Abstract
Mallotus furetianus is traditionally used as folk medicine on Hainan Island, China. Given the significance of the pharmacokinetic interaction between herbs and drugs, we investigated the effects of Mallotus furetianus ethanol extract (MFE) on CYP3A activity in rats. The major bioactive constituents of MFE, gallic acid (GA) and epigallocatechin gallate (EGCG), were analyzed by Reverse Phase High-Performance Liquid Chromatography (RP-HPLC) for MFE standardization. Rats were orally administered 320 mg/10mL/kg MFE or water (control) once a day for 7 days. Two hours after the last MFE treatment, the rats were euthanized, and the livers and small intestines were excised. The activity of CYP3A was measured in hepatic and intestinal microsomes, and the expression in hepatic and intestinal tissues was assessed by qRT-PCR and western blot. In the pharmacokinetics experiment, rats were administered MFE as described above. Two hours after the final dose of MFE, a 15 mg/kg midazolam solution was orally administered. Blood samples were collected before and after midazolam administration. Midazolam plasma concentration and 1-hydroxymidazolam formation in microsomes were measured by LC-MS/MS methods.CYP3A activity in hepatic microsomes exhibited a significant decrease in MFE treatment group, while no change was observed in intestinal microsomes. MFE treatment significantly increased CYP3A1 activity, as well as mRNA and protein expression in the small intestine but reduced these parameters in the liver. However, CYP3A2 remained unaffected by MFE treatment. Pharmacokinetics study showed that administration of MFE for 7 days significantly reduced the Cmax of midazolam from 919 ± 70 ng / mL to 708 ± 91 ng /mL. Conversely, t1/2 was increased from 0.45 ± 0.08 h in the control group to 0.69 ± 0.15 h in the MFE group.Collectively, our study indicated that MFE could modulate CYP3A1 activity and expression of CYP3A1, subsequently changing the metabolism of midazolam in rats.
Similar content being viewed by others
Introduction
The significance of pharmacokinetic interaction between herb and drugs has attracted considerable attention in recent years. By modulating the activity of metabolic enzymes or/and transporters, active constituents in the herb extracts may compromise the therapeutic efficacy of a drug or lead to unexpected side effects1. Drug interactions can occur atdifferent stages, including absorption, metabolism, excretion, and protein binding, with the main reason being changes in the activity of cytochrome P450 enzyme (CYP450). In clinical practice, over 90% of drugs are metabolized by CYP450 enzymes (including CYP1A, CYP2B, CYP2C, CYP2D, and CYP3A, etc.).Approximately 60% of these drugs are metabolized by CYP3A, which is mainly expressed in the liver and small intestine2. Recently, severalCYP3A substrates have been commonly used as “probes” to study the effects of other drugs or foods on CYP3A activity. Midazolam is a short-acting benzodiazepine sedative hypnotic drug that is primarily metabolized by CYP3A after oral administration. Sinceit is not a P-glycoprotein substrate, Midazolam is an ideal probe tostudy the effects of drugs or food on CYP3A activity, bothin vivo and in vitro3.
Mallotus furetianus is a tropical plant widely distributed on Hainan Island, the southernmost province of China. The leaves of Mallotus furetianus have been used traditionally in folk medicine, and is also popularly known as partridge teaand an aromatic beverage4. From Traditional Chinese Medicine perspective, it is believed that the leaves of Mallotus furetianus possess the ability to resolve food stagnation, clear summer-heat, strengthen the spleen, and nourish the stomach5. Extract of Mallotus furetianusis reported to exhibit biological activities such as choleretic effects6, anti-atherosclerotic properties7, anti-steatosis effects8, free radical scavenging, liver fibrosis protection9,10, and antiobesity effects11, indicating its potential in medicine development.
