Abstract
Turmeric leaf disease detection is essential to maintain the crop health and optimize the yield. Through early identification disease can be controlled and its relevant economic losses can be avoided. However, the existing methods for leaf disease detection exhibit limitations in extracting complex leaf features which leads to lower classification accuracy. Also, the existing models often struggle to process the fine details in turmeric leaves which further reduces the reliability in real-world applications. The objective of this research is overcome these limitations through a novel leaf disease detection model which incorporates Vision Transformer (ViT) with hybrid Falcon-Bowerbird Optimization (FBO). The proposed approach aimed to attain improved feature extraction abilities which enhances the overall performance of turmeric leaf diseases detection process. In the first step turmeric images are preprocessed histogram equalization to highlight the complex features like leaf texture and color intensity then the image is divided into non-overlapping patches. The Vision Transformer process each patch as a token through a self-attention mechanism so that the most relevant patches can be processed to extract the essential features. The Hybrid Falcon-Bowerbird Optimization further enhance the convergence speed and fine-tune the hyperparameters to attain improved detection performances. Using turmeric leaf disease dataset, the performance of the proposed model is evaluated through metrics like precision, recall, F1-score and accuracy. With an accuracy of 97.03%, the proposed model outperforms AlexNet which achieved 95.5%, and optimized MobileNetv3 which achieved 96.8%. The proposed hybrid optimized ViT model attained superior performance in turmeric leaf disease detection in terms of accuracy compared to existing techniques.
Similar content being viewed by others
Introduction
Turmeric is an important crop grown extensively across tropical regions. Turmeric is familiar for its medicinal, culinary, and cosmetic uses. Its vibrant yellow pigment, curcumin, holds high economic value particularly in the spice and pharmaceutical industries1. However, turmeric cultivation faces significant challenges that reduce crop yield and affect the quality of the product. Common diseases such as leaf spot, leaf blotch, and leaf blight not only delay the photosynthetic activity of the plants but also severely reduce their growth2. Diseases in turmeric plant often caused by fungi or bacteria and it spread quickly if the weather conditions favorable like high humidity and moderate temperatures. If the infections are not noticed in the initial stage, then the farmer will face huge economic loss. Thus, early and accurate identification of turmeric leaf diseases is essential to avoid these risks. Without precise results farmers need to provide huge chemical treatments which is again harmful and increases the cultivation cost. In another case the farmer might miss the optimal treatment period which leads to complete loss of crop and brings additional burden to farmer3. Mostly turmeric is grown in smallholder farms where farmers may not have easy access to advanced agricultural advice. Thus, an automated and accurate disease detection system help farmers with real time information. so that the farmers can aware of unwanted use of pesticides, reduce crop losses and get better economic outcomes.
Machine learning (ML) has been employed over a decade to handle various agricultural challenges. Traditional ML algorithms, such as decision tree, support vector machine (SVM), k-nearest neighbor (k-NN) and random forest are commonly used classification models for disease identification4. In plant leaf disease detection, ML models depend mainly on feature extraction techniques where relevant features such as texture, color, and shape are manually selected from image data to train the model. The classification model reported in5 utilized spectral imaging combined with ML classifiers to identify specific diseases based on leaf characteristics. In these approaches images are segmented to separate infected regions. Further texture-based descriptors like Gabor filters are applied to capture the visual differences between healthy and infected regions. Additionally, ML models have been designed to predict disease severity so that farmers can estimate the crop damage and adjust their intervention strategies accordingly6. Few hybrid approaches are also developed for disease classification in which multiple ML algorithms are combined to improve classification accuracy. Though ML models have achieved better performance in disease detection it mainly depends on the quality of input features and image preprocessing. This can be a limiting factor which affects the performance where image quality varies due to lighting.
Although ML models have been widely adopted in agricultural disease detection tasks, they face several limitations when applied to turmeric leaf disease detection. One of the main drawbacks is their dependence on manual feature extraction which requires domain expertise to identify relevant features in images. Also, this process is time-consuming and error-prone. Moreover, ML based leaf disease detection struggle with generalization issues on training data but lags in accuracy while using new data. This issue arises because traditional ML models do not learn hierarchical features or relationships within the data which leads to overfitting. Another limitation is their sensitivity to variations in image quality. Environmental factors such as lighting, angle, and noise can affects the accuracy of disease detection models and requires extensive preprocessing steps to standardize the input images. Furthermore, traditional ML algorithms require huge labeled data to attain optimal performance. However, in practical scenario annotated datasets for turmeric leaf diseases is not available. The ML models exhibit poor performance while processing the complex disease patterns. But deep learning models exhibit superior performance in image-based classification tasks7.
Unlike traditional ML models which depend on manual feature extraction, relevant features from image data are automatically learn through DL models8. This capability allows the DL model to process textures and edges. These features are then classified to differentiate the normal and disease affected leaves. Through the multiple convolutional layers, traditional DL models effectively handle the variations in the input9. Also, the DL models work well with different data and find the complex relationship between the features10. The DL model requires a minimum knowledge on feature processing whereas the ML models required better domain knowledge to extract and process the features. The adaptability and scalability of DL models make them suitable for real time disease detection11.
The motivation behind this research is obtained by analyzing the growing need for accurate and timely detection of turmeric leaf disease which ensures high yields and minimized economic losses. While traditional ML models have offered a basic approach to disease detection their limitations in feature extraction, sensitivity to environmental factors, and over-dependency on manually labeled data highlight the need for more advanced techniques. Recent advancements in DL have shown that these models outperform conventional ML methods in image recognition tasks including plant disease identification12,13,14. However, DL models are also having limitations like slow convergence, overfitting, and the need for extensive tuning of hyperparameters. Thus, a Vision Transformer with a novel hybrid Falcon-Bowerbird Optimization (FBO) is presented in this research work. The objective of this research is to enhance the classification accuracy using vision transformer and improve the convergence speed through the hyperparameter tuning using hybrid FBO.
The proposed work incorporates a hybrid Falcon-Bowerbird Optimization (FBO) to fine tune the hyperparameters of Vision Transformer. The Satin Bowerbird Optimization (SBO) in the hybrid model globally explores the hyperparameter space which avoid premature convergence. While the falcon optimization is included in the hybrid model for its fast convergence. Also, falcon optimization adjusts learning rates dynamically based on the gradient making it suitable for fine-tuning the Vision Transformer weights. The reason for combining these two algorithms is to provide a better balance between diverse hyperparameter configurations and the optimal solution. The contributions of the research work are summarized as follows.
-
Proposed a novel hybrid optimized Vision Transformer model to attain enhanced accuracy for turmeric leaf disease detection.
-
Presented a hybrid Falcon-Bowerbird Optimization (FBO) combining Satin Bowerbird Optimization for global hyperparameter tuning and Falcon Optimization for local weight fine-tuning. By optimizing the hyperparameter selection through SBO the proposed model can dynamically adjust key parameters such as patch size, embedding dimension, and number of transformer layers, reducing the need for manual tuning. Falcon Optimization algorithm fine-tunes the model weights using adaptive gradient methods to ensure quick convergence and improved performance over traditional optimizers.
-
Presented a detailed comparative analysis with existing deep learning models to validate the better performances.
The remaining discussions are arranged in the following order. “Related works” provides a detailed discussion on recent works in leaf disease detection. “Proposed work” provides the complete details of proposed optimized vision transformer model. “Results and discussion” presents the experimental results and discussion and finally “Conclusion” presents the conclusion.
Related works
A detailed literature review on recent works in leaf disease detection is presented in this section. A comparative analysis of ML and DL is presented in15 for plant disease detection and classification. The analysis considered ML algorithms such as DT, SVM and RF along with DL models like CNN, ResNet50 and MobileNetV2 algorithms. The experimentation using benchmark dataset reports the better performance of YOLOv5 over other models in object detection tasks. Whereas ResNet50 and MobileNetV2 exhibited the best trade-offs between accuracy and computational time for classification. However, the presented analysis did not consider any approaches to handle noisy, real-world data in early disease detection.
