Sci. Rep. 6, 31463 (2016).
Saigusa, D., Matsukawa, N., Hishinuma, E. & Koshiba, S. Identification of biomarkers to diagnose diseases and find adverse drug reactions by metabolomics. Drug Metab. Pharmacokinet. 37, 100373 (2021).
Hishinuma, E. et al. Wide-targeted metabolome analysis identifies potential biomarkers for prognosis prediction of epithelial ovarian cancer. Toxins 13, 461 (2021).
Consortium, G. P. et al. A global reference for human genetic variation. Nature 526, 68 (2015).
Chang, C. C. et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, s13742–s14015 (2015).
Galinsky, K. J. et al. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 98, 456–472 (2016).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Loh, P.-R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Grubbs, F. E. Sample criteria for testing outlying observations (University of Michigan, 1949).
Komsta, L. outliers: Tests for outliers R package version 0.15 (2022). https://CRAN.R-project.org/package=outliers.
R Core Team. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing (Vienna, Austria, 2021). https: //www.R-project.org/.
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164–e164 (2010).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Consortium, T. U. UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023).
Olivier, B. G., Rohwer, J. M. & Hofmeyr, J.-H.S. Modelling cellular systems with PySCeS. Bioinformatics 21, 560–561 (2005).
Kendall, M. G. A new measure of rank correlation. Biometrika 30(1–2), 81–93 (1938).
Bergsma, W. An independence test for continuous and categorical ordinal data based on a sign correlation related to Kendall’s Tau. Available at SSRN 1647863 (2010).
Muñoz-Pichardo, J. M. et al. Multiple ordinal correlation based on Kendall’s tau measure: A proposal. Mathematics 9(14), 1616 (2021).
Ji, S. et al. Conditional independence test by generalized Kendall’s tau with generalized odds ratio. Stat. Methods Med. Res. 27(11), 3224–3235 (2018).
Karjalainen, M. K. et al. Genome-wide characterization of circulating metabolic biomarkers. Nature 628, 130–138 (2024).
Lotta, L. A. et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat. Genet. 53, 54–64 (2021).
Matoba, N. et al. GWAS of 165,084 Japanese individuals identified nine loci associated with dietary habits. Nat. Hum. Behav. 4, 308–316 (2020).
Ishii, N. et al. Multiple high-throughput analyses monitor the response of Escherichia coli to perturbations. Science 316, 593–597 (2007).
Mochizuki, A. & Fiedler, B. Sensitivity of chemical reaction networks: A structural approach. 1. Examples and the carbon metabolic network. J. Theor. Biol. 367, 189–202 (2015).
Frosst, P. et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 10, 111–113 (1995).
Torell, F. et al. Multi-organ contribution to the metabolic plasma profile using hierarchical modelling. PLoS ONE 10, e0129260 (2015).
Okada, T. & Mochizuki, A. Law of localization in chemical reaction networks. Phys. Rev. Lett. 117, 048101 (2016).
Okada, T. & Mochizuki, A. Sensitivity and network topology in chemical reaction systems. Phys. Rev. E 96, 022322 (2017).
Yamauchi, Y., Hishida, A., Okada, T. & Mochizuki, A. Finding regulatory modules of chemical reaction systems. Phys. Rev. Res. 6, 023150 (2024).
He, L., Ding, Y., Zhou, X., Li, T. & Yin, Y. Serine signaling governs metabolic homeostasis and health. Trends Endocrinol. Metabol. 34, 361–372 (2023).
Wang, W. et al. Glycine metabolism in animals and humans: implications for nutrition and health. Amino Acids 45, 463–477 (2013).
Acknowledgements
The authors thank all participants of the TMM CommCohort Study. A complete list of ToMMo Study Group members is available in the Supplementary Material. This research was partially supported by the Research Support Project for Life Science and Drug Discovery (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under grant number JP23ama121019. This study used a supercomputer system provided by the Tohoku Medical Megabank Project (funded by AMED under grant numbers JP21tm0424601 and JP21tm0124005).
Author information
Authors and Affiliations
Consortia
Contributions
S. Kodate, M. S., I. N. M., and K. Kinoshita designed the study. S. Kodate, M. S., and K. Kinoshita wrote the manuscript. Simulation and structural sensitivity analyses were conducted by S. Kodate, and GWAS calculations were conducted by M. S. K. Kojima, who provided the MGWAS pipeline. Under the supervision of S. Koshiba, E. H. conducted sample treatment and MS metabolomics data collection. M. Y. and K. D. Y. reviewed the manuscript. K. Kinoshita supervised this study. S. Kodate and M. S. contributed equally to this study. All the authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kodate, S., Sato, M., Hishinuma, E. et al. Simulating metabolic pathways to enhance interpretations of metabolome genome-wide association studies. Sci Rep 15, 17035 (2025). https://doi.org/10.1038/s41598-025-01634-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01634-7