The solvent extraction of Mallotus furetianus contains various chemical constituents, such as polyphenols, organic acids and esters, polysaccharides, amino acids, terpenes, glycosides, and inorganic elements12,13,14. Hirai et al. have demonstrated that epigallocatechin gallate (EGCG), the primary polyphenolic compound found in Mallotus furetianus extract (MFE), irreversibly inhibits CYP3A4 activity15. However, it remains unclear whether MFE affects metabolism by regulating CYP3A.Considering the role of CYP3A in herb-drug interactions, understanding the effect of MFE on CYP3A enzyme activity and expression is of significant importance to ensure its safety as a folk medicine and beverage.
Rat CYP3A mainly includes five sub-enzymes, CYP3A1/CYP3A23,CYP3A2, CYP3A9, CYP3A18, and CYP3A62.These five sub-enzymes sharecharacterics of similar molecular weight, immunohistochemistry properties, and substrate specificities16. Among them, CYP3A1 and CYP3A2 are main isoformsin the rat liver, and rat CYP3A1 shares approximately 73% protein homology with human CYP3A4. Thus, this studyfocuses on CYP3A1 and CYP3A2, the key CYP3A proteins in rats17. Notably, rat CYP3A1 and CYP3A2 collectively perform functions analogous to human CYP3A4, making rat models valuable for preclinical drug metabolism studies. In the current study, we investigated the effects of MFE on CYP3A activity and expression in rats. We also examined how MFE affects CYP3A metabolism of its probe substrate midazolam. Our findings reveal the potential interaction between MFE and drugs through its effects on CYP3A1 and CYP3A2.
Materials and methods
Chemicals
Midazolam Maleate Tablets (15 mg) were purchased from Jiangsu Nhwa Pharmaceutical Co., Ltd.(Jiangsu, China), and 1-hydroxymidazolam was purchased from Shanghai Acmec Biochemical Co.,Ltd. (Shanghai, China). All other chemicals and reagents were of the highest grade commercially available.
Preparation of MFE
The leaves of Mallotus furetianusused in this study were collected from Hainan Island, China. The air-dried powdered leaves were extracted twice with 8 weight of 95% ethanol. After filtration, the solution was evaporated under reduced pressure to yield dry powder with an extraction efficiency of 11.7%. The dried extract (MFE) was stored in a sealed container at 4 °C until future use.
High-Performance liquid chromatography (HPLC)
Accurately weigh 124.1 mg of MFE, add 20 mL of 70% ethanol solution, and perform ultrasonic extraction for 30 min in a 55 °C water bath. Then the solution was centrifuged at 3000 rpm for 10 min. The supernatant was collected and filtered through a 0.45-µm membrane for reversed-phase HPLC (RP-HPLC) analysis. The analysis was performed using an LC-2010AHT HPLC system (Shimadzu, Japan) and a C18 chromatographic column (CAPCELL PAK C18 MG S-5, 4.6 mm × 250 mm, 5 μm). The mobile phase was a mixture of methanol (A) and 0.1% formic acid solution (B), with a gradient elution as follows: 0–8 min, 19% (A); 8–35 min, 50% (A); 35–36 min, 100% (A); 36–40 min, 19% (A). Detection was done at a wavelength set at 278 nm, a column temperature maintained at 30 °C, a flow rate set to 1.0 ml/min, and an injection volume of 10 µL.
Animals experiment and tissue sampling
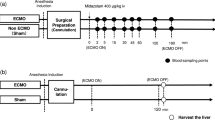
Male Sprague Dawley rats, aged 10 weeks and weighing 200–250 g, were supplied by the Experimental Animal Center of Hainan Medical University. All rats were housed in a temperature-controlled room with a 12-hour light/12-hour dark cycle, provided with a standard rodent diet and tap water. All experimental protocols were approved by the Animal Care and Use Committee of Hainan Medical university (NO.HYLL-2021-094) and ARRIVE guidelines (https://arriveguidelines.org). All methods were performed in accordance with the relevant guidelines and regulations. The anesthetic used in this experiment was Zoletil 50 ® 50, produced by VIRBAC, a French company. The powder was dissolved in sterile water in the provided vial at a final concentration of 50 mg/ml. Anesthetics were intraperitoneally injected into rats at a dose of 75 mg/kg and a volume of 1.5 ml/kg. CO2 inhalation was used for euthanasia.