Classifying plant leaf diseases using ML is presented in16 to enhance the classification performance compared to conventional methods. The presented model segments the input images into distinct regions to simplify the extraction of relevant features such as texture, shape, and color. These features are then used by machine learning algorithms like SVM and K-Means clustering. Experimental results exhibit the improved disease detection accuracy compared with conventional classification methods. However, the presented has limitations as it requires larger and more diverse datasets to validate the model’s performances.
Identifying and measuring the severity of leaf spot disease in turmeric plants is performed in17 through edge detection algorithms. The presented model capturing images of infected turmeric leaves and processing them to calculate the disease severity. Specifically, the disease intensity is computed based on the ratio of the infected area to the total leaf area using histogram-based color extraction and K-means clustering for segmentation. Edge detection algorithms further assist in identifying the affected regions. Experimental results indicate that the presented approach exhibit an accuracy of 81% using the histogram method and 70% through area calculation for leaf spot identification. However, the presented model utilized limited data samples which limits the model generalizability. Additionally, the presented model depends on accurate segmentation which is challenging under varying lighting conditions.
A robust approach for identifying and classifying turmeric plant leaf diseases is presented in18 using a combination of Linear Discriminant Analysis (LDA) and Adaptive Network-Based Fuzzy Inference System (ANFIS) algorithms. The presented converts the input image from RGB to HSI color space and then performs segmentation using the Spatial Fuzzy C-Means (SFCM) clustering algorithm. Feature extraction is done through Gray-Level Co-occurrence Matrix (GLCM) and the final classification is done through LDA-ANFIS classifier. Experimental results exhibit the presented model better classification accuracy over conventional Artificial Neural Networks (ANN) and SVM. However, the limited dataset affects the generalizability of the model which is considered as minor limitation of the presented model.
Early detection of diseases in turmeric plants is reported in19 using VGG-16 architecture. The presented model detects leaf diseases by extracting relevant features through VGG-16. A comparison made between the input images and the trained dataset. The trained model then provides the classification results including the type of disease and recommended precautionary measures. The experimental results of the proposed VGG-16 model exhibits its better accuracy which is better than AlexNet model. However, the presented model has limitations in terms of computational cost and requires fine tuning to handle more complex diseases. The deep learning model presented in20 performs early detection of major diseases in turmeric plants. The presented model includes Improved YOLOV3 tiny model with residual network to attain enhanced detection accuracy. The presented approach is designed to overcome limitations in traditional models by providing accurate detection of turmeric leaf diseases. Experimental results demonstrate the presented model better performance over traditional Yolo models and Faster R-CNN. However, the presented model performance depends on the dataset size which is considered as a minor limitation.
The hybrid model reported in21 for detecting rhizome rot disease in turmeric plants combines CNN with SVM. Through the CNN layers the hierarchical features are extracted and processed by the SVM to differentiate normal and diseased leaves. The presented hybrid CNN-SVM approach outperformed models such as CNN and VGG19 achieving the highest accuracy and faster execution time making it highly efficient for real-time applications. However, the presented model has limitations like data overfitting due to minimum training epochs. The hybrid approach presented in22 for plant leaf diseases detection incorporates deep CNN with Local Binary Pattern (LBP) features. The presented model pre-processes the leaf images and pass them through a deep CNN model for extracting high level features. These deep features are combined with LBP to capture local texture information. The combined feature set is then used for plant disease classification. The experimental results of presented model demonstrate the model ability to through superior classification performance compared to traditional CNN architectures like VGG16, AlexNet, and GoogleNet.
The hybrid model presented in23 combines the Segment Anything Model (SAM) and Fully Convolutional Data Description (FCDD) for detecting plant diseases in field images. The presented model utilizes SAM to segment all objects within an input image so that key elements such as leaves, stems, and other background components can be identified. Then FCDD is applied to differentiate actual plant leaves and background objects. The final step performs classification which classifies the identified region of interest (ROI). Experimental results demonstrated significant improvements over existing models with higher accuracy and effectively addresses the challenges occurs in disease detection due to complex backgrounds and varying lighting conditions in field images. However, the presented model has limitations as it needs more effective procedure to differentiate densely green backgrounds and actual leaves.
The deep learning model presented in24 combines the YOLOv7 detection algorithm with an enhanced optimization framework for leaf image processing. The presented model incorporates Contrast-Limited Adaptive Histogram Equalization (CLAHE) for image preprocessing to enhance the image contrast. Followed by YOLOv7 model is used for accurate detection of diseased regions in plant leaves. Finally, the features are extracted and classified using ShuffleNetV2 which categorizes the detected diseases. Also, the parameters of classifier are optimized by incorporating an Enhanced Rat Swarm Optimizer (ERSO). The presented model additionally includes Red Fox Optimization (RFO) algorithm to segment the diseased regions. The better accuracy over CNN models is presented through the experimental results. However, the presented model has limitations in terms of computational complexity due to the incorporation of multiple optimization and learning models.
Early detection of leaf diseases detection is presented in25 by integrating Internet of Things (IoT) with Deep Learning (DL) model. The presented multi-model fusion network includes CNN with IoT devices to collect the environmental data and perform real-time disease detection. The fusion architecture consists of three sub-networks such as RL-block for coarse-grained feature extraction and a set of PL-blocks for fine-grained and environmental feature extraction. The model is trained using leaf images which affected by common rust, blight, and gray leaf spot. The analysis is then enhanced with data from IoT sensors which monitor environmental conditions like temperature, humidity, and soil moisture. The final decision is obtained through a decision-level fusion model that combines image and environmental features. Experimental results show that the presented MMF-Net exhibit a high accuracy after fusing multi-contextual features and outperforms traditional CNN models like AlexNet and VGG. However, the experimentation depends on the data from IoT devices which may face connectivity issues, and the need for further refinement to handle more complex environmental variables. Future work could explore the inclusion of additional environmental factors and real-time edge computing to enhance scalability and robustness.
A hybrid model presented in26 combines the Capsule Network (CapsNet) and Residual Network (ResNet) for plant leaf disease classification. The presented model includes modified ResNet with small convolution kernels in the first layer to enhance fine-grained feature extraction and a channel attention mechanism to focus on key features. Then CapsNet is used to preserve positional information which is often lost in traditional pooling operations. The fusion of these two networks allows the model to capture both detailed features and the spatial structure of plant disease spots. Experimentation using benchmark leaf disease dataset exhibit the better performance over traditional models like AlexNet, GoogLeNet, and VGG16. However, high computational cost and inference time of presented model may introduce challenges while implanting in real-time applications. The leaf disease detection model presented in27 combines residual connections and Xavier weight initialization to enhance the ability to learn complex features of diseased leaves in noisy environmental conditions. The presented LeafNet architecture undergoes extensive training utilizing both Cross-Entropy Loss and Focal Loss functions to fine-tune the detection performance. Experimental evaluation of the presented model exhibits the superior performance over DenseNet161, VGG16, and MobileNetV2 models. Table 1 summarizes the research works available in the literature related to the proposed work.
Research gaps
From the literature analysis summary presented in Table 1, several key research gaps in the field of leaf disease detection are identified and summarized in this section. A common limitation across which observed in existing works is its dependence on small or limited datasets. This restricts the classification or detection model generalizability and robustness in real-world scenarios. Though the existing methods exhibits its high accuracy in controlled environments it brings challenges to attain the same while performing real-time detection and classification, particularly when dealing with large-scale agricultural systems. Existing deep learning models and hybrid approaches have demonstrated its better performance but their computational complexity and inference time are considered as major limitations which makes these methods less feasible for real-time applications. Another identified gap is the limited testing of these models across multiple plant species and diseases. Most studies focus on one or a few specific crops which restricts the broader application of the models. Additionally, there is a need for better segmentation techniques to differentiate between diseased and non-diseased areas more accurately particularly in complex backgrounds. The existing models addressed the need for optimizing models for resource-constrained environments where computational resources are limited.