For the CYP3A activity and gene expression study, rats were divided into two groups (n = 5 per group). The experimental groups were orally administered MFE (320 mg/kg body weight in 10 mL water) or water (control) once a day for 7 days. Two hours after the final administration, rats were euthanized and the liver and small intestine were excised. Hepatic and intestinal microsomes were prepared by a conventional fractional centrifugation method described previously18. Briefly, tissues were weighed, cut into smaller pieces, and mixed with phosphate-buffered saline (PBS) in a 1:3 weight-to-volume ratio. The mixture was homogenized using an automatic homogenizer, then centrifuged at 10,000 g for 15 min. The supernatant was collected and further centrifuged at 100,000 g for 60 min. The resulting pellet was re-suspended in 100 mM potassium phosphate buffer and stored at − 80 °C until use. All procedures were conducted at temperatures below 4 °C, and protein concentrations were measured by the Lowry method.
In the pharmacokinetics experiment, rats were randomized into two groups (n = 5 per group), and administered MFE as described above. Two hours after the final dose of MFE, a 15 mg/kg midazolam solution was orally administered. To prepare midazolam solution, eight tablets of midazolam maleate were finely ground, dissolved in 60 mL of 5% Tween-80 solution, and filtered through a 0.45 μm membrane filter. The filtrate was analyzed for midazolam content according to the Chinese Pharmacopoeia (2020 edition), then adjust the solution to a final concentration of 1.5 mg/mL. Blood samples (0.2 mL) were collected from the jugular vein at baseline and 15, 30, 45, 60, 90, 120, 180, and 240 minutespost-midazolam administration using a catheterization technique. The procedure was performed under Zoletil 50® anesthesia (75 mg/kg, intraperitoneally) to minimize stress and discomfort in the animals. The samples were centrifuged at 15,000 g for 10 min at 4 °C, and the collected plasma was stored at − 80 °C until analysis.
CYP3A activity assay
The activity of CYP3A was measured in hepatic and intestinal microsomes prepared from rats treated with MFE for seven days. The reaction mixtures consisted of 80 mM phosphate buffer (pH 7.4), hepatic (100 µg) or intestinal (200 µg) microsomes protein, 10µM midazolam, and 1 mM nicotinamide adenine dinucleotide phosphate (NADPH) in a final volume of 200 µL. After incubation at 37 °C for 30 min, the reactions were quenched by adding 100 µL cold acetonitrile to measure the 1-hydroxymidazolam formation as a marker of CYP3A activity, as described previously19. The activities of CYP3A1 and CYP3A2 were measured using ELISA kits (Aviva Systems Biology, USA) according to the manufacturer’s instructions.
Pharmacokinetic analysis
The pharmacokinetic parameters of midazolam were calculated by non-compartmental analysis using the add-in program PKSolver for Microsoft Excel. The area under the curve (AUC) was calculated using the log-linear trapezoidal method, the apparent elimination rate constant (ke) was determined from the slope of the elimination phase, and the elimination half-life (t1/2) was calculated as ln2/ke. The maximum plasma concentration (Cmax) and the time to reach the maximum plasma concentration (Tmax) were directly obtained from the observed data.
Liquid chromatography with tandem mass spectrometry (LC-MS/MS)
Midazolam plasma concentration and 1-hydoxymidazolam formation in microsomes were measured by LC-MS/MS methods. Each 50 µL plasma sample was extracted with 440 µL acetonitrile for 2 min in the presence of 10 µL diazepam (50ng/mL) as an internal standard. After centrifuging at 15,000 g for 10 min at 4 °C, 2 µL of the supernatant was used for LC-MS/MS analysis. For determination of 1-hydroxymidazolam formation in microsomes, a 200 µL reaction mixture was quenched by 100 µL cold acetonitrile containing diazepam as an internal standard. After vortexing for 2 min, the mixture was then centrifuged at 15,000 g for 10 min at 4 °C. Two microliters of the supernatant was used for measuring 1-hydroxymidazolam.