Proposed work
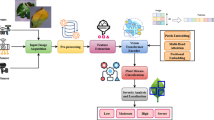
The proposed work focuses on optimizing a Vision Transformer (ViT) model for turmeric leaf disease detection by utilizing a novel hybrid optimization strategy, Falcon-Bowerbird Optimization (FBO). To effectively address the complex visual features present in turmeric leaf diseases, the proposed approach is developed to overcome the shortcomings of earlier techniques that either depend on handcrafted features or struggled with generalization across diverse disease classes. Traditional ML models, although interpretable have limitations in terms of learning the features. Also, it requires hierarchical representations for complex image data. while conventional DL models like CNN often focus on localized patterns and lack the ability to model long-range dependencies. To overcome these limitations, the Vision Transformer (ViT) was incorporated due to its capacity to process image patches independently and capture global context through its self-attention mechanism. The Vision Transformer selected for its ability to handle image patches and capture long-range dependencies through self-attention mechanisms. The hybrid approach combines Satin Bowerbird Optimization (SBO) and Falcon Optimization algorithm. The reason for selecting Satin Bowerbird Optimization is to effectively explore the hyperparameter space by imitating the competitive and creative behavior of bowerbirds. This ensures better diversity and avoids premature convergence. Whereas Falcon Optimization use adaptive gradient-based techniques to fine-tune model parameters quickly and accurately. This ensures faster convergence and improved accuracy. The proposed approach begins by preprocessing turmeric leaf images into patches. Then using vision transformer with FBO process the input image and provides better performance in turmeric leaf disease detection. This combination improves the model capability to detect complex patterns in diseased turmeric leaves making it highly effective for the classification task. Figure 1 provides the overview of the proposed turmeric leaf disease detection model.
Preprocessing
The preprocessing phase of the proposed turmeric leaf disease detection system is essential for enhancing the input images. In this preprocessing step, the images are processed to improve important features clarity such as leaf texture and color intensity. The preprocessing steps is essential for accurate disease detection. The proposed model preprocessing includes histogram equalization to improve contrast and patch division to divide the image into smaller, non-overlapping patches for subsequent processing by the Vision Transformer. The histogram equalization is technique used to enhance the image contrast by redistributing the intensity values. While preprocessing the turmeric leaves, the main goal is to make the texture and color variations into distinct so that the Vision Transformer can easily identify diseased regions. Consider the image \(\:I\) and the pixel intensity levels range from \(\:\text{0}\) to \(\:(L\:-\:1)\) in which \(\:L\) indicates the number of possible intensity levels. The probability distribution of pixel intensities is represented as \(\:p\left(i\right)\) where \(\:x\) is the intensity level and \(\:p\left(i\right)\) represents the probability that a randomly selected pixel in the image has intensity \(\:i\). The cumulative distribution function (CDF) is formulated as
The CDF \(\:c\left(x\right)\) gives the cumulative probability of pixel intensities up to level \(\:x\), effectively stretching the intensity range to improve the contrast. The transformed intensity value \(\:\:y\) for each pixel is calculated as follows.
This ensures that the intensity values are redistributed across the available range and making the darker areas lighter and vice versa. This improves the overall contrast of the input image. After applying histogram equalization, the output image highlights the texture and even mild differences in color. Once the contrast of the image is improved, the next step is to divide the image into non-overlapping patches. Since the Vision Transformer operates on patches of the image instead of utilizing the entire image patches are generated in the next step. Each patch is treated as an individual token in the transformer model and these patches are processed independently to capture local as well as global features of the image. Consider the histogram enhanced image \(\:I\) have dimensions \(\:W\times\:H\:\) in which width is indicated as \(\:W\) and height is indicated as \(\:H\). The image is divided into \(\:N\) non-overlapping patches, where each patch has dimensions \(\:(P\times\:P)\). Then the number of patches \(\:N\) is calculated as follows
In each patch \(\:{I}_{i,j}\) is extracted from the image, where \(\:\:i\) and \(\:j\)denote the position of the patch in the grid. The total number of patches is determined by the width and height of the image. The patches are indexed as \(\:{I}_{i,j}\) and the complete set of patches are formulated as
Further each patch \(\:{I}_{i,j}\) is then flattened into a one-dimensional vector and this transformation is necessary because the Vision Transformer process these flattened vectors as tokens. The flattening operation is mathematically formulated as
The flattened vector consists of the pixel values of the patch in a sequential format, and each vector has a length of \(\:{P}^{2}\). This flattened representation of the patch is essential for further processing in the Vision Transformer, where each patch is treated as a token that passes through the transformer’s layers for feature extraction. The histogram equalization in the preprocessing step focuses on contrast improvement, the proposed work includes additional feature enhancement techniques such as edge detection and texture-enhancing filters to the patches to make the disease-related patterns more visible. To perform edge detection, Sobel operator is used which highlights the boundaries of diseased regions. The Sobel operator calculates the gradient of pixel intensities in the \(\:x\) and \(\:y\) directions which is mathematically formulated as follows.
where \(\:\left(*\right)\) indicates the convolution operation. The resulting edge gradient magnitude is formulated as
The gradient magnitude \(\:G\) highlights the edges to detect the boundaries of diseased areas in the leaves. Similarly, texture enhancing filters are utilized to highlight the unique texture patterns in healthy and diseased turmeric leaves. These filters are utilized to highlight specific frequency components in the image. In the proposed work Gabor filter is used as texture enhancing filter which is mathematically expressed as
where the wavelength of the sinusoidal factor is indicated as \(\:{\uplambda\:}\), orientation is indicated as \(\:\theta\:\), phase offset is indicated as \(\:\psi\:\), standard deviation of the Gaussian is indicated as \(\:\sigma\:\), and spatial aspect ratio is indicated as \(\:\gamma\:\). This filter highlights the specific texture frequencies in the leaf and allows the Vision Transformer to capture complex patterns that indicates the disease. By improving the image quality and segmenting the input into patches the Vision Transformer is able to process the turmeric leaf images more effectively and provide better performance in disease detection process.
Vision transformer (ViT)
The Vision Transformer (ViT) in the proposed turmeric leaf disease detection model process the image patches obtained in the preprocessing step. The first step in the Vision Transformer is to embed the patches into a lower-dimensional space. Given an input image \(\:I\) that has been preprocessed and divided into \(\:N\) non-overlapping patches, each patch \(\:{I}_{i,j}\) of size \(\:(P\times\:P)\) pixels are flattened into a 1D vector. This vector is then projected into a fixed embedding dimension using a linear transformation. Mathematically, this embedding process is formulated as
where \(\:{f}_{i,j}\) indicates the resulting embedded vector for the patch located at position \(\:\left(i,j\right)\), \(\:{W}_{e}\) indicates the learned embedding matrix, and \(\:{b}_{e}\) indicates the bias term. The flattening operation converts the 2D image patch into a 1D vector and the embedding matrix projects this 1D vector into a lower-dimensional space so that each patch will be treated as a token. So that each patch is represented as a token with a fixed-dimensional feature vector that captures the essential information of the patch. Followed by patch encoding, positional encoding is performed in vision transformer. Since transformers do not capture positional information, positional encoding is introduced in the vision transformer to retain the spatial structure of the image. Without positional encoding the transformer treat the patches as independent objects and ignore their relative positions. Thus, the positional encoding is included to each token after embedding and it allows the model to understand the spatial relationships between patches. Positional encoding uses a combination of sine and cosine functions of different frequencies to generate a unique encoding for each patch. For a patch at position \(\:\left(pos\right)\) the positional encoding for the \(\:{2k}^{th}\) dimension is obtained through the formulation as follows
Similarly, for the \(\:{\left(2k+1\right)}^{th}\) dimension, the positional encoding is formulated as
where \(\:pos\) indicates the position index of the patch in the sequence, \(\:k\) indicates the dimension index, and \(\:d\) indicates the total dimensionality of the embedded token. The use of sine and cosine ensures that the positional encodings for neighboring patches are close in the encoding space and preserves the spatial locality. After this, the positional encoding is added to the embedded token which is mathematically expressed as
where \(\:{z}_{i,j}\) is the positional-encoded token.