The separation of midazolam, 1-hydroxymidazolam, and internal standard diazepam was achieved by using an Agilent C18 column (50 × 4.6 mm, 2.7 μm) with a mobile phase gradient (A: 2 mmol·L-1 ammonium acetate solution containing 0.1% formic acid; B: acetonitrile solution containing 0.1% formic acid). The gradient elution was as follows: 0.01–2.20 min, 30–45% (B); 2.20–4.00 min, 45–90% (B); 4.00–7.00 min, 90–90% (B); 7.00–7.10 min, 90 − 30% (B); 7.10–10.00 min, 30–30% (B). The flow rate was 0.3 mL/min, and the total run time was 10 min.Quantification of midazolam (m/z 326.1 → 291.1) and 1-hydroxymidazolam (m/z 342.11 → 324.1) was performed using diazepam (m/z 285.1 → 154.0) as aninternal standard with a dwell time of 100 ms. The extracted ion chromatograms of midazolam, 1-hydroxymidazolam, and diazepam reference standards are shown in supplementary materials (Supplementary materials 1–7).
Real-time polymerase chain reaction (PCR) analysis
Approximately 20 mg of liver and intestinal tissues from sacrificed rats was used to determine the expression of CYP3A mRNA. Total RNA was isolated using Total RNA Extraction Kit (R1200, Solarbio) according to the manufacturer’s protocol. The quantity and quality of RNA samples were determined by the 260:280 nm absorbance ratio. The RNA samples were reverse transcribed with iScript™ cDNA Synthesis Kit (1708890, Bio-Rad Laboratories). Real-time PCR was performed using an Applied Biosystems 7500 real-time PCR system. The relative expression was calculated using the 2−ΔΔCT method, and normalized to mRNA levels of β-actin. Primer sequences are listed in Table 1.
Western blot analysis
Hepatic and intestinal tissues were homogenized using a Histocyte Total Protein Extraction Kit (P1250-50, Applygen) containing protease inhibitors (P8340, Sigma-Aldrich). After centrifugation at 12,000 g, 4 °C for 10 min, the protein concentration in the supernatant was determined using a Protein Assay Dye Reagent Concentrate (5000002, Bio-Rad Laboratories). Total protein was fractionated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane by Mini Trans-Blot cell apparatus (Bio-Rad Laboratories) according to the manufacturer’s protocol. The membrane was blocked with 5% skim milk in TBST for 1 h and incubated with primary antibodies of anti-CYP3A1 (ab22724,Abcam), anti-CYP3A2 (P3A2PT, Detroit R&D), or anti-GAPDH (TA-08, Zhongshan Jinqiao Bio) overnight at 4 °C. After washing 3 times with TBST for 15 min total, the membrane was incubated with an IRDye second antibody (LI-COR) at room temperaturefor 1 h. After three additional washes with TBST, immunoblots were visualized using an Odyssey infrared imaging system (LI-COR Biosciences).
Statistical analysis
Experimental data are expressed as mean ± standard deviation (SD). Statistical analyses was done using GraphPad Prism 5.0 (GraphPad Software Inc., Boston, MA, USA). A paired t-test was used to analyze the differences between experimental groups. P < 0.05 was considered statistically significant.
Results
Standardization of MFE
MFE was standardized using RP-HPLC analysis using the external standard method. The components EGCG and gallic acid (GA) are identified by comparing their retention times (Rt) with those of the standard mixture. A series of mixed standard concentrations was used for linear regression, and the content of each component was calculated based on the linear regression equation.The retention times for GA and EGCG are 12.212 min and 32.309 min, respectively. The linear regression equations for GA and EGCG are Y = 3 × 106 X + 18,451, R² = 0.999 and Y = 1 × 106 X + 15,719, R² = 0.999, respectively. GA and EGCG show a strong linear relationship in the concentration ranges of 0.05 to 1.60 mg/mL and 0.06 to 1.92 mg/mL, respectively (Fig. 1A and B). Based on the chromatographic peak areas in Fig. 1B, the content of GA and EGCG in MFE were calculated as 10.62% and 31.15%, respectively.
Effects of MFE treatment on CYP3A activity
After orally administered with 320 mg/10 mL/kg of MFE for 7 days, hepatic and intestinal tissues were excised from rats and CYP3A activities were analyzed. Compared to the control group, rats treated with MFE for 7 days exhibited an increase in CYP3A activity in intestinal microsomes (Fig. 2A). Conversely, MFE treatment significantly reduced CYP3A activity in the liver (Fig. 2B).To further investigate the subtype specificity of CYP3A activity modulation by MFE in the small intestine and liver, we conducted ELISA experiments to distinguish whether CYP3A1 or CYP3A2 activity was affected. The results showed that MFE increased CYP3A1 activityin intestinal microsomes and reduced CYP3A1 activity in the liver compared to the control group (Fig. 2C, D), indicating the regulatory effect of MFE on CYP3A1 activity in the liver and small intestine of rats.
Effects of MFE on CYP3A activity in the small intestine and liverof rats. (A, B) Rats were treated with MFE at 320 mg/10 mL/kg or water at 10 mL/kg for 7 days. The activity of CYP3A was evaluated by the formation rate of 1-hydroxymidazolam.(C, D) CYP3A1 and CYP3A2 assay. Bars represent the mean ± S.D. (n = 5). * p < 0.05,** p < 0.01 compared with the control group.
Effects of MFE treatment on CYP3A expression
We investigated the effects of MFE treatment on CYP3A expression by measuring mRNA levels of CYP3A1 and CYP3A2 in rat. As shown in Fig. 3A and B, compared to the control group, MFE significantly increased CYP3A1 mRNA level in small intestine of rat while decreasing CYP3A1 mRNA level in the liver. To further confirm the induction of CYP3A1 by MFE treatment, we conducted a western blot analysis (Fig. 3C-E). Consistent with the mRNA results, MFE significantly elevated the protein level of CYP3A1 in small intestine while decreasing its liver expression (Fig. 3C-E). We also assessed the effects of MFE treatment on CYP3A2 expression at the mRNA and protein levels in the liver and small intestine. As depicted in Fig. 3A, B,F, and G, MFE had no effects on CYP3A2 expression.
Effects of MFE on the expression of CYP3A. Rats were treated with MFE at 320 mg/10 mL/kg or water at 10 mL/kg for 7 days. The mRNA levels of CYP3A1 and CYP3A2 in the small intestine (A) and liver (B) were detected by real-time PCR (n = 5). Western blot was performed to determine the CYP3A1 and CYP3A2 protein levels in the small intestine (C, D,F) and liver (E, G)(n = 4). Bars represent the mean ± S.D. * p < 0.05,** p < 0.01 compared with the control group.
Effects of MFE pre-treatment on Midazolam pharmacokinetics
Following a 7-day pre-treatment with MFE, plasma concentration of midazolam was determined using the LC-MS/MS method. The plasma concentration-time profiles and the pharmacokinetic parameters of midazolam are presented in Fig. 4; Table 2, respectively. Compared to the control group, MFE pre-treatment significantly reduced Cmax of midazolam, decreasing it from 919 ± 70 ng/mL to 708 ± 91 ng/mL. Conversely, MFE pre-treatment significantly increased t1/2 of midazolam, which was 0.45 ± 0.08 h in the control group, to 0.69 ± 0.15 h in the MFE group. The AUC0−∞ of midazolam in rats showed no significant difference between the two groups; however, it was 12% higher in the MFE pre-treatment group when compared to the control group. Additionally, the Tmax in the MFE-treated group was significantly reduced to 15 min, compared to 30 min in the control group, indicating potential alterations in midazolam absorption dynamics.