Self-attention mechanism
Once the tokens are embedded and enhanced with positional information the Vision Transformer applies a self-attention mechanism to capture relationships between all patches in the image. Self-attention enables the model to weigh the importance of each token relative to every other token and allows the model to focus on the most relevant patches. For each patch token three different vectors are computed such as the query \(\:Q\), key \(\:K\), and value \(\:V\). These vectors are obtained by multiplying the token with learned weight matrices which is mathematically formulated as follows.
where \(\:{W}_{Q}\), \(\:{W}_{K}\), and \(\:{W}_{V}\) indicates the learned projection matrices for the query, key, and value, respectively. The attention score \(\:{{\upalpha\:}}_{i,j}\) between token \(\:i\) and token \(\:j\) is obtained as the scaled dot product of the query and key vectors which is formulated as follows
where \(\:{d}_{k}\) indicates the key vectors dimensionality, and attention weights \(\:{{\upalpha\:}}_{i,j}\) is normalized using the SoftMax function. These weights determine the contribution of each token to the self-attention mechanism output. The output of the self-attention layer for token \(\:i\) is then computed as a weighted sum of the value vectors \(\:{V}_{j}\) which is formulated as follows
This allows the model to aggregate information from all tokens in the sequence and focus on the most relevant patches, enabling it to capture complex inter-patch relationships.
Multi-head attention
Further multi-head attention is used to enhance the vision transformer ability in capturing different patterns and relationships between tokens. In this mechanism, the self-attention process is repeated multiple times in parallel with each head focusing on different characteristics of the token. Then each attention head output is concatenated and linearly projected to the original dimension using a learned weight matrix \(\:{W}_{o}\). The multi-head attention layer final output is formulated as
where \(\:h\) indicates the number of attention heads, and each head processes different projections of the key, query, and value vectors. Multi-head attention process image features simultaneously and improves the robustness of the feature extraction process.
Feed-forward network
After the multi-head attention layer, the output tokens are passed through a feed-forward network (FFN) to introduce non-linearity and further refine the feature representation. The FFN consists of a non-linear activation function (ReLU) in between two linear layers. The output of the FFN for each token \(\:{O}_{i}\) is obtained as follows
where \(\:{W}_{1}\) and \(\:{b}_{1}\) indicates the weights and bias for the first linear transformation and e ReLU indicates the activation function. The FFN final output is then computed as follows
where \(\:{W}_{2}\) and \(\:{b}_{2}\) indicates the parameters of the second linear transformation. This network allows the model to capture non-linear relationships between the features extracted from the image patches and improves the classification performance.
Classification layer
The final step of the Vision Transformer involves collecting the outputs from all patches to make a classification decision. The output tokens from the FFN are then fed into a classification layer that maps the feature representation to the predefined classes. The classification layer in the proposed model utilizes a SoftMax function to convert the raw scores into probabilities for each class which is mathematically formulated as
where \(\:{s}_{i}\) indicates the score for class \(\:i\), and \(\:\widehat{{y}_{i}}\) indicates the predicted probability for class \(\:i\). The class with the highest probability is selected as the model final prediction. The Vision Transformer in the proposed work provides a better performance in turmeric leaf disease detection by processing image patches. Unlike convolutional models that focus on local patterns, the proposed vision transformer learns complex relationships through its self-attention mechanism. By combining multi-head attention and feed-forward layers the proposed vision transformer fine tunes the feature and make it suitable for leaf disease detection.
Hybrid Falcon–Bowerbird optimization (FBO)
The Hybrid Falcon-Bowerbird Optimization (FBO) proposed in this research work combines two optimization algorithms such as Satin Bowerbird Optimization (SBO) and Falcon Optimization. Specifically, SBO is selected for global exploration and Falcon Optimization is selected for local exploitation. The hybrid approach ensures that the proposed model attain diverse search capabilities and converge quickly on optimal solutions. The hybrid FBO used in the proposed work optimizes the hyperparameters and fine-tune the weights of the Vision Transformer used for turmeric leaf disease detection.
Satin Bowerbird optimization (SBO) for global exploration
The Satin Bowerbird Optimization imitates the male satin bowerbird’s behavior which build elaborate bowers to attract mates. In the optimization process each bowerbird represents a candidate solution for the Vision Transformer. The goal of each bowerbird is to improve its solution through competition with other bowerbirds learning from better-performing solutions. Consider \(\:{s}_{i}^{\left(t\right)}\) be the solution of the \(\:{i}^{th}\) bowerbird, which consists of a set of hyperparameters such as number of transformer layers, learning rate, patch size and attention heads. The fitness function \(\:F\left({s}_{i}^{\left(t\right)}\right)\) evaluates the quality of each solution based on the classification accuracy of the vision transformer model. The SBO operates in iterative steps and each bowerbird updates its solution by learning from the best-performing bowerbird in the current population. Mathematically the update formula for the solution of bowerbird \(\:i\) is formulated as
where \(\:{s}_{best}^{\left(t\right)}\) is the best solution found in the population at iteration \(\:t\), \(\:{\lambda\:}_{sbo}\) indicates the scaling factor that controls how much influence the best solution has on the current bowerbird’s update. The factor \(\:({\upgamma\:}\cdot\:N\left(\text{0,1}\right))\) introduces a random perturbation to maintain population diversity and avoid premature convergence, \(\:N\left(\text{0,1}\right)\) indicates normal distribution which ensures degree of randomness is maintained during exploration. This process allows bowerbirds to explore the solution space productively and adjust their strategies based on the performance of the best bowerbird while ensuring diversity through random variations. Also, this exploration ensures that the model can cover a wide range of possible hyperparameter configurations and making the optimization robust against poor initial guesses. As the algorithm progresses the poor-performing bowerbirds gradually improve their solutions by imitating the strategies of better-performing ones and leads to a gradual improvement in the overall population of solutions.
Falcon optimization–local exploitation
By using SBO global exploration, a set of promising solutions are identified and then using Falcon Optimization local exploitation is performed in the proposed hybrid model. Falcons are precision-based optimization agents that quickly fine tune the best solutions identified by the bowerbirds. The main objective of utilizing Falcon Optimization is to ensure that the best solutions converge quickly to an optimal configuration by using gradient-based optimization techniques. In the proposed model, AdaBelief is used as gradient model. Consider \(\:\theta\:\) represent the set of weights and parameters of the Vision Transformer model. The Falcon Optimization objective is to minimize the loss function \(\:L\left(\theta\:\right)\) which represents the error between the true labels and predicted labels in the turmeric leaf classification task. The weight update at each iteration \(\:t\) is mathematically formulated as follows
where \(\:{{\uptheta\:}}_{t}\) indicates the updated parameters at iteration \(\:t\), \(\:{\eta\:}_{t}\) indicates the learning rate at iteration \(\:t\), \(\:{\nabla\:}_{{\uptheta\:}}L\left({\uptheta\:}\right)\) indicates the loss function gradient. The AdaBelief optimizer which is used in Falcon Optimization adjusts the learning rate adaptively based on the belief in the current gradient direction. Also, AdaBelief improves upon traditional gradient-based optimizers like Adam by considering the difference between predicted and actual gradients. The update rule for AdaBelief is mathematically formulated as
where \(\:{m}_{t}\) indicates the moving average of the gradients (first moment), \(\:{s}_{t}\) indicates the moving average of the squared gradient differences (second moment), \(\:{{\upbeta\:}}_{1}\) and \(\:{{\upbeta\:}}_{2}\) indicates the exponential decay rates for the first and second moments respectively and \(\:\text{ϵ}\) indicates a small constant added for numerical stability. The AdaBelief optimizer refines the model weights by dynamically adjust the learning rate considering the smoothness of the loss and allows the falcon to quickly converge to an optimal solution and avoiding overfitting or trap into local optimal solution.