Discussion
In this study, we investigated the effects of MFE on the activity of CYP3A1 in rats to explore the potential herb-drug interactions when Mallotus furetianus is used as a folk medicine. Its potentially beneficial effects on several disease models have been reported6,7,8,9,10,11, however the pharmacokinetic profile of MFE particularly on CYP3A activities has yet to be determined. Our results suggested that MFE treatment decreases CYP3A1 activity in the rat liver. Polyphenols are the primary components of MFE, making up approximately 47% of the extract’s dry weight20. EGCG is the predominant polyphenol present in MFE. Our results show that the EGCG content in MFE is about 30%, consistent with the findings reported by Li et al.21. Previous studies have suggested that EGCG moderately inhibits CYP3A activity in human liver microsomes and alters the pharmacokinetics of nicardipine in rats by inhibiting CYP3A activity15,22. These findings align with the inhibitory effects on CYP3A in the liver observed in our current study, suggesting that EGCG is likely to be the chemical basis underlying the CYP3A inhibitory effects of MFE.
While MFE significantly inhibited CYP3A activity in rat liver microsomes, we found that CYP3A activity was notably increased in the intestinal microsomes by MFE treatment. One possible explanation is that EGCG, being a weak inhibitor of CYP3A, may not achieve sufficient concentration in the intestine following MFE administration, thus remaining below the threshold required to effectively inhibit CYP3A activity. This hypothesis requires further research, perhaps byquantifying EGCG levels in the intestine. Another more likely explanation is that MFE may alter CYP3A expression in rats, leading to higher content and increased activity. The primary mechanism of CYP3A induction is receptor-mediated transcriptional activation. PXR, predominantly expressed in the liver and intestine, is well-known for regulating CYP3A4 expression by binding to sites upstream of the gene23. It has been reported that dietary polyphenols can alter PXR-DNA binding ability, thereby modulating the expression and activity of CYP3A in rats24,25,26,27. Through ELISA, Real-time PCR and western blot analysis, our studies demonstrated that MFE significantly increasedactivity and expression of CYP3A1 in the small intestine, likely explaining the increased CYP3A activity. Additionally, our study showed that MFE treatment resulted in a significant decrease in activity and expression of CYP3A1 in the liver, possibly through both the inhibition of CYP3A1 activity and the downregulation of its expression.
Midazolam is a well-established substrate of CYP3A and is commonly used to assess its activity both in vitro and in vivo. Our study indicates that MFE can modify the metabolic profile of midazolam. Administering MFE for 7 days results in a significant reduction in the Cmax and a decrease in Tmax of midazolam, suggesting an accelerated absorption process. Since the small intestine contributes to the first-pass metabolism of midazolam via mucosal CYP3A428, the change in Cmax may be attributed to increased CYP3A activity in the small intestine, subsequently enhancing the first-pass effect of midazolam. On the other hand, the t1/2 of midazolam was prolonged by MFE pre-treatment, suggesting the inhibition of CYP3A enzymes in the liver impairs midazolam elimination in rats. Although the AUC0−∞ did not change significantly, this is likely due to the combined effect of the increased t1/2 and decreased Cmax. If midazolam were administered via the intravenous route, the induction of CYP3A in the small intestine will no longer affect the value of Cmax, potentially leading to a significant increase in AUC0−∞. To confirm this hypothesis, further studies comparing the pharmacokinetics of intravenous vs. oral midazolam administration following MFE treatment are required. Such studies would help clarify the relative contributions of hepatic and intestinal CYP3A modulation to the observed pharmacokinetic changes.
As a commonly used folk medicine and beverage, Mallotus furetianus has recently been studied exploring its health benefits and chemical constituents. However, its impact on drug-metabolizing enzymes and potential herb-drug interactions has not received sufficient attention. Our study is the first to provide insights into the effects of MFE on CYP3A1 activity and its subsequent effects on midazolam metabolism. In this study, we administered MFE at a dose of 320 mg/kg to rats, which is equivalent to an intake of about 30 g of Mallotus furetianus leaves in human. Thisis a common dose when Mallotus furetianusis used as folk medicine. The study by Lin et al. on its protective effects on liver fibrosis also describes similar dosage10.