The proposed hybrid Falcon-Bowerbird Optimization (FBO) balances global exploration and local exploitation to optimize both the hyperparameters and weights of the Vision Transformer. The Satin Bowerbird Optimization ensures that the solution space is thoroughly explored and allows the model to search for diverse fields. Once the best candidates are identified Falcon Optimization performs precise adjustments to ensure fast convergence. The final optimized hyperparameters and weights are selected after several iterations of this hybrid optimization process. By combining the strengths of both global exploration which obtained through the bowerbirds and local exploitation which obtained through the falcon optimization the proposed model achieves high accuracy and faster convergence.
The hybrid approach utilizes SBO to introduces diversity and creativity in the search space and Falcon Optimization focuses on exploiting the most promising solutions using a AdaBelief optimizer. The AdaBelief optimizer ensures that the falcons can adjust the learning rate considering the loss and provides faster and more accurate convergence compared to traditional optimizers. The combination of exploration and exploitation ensures that the Vision Transformer achieves an optimal configuration for the turmeric leaf disease detection process and provides improved classification accuracy, faster convergence compared to traditional disease detection methods. The proposed hybrid optimized Vision Transformer summarized pseudocode is presented as follows.

Pseudocode for the proposed hybrid optimized visiontransformer for turmeric leaf disease detection

Results and discussion
The experimentation for the proposed hybrid optimized Vision Transformer model for turmeric leaf disease detection is performed using Python tool. A turmeric leaf dataset comprising 1,000 original image samples is prepared manually from the turmeric fields and through data augmentation techniques such as flipping, rotation, and scaling, the dataset is improved with 5,000 images. This augmentation step ensures that the model utilizes a wide range of samples during training and making it more robust to variations in leaf appearance caused by different angles, lighting, or environmental factors. The entire dataset was split into training and testing sets, with 70% of the sample images (3,500 samples) are used for training the Vision Transformer model and the remaining 30% (1,500 samples) are used for testing. The training process involved hyperparameter tuning through FBO to select optimal configurations such as patch size, embedding dimension, learning rate and fine-tuned the model weights using AdaBelief. The performance of the model is evaluated on the test set in terms of accuracy, precision, recall, and F1-score to provide a comprehensive evaluation of the proposed model ability in detecting turmeric leaf diseases. Table 2 presents the details of simulation hyperparameters used in the proposed model experimentation.
The dataset used in the experimentation includes images of turmeric leaves affected by four diseases such as Leaf Spot, Leaf Blight, Leaf Rot, and Leaf Curl. The original dataset consisted of 315 samples for Leaf Spot, 226 for Leaf Blight, 290 for Leaf Rot, and 169 for Leaf Curl. To enhance the model training capabilities and avoid overfitting data augmentation techniques were applied which increases the total number of samples to 1575 for Leaf Spot, 1130 for Leaf Blight, 1450 for Leaf Rot, and 845 for Leaf Curl. For training and testing purposes, the dataset was split in a 70:30 ratio. This provided 1103 training and 473 testing samples for Leaf Spot, 791 training and 339 testing samples for Leaf Blight, 1015 training and 435 testing samples for Leaf Rot, and 591 training and 254 testing samples for Leaf Curl, respectively. Complete details about the dataset are presented in Table 3.
Figure 2 depicts the training and test accuracy of proposed model for 150 epochs. Initially, the training and testing accuracy values increase gradually in which the test accuracy follows the training accuracy. By 20th epoch the training and testing accuracy crosses approximately 95% indicating the model effectiveness in learning in the early stages of training. As the number of epochs increases the accuracy improves and reached a maximum of around 97.90% for training and 97.03% for testing. This indicates that the proposed model is well-trained and generalizes as well to unseen data. The relatively small gap between the training and testing accuracy curves suggests minimal overfitting as both accuracies converge toward close values by the end of training. The proposed model demonstrates its high reliability and effectiveness in classifying turmeric leaf diseases as the testing accuracy remains stable with minimal fluctuations which reflects a robust learning process and good generalization performance.
Figure 3 depicts the proposed model training and testing loss which decreases over the course of training. Initially, both training and testing losses start at around 0.41 after few epochs the loss values reduces gradually which indicates the proposed model ability in quickly adjusting its weights to reduce errors. By epoch 20, the loss values for both training and testing have dropped below 0.36. After 20th epoch the loss becomes gradual in training as well as testing and stabilizing around 0.35 as the number of epochs increases. Specifically, the loss curves for training and testing remain close to each other throughout the training process which indicates that the proposed model avoids overfitting. The small gap between training and testing loss further highlights that the model maintains a better balance between fitting the training data and testing data which is essential for turmeric leaf disease detection.
The training confusion matrix of the proposed model given in Fig. 4, demonstrates the proposed model accurate classification performances across all four classes of turmeric leaf diseases like Leaf Spot, Leaf Blight, Leaf Rot, and Leaf Curl. For the Leaf Spot class, out of 1103 total samples the proposed model correctly classified 1078 samples and misclassified 10 samples as Leaf Blight, 15 as Leaf Rot, and 5 as Leaf Curl. Similarly, for Leaf Blight, out of 791 samples, the proposed model correctly predicted 778 samples with 12 misclassifications. For the Leaf Rot class, out of 1015 samples, 993 samples are correctly predicted with 15 misclassified as Leaf Spot, 1 as Leaf Blight, and 1 as Leaf Curl. The Leaf Curl obtains 580 correct classifications out of 591 total samples with 10 misclassified as Leaf Spot, 1 as Leaf Blight, and 1 as Leaf Rot. The small number of misclassifications across all classes especially with Leaf Blight and Leaf Curl highlights the model better performance in differentiating between various diseases during training which results in high precision and recall values.
Similarly in the testing confusion matrix given in Fig. 4, the proposed model exhibits its reliable performance with slightly larger number of misclassifications compared to the training process. For Leaf Spot, 463 out of 473 samples were correctly identified with 5 misclassified as Leaf Blight, 3 as Leaf Rot, and 2 as Leaf Curl. In the case of Leaf Blight 327 out of 339 samples were accurately predicted with 6 misclassified as Leaf Spot and 5 as Leaf Curl. In case of Leaf Rot 424 correct predictions are made from 435 samples, with only 4 misclassified as Leaf Blight and 2 as Leaf Spot. In the last, Leaf Curl exhibit 245 correct classifications out of 254 total samples with minor misclassifications into the other categories. These test results highlight the proposed model generalization ability and consistency with training and maintaining better accuracy while dealing disease data.
Table 4 presents the proposed model training performance metrics for all the four turmeric leaf disease categories such as Leaf Spot, Leaf Blight, Leaf Rot, and Leaf Curl. For Leaf Spot, the proposed model achieved a precision of 0.9775 indicates that nearly 97.75% of the predicted samples as Leaf Spot were correct. The recall of 0.9773 indicates that the model was able to identify 97.73% of all actual Leaf Spot samples leading to an f1-score of 0.9774, which balances precision and recall effectively. The proposed model specificity for Leaf Spot is 0.9786 which reflects the proportion of true negatives that were correctly identified. The accuracy of proposed model during training is attained as 0.9773 which exhibits overall performance for this class. For the Leaf Blight class, the model achieved even higher performance with a precision of 0.9795 and a recall of 0.9836 indicating that the model can accurately predict the majority of Leaf Blight samples. The f1-score of 0.9815 shows a high balance between precision and recall. The specificity is 0.9784 ensuring that non-Leaf Blight samples were correctly identified while the accuracy was the highest among all classes as 0.9836. For Leaf Rot the proposed model exhibit better performance and attained a precision of 0.9779 and a recall of 0.9793. The f1-score of 0.9786 exhibit the proposed model performance in balancing both false positives and false negatives. The specificity of the proposed model for leaf rot is 0.9792 and the accuracy of 0.9793 reflects the strong classification performance. Finally for Leaf Curl, precision of 0.9746 and recall of 0.9797 of proposed model is slightly less when compared to others. The f1-score of 0.9771 and specificity of 0.9791 show that the model effectively differentiates leaf curl. With an accuracy of 0.9797, the proposed model exhibits better performance in leaf curl compared to all classes in turmeric leaf disease classification.