The CYP3A gene is responsible for midazolam metabolism in both humans and rats, and it also shares similar regulatory mechanisms in gene expression and activity29. Human CYP3A enzymes, predominantly CYP3A4, and rat CYP3A isoforms, particularly CYP3A1 and CYP3A2, metabolize a broad spectrum of drugs, catalyzing similar oxidative reactions, such as hydroxylation, demethylation, and epoxidation. CYP3A4 accounts for approximately 30% of total human liver CYP450 content and is the most abundant hepatic CYP450 isoform involved in the metabolism of over 50% of clinical drugs30. Similarly, rat hepatic CYP3A enzymes, especially CYP3A1, have been extensively studied in non-clinical drug metabolism research, with findings frequently applied to predict drug metabolism changes in human clinical contexts31. CYP3A1, the rat orthologue of human CYP3A4, demonstrates about 73% amino acid homology32, making it the most metabolically relevant rat isoform for comparative studies33. Rat CYP3A2, which exhibits approximately 88% sequence identity with CYP3A1, is also considered important due to its homology with human CYP3A4534. However, subtle yet important differences exist; CYP3A1 expression is inducible and closely resembles the inducible nature of human CYP3A4, whereas CYP3A2 is predominantly expressed constitutively in male rats, influenced significantly by hormonal regulation35. These species-specific and sex-specific variations can lead to quantitative differences in enzyme activity and substrate affinity, potentially impacting drug pharmacokinetics and toxicity predictions. Therefore, further studies involving human primary hepatocytes from both sexes are required to elucidate the effects of Mallotus furetianus on human CYP3A4 activities and potential interactions with prescription medications.
While our findings in rats provide valuable mechanistic insight into the modulation of CYP3A1 by MFE, other limitations should be acknowledged. First, the use of a single dose, although representative of traditional medicinal usage of MFE, limits our ability to define dose–response relationships or therapeutic thresholds for enzyme modulation. Additionally, the relatively small sample size (n = 5 per group) may constrain statistical power and increase variability in the pharmacokinetic and molecular readouts. Our experimental design also did not address potential sex-based differences, as only male animals were included. Moreover, although we focused on CYP3A1 and CYP3A2 due to their relevance as orthologs of human CYP3A4, we did not assess the potential effects of MFE on other drug-metabolizing enzymes or transporters, which may contribute to herb–drug interactions. Therefore, our findings should be interpreted as preliminary and hypothesis-generating. Future studies including investigation supported by multiple doses, sex-balanced and larger sample sizes, broader enzyme profiling, and human-derived models to fully define the clinical implications of MFE consumption.
In summary, this study demonstrates that Mallotus furetianus extract exerts differential regulatory effects on CYP3A1 in the liver and small intestine, altering the metabolism of the CYP3A substrate midazolam in rats. These findings highlight a potential for herb–drug interactions when co-administering MFE with CYP3A-metabolized medications. While preliminary, our study suggests that MFE could modulate the activity and expression of CYP3A1, altering the metabolism of midazolam in rats.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Ased, S., Wells, J., Morrow, L. E. & Malesker, M. A. Clinically significant Food-Drug interactions. Consult Pharm. 33 (11), 649–657 (2018).
Hakkola, J., Hukkanen, J., Turpeinen, M. & Pelkonen, O. Inhibition and induction of CYP enzymes in humans: an update. Arch. Toxicol. 94 (11), 3671–3722 (2020).
Klyushova, L. S., Perepechaeva, M. L. & Grishanova, A. Y. The role of CYP3A in health and disease. Biomedicines 10 (11), 2686 (2022).
Wu, K. Liu Y,QiuX,etal. Development of quality standard for Mallotus furetianus (Bail) Muell-Arg.; a native traditional medicine in Hainan. Chin. J. Trop. Agric. 43 (01), 102–106 (2023).
Zhou, S. J., Zhang, M. & Lin, D. B. Impact of Zhegu tea on the hypolipidemic effect and intestinal flora in patients with hyperlipidemia of blockage of turbid phlegm syndrome. J. Guangzhou Univ. Tradit Chin. Med. 40 (02), 300–308 (2023).