The proposed model test performance metrics for all the four classes of turmeric leaf diseases are presented in Table 5. For Leaf Spot, the proposed model achieved a precision of 0.9731 indicating that 97.31% of samples predicted as Leaf Spot were correct. The recall was slightly higher at 0.9771 indicates that the proposed model correctly identified 97.71% of all actual Leaf Spot samples which further leads to an f1-score of 0.9751. The proposed model specificity for Leaf Spot was 0.9784 which exhibit the model ability to correctly classify non-Leaf Spot samples through the accuracy of 0.9771. For Leaf Blight, the model performed well with a precision of 0.9745 and recall of 0.9834 indicates that the model identified 98.34% of the true Leaf Blight samples. This balance between precision and recall resulted in a strong f1-score of 0.9789. The specificity for Leaf Blight was 0.9781 which demonstrates the proposed model ability to minimize false positives and it is validated through the accuracy of 0.9649. In the case of Leaf Rot the model achieved a precision of 0.9764 and a recall of 0.9784 which leads to an f1-score of 0.9774 and exhibit a balanced performance in detecting true positives and minimizing false positives. The specificity was 0.9790 and the accuracy as 0.9747, reflecting the proposed model reliability in classifying this class. Finally, for Leaf Curl, the proposed model exhibit 0.9702 as precision and 0.9794 as recall which leads to an f1-score of 0.9747. The specificity for Leaf Curl was 0.9776 and the accuracy was 0.9646 indicating the better performance. These results demonstrate the proposed model ability to generalize well with high precision, recall, and specificity across all classes during testing confirms its efficiency in turmeric leaf disease detection.
The precision-recall analysis given in Fig. 5 for the proposed model training depicts the better balance between precision and recall for four turmeric leaf disease classes. The precision recall curve exhibits high precision across all classes with slight variations as recall increases. For Leaf Spot the average precision (AP) achieved is 0.9775 indicating that 97.75% of samples predicted as Leaf Spot were correct. The Leaf Blight class shows a slightly higher AP of 0.9795 highlighting the model excellent performance in terms of precision and recall balance. In Leaf Rot the strong classification performance of proposed model is exhibited with an AP of 0.9779 which reflects the minimal false positives and negatives. Leaf Curl attained the lowest AP among the four classes as 0.9746, but it still shows a highly effective model performance. As recall increases towards 1 there is a slight drop in precision for all classes, particularly for Leaf Spot and Leaf Curl but the model maintains high precision above 90% even at higher recall values. This suggests the model is capable of identifying most positive cases across all classes while minimizing false positives and demonstrates better training performance and reliability in classifying turmeric leaf diseases.
The precision-recall analysis is given in Fig. 6 for the proposed model testing for four turmeric leaf disease classes. For Leaf Spot, the average precision (AP) is achieved as 0.9731 which indicates a slight drop in precision compared to the training phase but still reflecting strong classification performance. The proposed model constantly exhibits high precision until recall approaches 0.9 after which a slight decline is observed. For Leaf Blight, the AP is 0.9745 which exhibit a well-balanced performance in terms of precision and recall, with only minor variations across different recall values. The Leaf Rot class which achieves an AP of 0.9764 indicates the model accurately identifies the majority of positive samples with minimal false positives. Leaf Curl while having the lowest AP at 0.9702 exhibit strong overall precision and recall with precision remaining above 90% until recall nears 0.8. Across all classes the model maintains high precision above 95% for most recall values with only minor drops in precision at higher recall levels. These results indicate that the model generalizes well to unseen data maintaining better performance and minimal degradation compared to the training phase.
The overall training and testing performance of the proposed model presented in Table 6 demonstrates its effectiveness in classifying turmeric leaf diseases with a high degree of accuracy. In terms of precision, the model achieved 0.9774 during training indicating that 97.74% of the predictions made by the model were correct. In the testing phase precision slightly decreased to 0.9736 which suggests that the model continues to perform well on unseen data and maintained minimal false positives. Recall for training was 0.9799 which reflects the proposed model ability to correctly identify 97.99% of all actual positive samples with testing recall being almost identical at 0.9796. This indicates a high capability for detecting true positives and ensures strong model generalization between training and testing. The F1-score of 0.9786 in training exhibit the proposed model balance between precision and recall, with a slight drop in the testing F1-score to 0.9766. This small variation indicates consistent performance across both phases and further supports the proposed model ability to minimize both false positives and false negatives. Specificity, which measures the ability of the model to correctly identify negative samples was 0.9788 during training and 0.9783 in testing which demonstrates that the proposed model effectively differentiates diseased and non-diseased samples with minimal errors. Finally, the accuracy of the proposed model reached 0.9790 during training and 0.9703 during testing. While there is a small drop in testing accuracy but the overall performance remains high. These results highlight the proposed model robustness and reliability in classifying turmeric leaf diseases and ensures consistent performance when applied to unseen data.
The precision analysis given in Fig. 7 compares the proposed model performance with existing models such as SVM, Decision Tree (DT), Random Forest (RF), XGBoost (XGB), CNN, AlexNet, and MobileNetV3 over 160 epochs. The proposed model consistently outperforms the others reaching a precision of 0.984 by the 160th epoch. Whereas the existing models MobileNetV3, achieved a precision of 0.971 which is low compared to proposed. In contrast to proposed model, existing DT and SVM show much lower precision 0.948 and 0.956 respectively which indicates its weaker classification ability compared to the proposed model. AlexNet and MobileNetV3 exhibit strong early performance with AlexNet reaching 0.968 at 160 epochs, but they saturate much earlier than the proposed model. CNN and RF models demonstrate an average performance reaching precision values of 0.961 and 0.952, respectively. But these models fail to match the precision of the proposed model as training continues. The proposed model ability to consistently outperform all others compared to proposed hybrid optimized Vision Transformer model.
The recall analysis given in Fig. 8 compares the performance of the proposed model against existing models such as SVM, DT, RF, XGBoost, CNN, AlexNet, and MobileNetV3, across 160 epochs. The proposed model demonstrates its superior performance with a maximum recall of 0.9796 by the 160th epoch which is better than all other models. The existing MobileNetV3 which exhibit recall of 0.971 and AlexNet follows with 0.962 which is lesser than the proposed model. The proposed model recall curve shows consistent improvement throughout the testing indicating its capability to identify the majority of true positives with minimal false negatives. Early in the testing process (around 40 epochs), most models, including RF and XGBoost, exhibit recall around 0.94, but as epoch increases the proposed model and MobileNetV3 show distinct gains. Models like DT and SVM exhibit much lower recall values and reaches maximum recall of 0.933 and 0.941 respectively which is less compared to proposed. The existing CNN and RF exhibit moderate results with recall values of 0.957 and 0.952 respectively which is lesser compared to the proposed model. The merit of the proposed model present in its ability in continuously improving its recall throughout testing and ensures higher sensitivity as well as better classification performance.
The F1-Score analysis of the proposed and existing models given in Fig. 9 depicts that better performance over existing methods. The proposed model consistently outperforms all other models achieving better F1-score of 0.978 at 160 epochs. MobileNetV3 which exhibit F1-score of 0.971, followed by AlexNet at 0.965 which is lesser than the proposed model. The proposed model demonstrates continuous improvement in its F1-score reflecting its ability to balance precision and recall effectively making it well-suited for dealing with both false positives and false negatives. In the initial testing process, XGBoost and RF exhibit low F1-scores between 0.92 and 0.94, whereas RF reached to 0.950. However, the existing model recall performances are lower compared to the proposed model. CNN exhibits moderate performance, peaking at an F1-score of 0.961 while SVM and DT demonstrate the lowest scores struggling to exceed 0.941 and 0.935 respectively. However, the f1-scores of these models are lower when compared to the proposed model F1-score.
The specificity analysis given in Fig. 10 compares the performance of the proposed model with existing models across 160 epochs. Specificity measures the model’s ability to correctly identify negative cases or true negatives which is essential for minimizing false positives in classification tasks. The proposed model consistently demonstrates superior performance achieving a specificity of 0.989 at the 160th epoch. MobileNetV3 which follows closely with a specificity of 0.984 while AlexNet reaches 0.982. The graph shows that the proposed model steadily improves from an initial specificity of 0.943 at 20 epochs and consistently outperforming all other models throughout the testing process. The specificity for models like XGBoost and CNN rises gradually reaches 0.972 and 0.970 respectively. However, the results achieved by the proposed model are much better than the existing models. Models like RF and SVM exhibit relatively lower specificity reaches around 0.965 and 0.962 respectively which indicates a better performance in misclassifying the negative samples compared to the proposed model. DT demonstrates the lowest performance among the models, with a specificity of 0.960 which is lesser than the proposed. The advantage of the proposed model present in its consistent improvements in identifying true negatives while maintaining low false-positive rates making it highly reliable for leaf disease detection process.
The accuracy analysis given in Fig. 11 highlights the superior performance of the proposed model over existing models like SVM, DT, RF, XGBoost, CNN, AlexNet, and MobileNetV3 over 160 epochs. The proposed model exhibits better performance than existing with a high accuracy of 0.978 by the 160th epoch. The existing MobileNetV3 which exhibit accuracy of 0.971 and AlexNet which attained accuracy of 0.965 is lower compared to the proposed model. In contrast, models like XGBoost, CNN, and RF show moderate accuracy of 0.952, 0.961, and 0.950 respectively. The existing SVM and DT exhibit much lower accuracies of 0.941 and 0.935 respectively indicates the model effectiveness in capturing complex patterns within the dataset. The results validate the proposed model’s superior performance and also indicates the use of hybrid optimization techniques and the Vision Transformer architecture in turmeric leaf disease detection.
The overall performance comparison presented in Table 7 for the proposed hybrid optimized vision transformer model and existing models such as SVM, DT, RF, XGBoost, CNN, AlexNet, and MobileNetV3 highlights the superiority of the proposed model across all metrics. The proposed model achieved the highest precision at 0.9736, which overcomes the MobileNetV3 which exhibit 0.971. While the lowest precision was observed in the DT model at 0.948. In terms of recall, the proposed model outperformed all others with 0.9796, demonstrating its effectiveness in correctly identifying positive cases. The F1-score which balances precision and recall was also highest for the proposed model at 0.9766 followed by MobileNetV3 at 0.969 and AlexNet at 0.965. This indicates the proposed model superior balance between precision and recall. In terms of specificity, the proposed model achieved 0.974 which highlights its ability in correctly identify negative cases. The lowest specificity exhibited by DT at 0.958 which indicates more false positives compared to other models. Finally, the proposed model demonstrated the highest overall accuracy at 0.9703, outperforming MobileNetV3, which exhibit 0.968. Also, the proposed model accuracy is significantly higher than the models like SVM and RF which exhibit 0.922 and 0.916, respectively. These results confirm the reliability of the proposed model for turmeric leaf disease detection and exhibit its clear advantage over existing models in terms of classification performance across all metrics.
Conclusion
A novel hybrid optimized Vision Transformer is presented in this research work for turmeric leaf disease detection. The model combines the capabilities of vision transformer for effective feature extraction with the Falcon-Bowerbird optimization (FBO) algorithm to enhance the convergence while optimizing model parameters. Experimental analysis was performed on turmeric leaf diseases dataset and the proposed model demonstrated superior performance across various metrics. Achieving a precision of 0.9736, recall of 0.9796, F1-score of 0.9766, and an accuracy of 0.9703 the proposed model outperforms in turmeric leaf disease detection compared to traditional models such as SVM, DT, RF, and advanced architectures like CNN, AlexNet, and MobileNetV3. The comparative analysis exhibits the proposed model ability in maintain high specificity and minimize false positives making it highly effective for practical application in disease detection. However, the proposed model was tested on turmeric leaf disease images and future research should consider extending the approach to other plant species or diseases to broaden its applicability. Additionally, the FBO optimization improved performance but it increases the computational cost of vision transformer training which is the minor limitation. Future work could focus on optimizing computational efficiency by exploring transfer learning techniques.
Data availability
All data generated or analysed during this study are included in this published article.
Abbreviations
- \(\:p\left(i\right)\) :
-
Probability distribution of pixel intensities
- \(\:x\) :
-
Intensity level
- \(\:c\left(x\right)\) :
-
Cumulative distribution function
- \(\:y\) :
-
Transformed intensity value
- \(\:W\times\:H\) :
-
Width × height
- \(\:{I}_{i,j}\) :
-
Patch
- \(\:i\) and \(\:j\) :
-
Position of the patch in the grid
- \(\:{I}_{\text{patch}}\) :
-
Set of patches
- \(\:G\) :
-
Gradient magnitude
- \(\:{G}_{x}\) and \(\:{G}_{y}\) :
-
Gradient of pixel intensities in the \(\:x\) and \(\:y\) directions
- \(\:\lambda\:\) :
-
Wavelength of the sinusoidal factor
- \(\:\theta\:\) :
-
Orientation
- \(\:\psi\:\) :
-
Offset
- \(\:\sigma\:\) :
-
Standard deviation of the Gaussian
- \(\:\gamma\:\) :
-
Spatial aspect ratio
- \(\:{f}_{i,j}\) :
-
Embedded vector for the patch located at position \(\:\left(i,j\right)\)
- \(\:{W}_{e}\) :
-
Learned embedding matrix
- \(\:{b}_{e}\) :
-
Bias term
- \(\:pos\) :
-
Position index of the patch in the sequence
- \(\:k\) :
-
Dimension index
- \(\:d\) :
-
Total dimensionality of the embedded token
- \(\:{z}_{i,j}\) :
-
Positional-encoded token
- \(\:Q,\:K,\:V\) :
-
Query, Key, Value
- \(\:{W}_{Q}\), \(\:{W}_{K}\), \(\:{W}_{V}\) :
-
Learned projection matrices for the query, key, and value
- \(\:{{\upalpha\:}}_{i,j}\) :
-
Attention score between token \(\:i\) and \(\:j\)
- \(\:{d}_{k}\) :
-
Key vectors dimensionality
- \(\:{V}_{j}\) :
-
Weighted sum of the value vectors
- \(\:h\) :
-
Number of attention heads
- \(\:{W}_{o}\) :
-
Output weight matrix
- \(\:{W}_{1}\), \(\:{b}_{1}\) :
-
Weights and bias for the first linear transformation
- \(\:{W}_{2}\), \(\:{b}_{2}\) :
-
Weights and bias for the second linear transformation
- \(\:{s}_{i}\) :
-
Score for class \(\:i\)
- \(\:\widehat{{y}_{i}}\) :
-
Predicted probability for class \(\:i\)
- \(\:{s}_{i}^{\left(t\right)}\) :
-
Solution of the \(\:{i}^{th}\) bowerbird
- \(\:{s}_{best}^{\left(t\right)}\) :
-
Best solution found in the population
- \(\:{\lambda\:}_{sbo}\) :
-
Indicates the scaling factor in the optimization
- \(\:{{\uptheta\:}}_{t}\) :
-
Updated parameters at iteration \(\:t\)
- \(\:{\eta\:}_{t}\) :
-
Learning rate at iteration \(\:t\)
- \(\:{\nabla\:}_{{\uptheta\:}}L\left({\uptheta\:}\right)\) :
-
Loss function gradient
- \(\:{m}_{t}\) :
-
Moving average of the gradients
- \(\:{s}_{t}\) :
-
Moving average of the squared gradient differences
- \(\:{{\upbeta\:}}_{1}\) and \(\:{{\upbeta\:}}_{2}\) :
-
Exponential decay rates for the first and second moments
- \(ε\) :
-
Constant
References
Sharifi-Rad, J. et al. Turmeric and its major compound Curcumin on health: bioactive effects and safety profiles for food, pharmaceutical, biotechnological and medicinal applications. Front. Pharmacol. 11, 1–23. https://doi.org/10.3389/fphar.2020.01021 (2022).
Archana Jaina, S., Sarsaiya, Q., Wua, Y., Lu, J. & Shi A review of plant leaf fungal diseases and its environment speciation. Bioengineered 10 (1), 409–424. https://doi.org/10.1080/21655979.2019.1649520 (2019).
Ajaikumar, B. et al. Role of turmeric and Curcumin in prevention and treatment of chronic diseases: lessons learned from clinical trials. ACS Pharmacol. Transl. Sci. 14 (6), 447–518 (2023).
Chittabarni Sarkar, D., Gupta, U., Gupta, B. B. & Hazarika Leaf disease detection using machine learning and deep learning: review and challenges. Appl. Soft Comput. 145, 1–28. https://doi.org/10.1016/j.asoc.2023.110534 (2023).
Liu, K. H., Yang, M. H., Huang, S. T. & Lin, C. Plant species classification based on hyperspectral imaging via a lightweight convolutional neural network model. Front. Plant Sci. 13, 1–17. https://doi.org/10.3389/fpls.2022.855660 (2022).
Anuja Bhargava, A. et al. Plant leaf disease detection, classification, and diagnosis using computer vision and artificial intelligence: A review. IEEE Access. 12, 37443–37469. https://doi.org/10.1109/ACCESS.2024.3373001 (2024).
Samuel Manoharan, P. R. & Michael Angelo, J. R. S. A novel hybrid machine learning approach for traffic sign detection using CNN-GRNN. J. Intell. Fuzzy Syst. 44, 1283–1303. https://doi.org/10.3233/JIFS-221720 (2022).
Gopal, S. M. J., Sangeetha, S. T., Muthu, S. S. & Patel, M. S. A hybrid fuzzy based cross neighbor filtering (HF-CNF) for image enhancement of fine and coarse powder scanned electron microscopy (SEM) images. J. Intell. Fuzzy Syst. 42 (6), 6159–6169. https://doi.org/10.3233/JIFS-212561 (2022).
Doutoum, A. S. & Tugrul, B. A review of leaf diseases detection and classification by deep learning. IEEE Access. 11, 119219–119230. https://doi.org/10.1109/ACCESS.2023.3326721 (2023).
Li, L., Zhang, S. & Wang, B. Plant disease detection and classification by deep learning—A review. IEEE Access. 9, 56683–56698. https://doi.org/10.1109/ACCESS.2021.3069646 (2021).
Wasswa Shafik, A., Tufail, A., & Namoun, L. C. A systematic literature review on plant disease detection: motivations, classification techniques, datasets, challenges, and future trends. IEEE Access. 11, 59174–59203. https://doi.org/10.1109/ACCESS.2023.3284760 (2023).
Shovon, M. S. H. et al. PlantDet: A robust multi-model ensemble method based on deep learning for plant disease detection. IEEE Access. 11, 34846–34859. https://doi.org/10.1109/ACCESS.2023.3264835 (2023).
Vishnoi, V. K., Kumar, K., Kumar, B., Mohan, S. & Khan, A. A. Detection of Apple plant diseases using leaf images through convolutional neural network. IEEE Access. 11, 6594–6609. https://doi.org/10.1109/ACCESS.2022.3232917 (2023).
Asad Khattak, M. U. et al. Automatic detection of Citrus fruit and leaves diseases using deep neural network model. IEEE Access. 9, 112942–112954. https://doi.org/10.1109/ACCESS.2021.3096895 (2021).
Vasileios Balafas, E., Karantoumanis, M., Louta, N. & Ploskas Machine learning and deep learning for plant disease classification and detection. IEEE Access. 11, 114352–114377. https://doi.org/10.1109/ACCESS.2023.3324722 (2023).
Anita Shrotriya, A. K., Sharma, S., Prabhu, A. K. & Bairwa An approach toward classifying plant-leaf diseases and comparisons with the conventional classification. IEEE Access. 12, 117379–117398. https://doi.org/10.1109/ACCESS.2024.3411013 (2024).
Manjula, R., Chougala, A. C. & Ramachandra Measurement of leaf spot disease severity in turmeric plant. Int. J. Res. Eng. Sci. Manag. 3 (12), 164–167. https://doi.org/10.47607/ijresm.2020.429 (2020).
Thangavel.R, K. M. A robust approach for identification and classification of turmeric plant leaf diseases using LDA-ANFIS algorithm. Int. J. Res. Appl. Sci. Eng. Technol. 11 (3), 1474–1481. https://doi.org/10.22214/ijraset.2023.49717 (2023).
Laxmi, U. S., Kathawate, G. & Samata Gadde, S. W. B. Recent turmeric plants agronomy analysis and methodology using artificial intelligence. Int. J. Bot. Stud. 7 (2), 233–236 (2022).
Devi Priya, D. V. & Anitha.N, R. Early detection of major diseases in turmeric plant using improved deep learning algorithm. Bull. Pol. Acad. Sci. Tech. Sci. 70 (2), 1–10. https://doi.org/10.24425/bpasts.2022.140689 (2022).
Patil, S. A., Mahadik, S. R. & Bhosale, A. G. Rhizome rot disease recognition in turmeric plant using CNN-SVM hybrid algorithm. Afr. J. Biomed. Res. 27 (3), 295–305. https://doi.org/10.53555/AJBR.v27i3S.1972 (2024).
Khalid, M. et al. Multi-class classification of plant leaf diseases using feature fusion of deep convolutional neural network and local binary pattern. IEEE Access. 11, 62307–62317. https://doi.org/10.1109/ACCESS.2023.3286730 (2023).
Emmanuel Moupojou, F. et al. Segment anything model and fully convolutional data description for plant multi-disease detection on field images. IEEE Access. 12, 102592–102605. https://doi.org/10.1109/ACCESS.2024.3433495 (2024).
Chunduri, M. & Jubilson, E. A. YR2S: efficient deep learning technique for detecting and classifying plant leaf diseases. IEEE Access. 12, 3790–3804. https://doi.org/10.1109/ACCESS.2023.3343450 (2024).
Rubina, R., Aslam, W., Aziz, R. & Aldehim, G. An early and smart detection of corn plant leaf diseases using IoT and deep learning Multi-Models. IEEE Access. 12, 23149–23162. https://doi.org/10.1109/ACCESS.2024.3357099 (2024).
Zhang, X., Mao, Y., Yang, Q., & Zhang, X. A plant leaf disease image classification method integrating capsule network and residual network. IEEE Access. 12, 44573–44585. https://doi.org/10.1109/ACCESS.2024.3377230 (2024).
Paramanandham, N., Sundhar, S., & Priya, P. Enhancing disease detection with weight initialization and residual connections using leafnet for groundnut leaf diseases. IEEE Access. 12, 91511–91526. https://doi.org/10.1109/ACCESS.2024.3422311 (2024).
Funding
The author did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
All the authors contributed to this research work in terms of concept creation, conduct of the research work, and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Selvaraj, R., Devasena, M.S.G. A novel attention based vision transformer optimized with hybrid optimization algorithm for turmeric leaf disease detection. Sci Rep 15, 17238 (2025). https://doi.org/10.1038/s41598-025-02185-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-02185-7