Song, J. & Zhang, Z. Research progress on chemical components and Pharmacological action of Mallotus furetianus. Chin. J. Hosp. Pharm. 36 (02), 152–156 (2016).
Yueli, L., Liqun, W., Haitao, W., Lianbo, L. & Xinan, Y. Comparison of anti-atherosclerotic effects of two different extracts from leaves of Mallotus furetianus. Asian Pac. J. Trop. Med. 4 (11), 878–882 (2011).
Huang, X. et al. Anti-steatosis compounds from leaves of Mallotus furetianus. Nat. Prod. Res. 32 (12), 1459–1462 (2018).
Yoshikawa, E., Matsui-Yuasa, I., Huang, X., Kobayashi, Y. & Kojima-Yuasa, A. Mallotus furetianus extract protects against ethanol-induced liver injury via the activation of the cAMP-PKA pathway. Food Sci. Nutr. 8 (7), 3936–3946 (2020).
Lin, D. et al. Efficacy and mechanism of Mallotus furetianus müll. Arg. Extract on nonalcoholic fatty liver disease. Evid. Based Complement. Alternat Med. 2022, 4897463 (2022).
Nakano, T. et al. The suppression of the differentiation of adipocytes with Mallotus furetianus is regulated through the posttranslational modifications of C/EBPβ. Food Sci. Nutr. 11 (10), 6151–6163 (2023).
Lin, L. B. et al. [Studies on chemical constituents in leaves of Mallotus furetianus I]. Zhongguo Zhong Yao Za Zhi. 31 (6), 477–479 (2006).
ChenDL, Zheng, W. & FengJ,Liu, Y. Y. Chemical constituents from Mallotus furetianus. Chin. Tradit Herb. Drugs. 48 (23), 4851–4855 (2017).
Li, S. et al. Screening and identification of natural α-glucosidase and α-amylase inhibitors from Partridge tea (Mallotus furetianus Muell-Arg) and in Silico analysis. Food Chem. 388, 133004 (2022).
Hirai, T., Nishimura, Y. & Kurata, N. etal. Effect of Benifuuki tea on cytochrome P450-mediated metabolic activity in rats. Vivo 32 (1), 33–40 (2018).
Matsubara, T. et al. Isolation and characterization of a new major intestinal CYP3A form, CYP3A62, in the rat. J. Pharmacol. Exp. Ther. 309 (3), 1282–1290 (2004).
Moon, Y. J. et al. Effects of acute renal failure on the pharmacokinetics of Chlorzoxazone in rats. Drug MetabDispos. 31 (6), 776–784 (2003).
Li, L. & Chen, J. The Experimental Technologies of Modern Toxicology, Principles and Methodspp. 110–111 (Chemical Industry, 2006).
Li, W. L., Xin, H. W. & Su, M. W. Inhibitory effects of continuous ingestion of schisandrin A on CYP3A in the rat. Basic. Clin. PharmacolToxicol. 110 (2), 187–192 (2012).
DuanZ,Li, P. & He, A. Antioxidant and bacteriostasis activity of flavanoid from Mallotus oblongifolius by different extraction methods. Chin. J. Trop. Crops, 42(3), 847–853 (2021).
Li, Y. J. & Wang, Y. G. Y. J. MaX.Y,ZhangYY. Antibacterial activity and composition analysis of Mallotus oblongifolius extraction. Sci. Technol. Food Ind. 35 (10), 202–204 (2013).
Choi, J. S. & Burm, J. P. Effects of oral Epigallocatechin gallate on the pharmacokinetics of Nicardipine in rats. Arch. Pharm. Res. 32 (12), 1721–1725 (2009).
Lolodi, O., Wang, Y. M., Wright, W. C. & Chen, T. Differential regulation of CYP3A4 and CYP3A5 and its implication in drug discovery. Curr. Drug Metab. 18 (12), 1095–1105 (2017).
Kluth, D., Banning, A., Paur, I., Blomhoff, R. & Brigelius-Flohé, R. Modulation of pregnane X receptor- and electrophile responsive element-mediated gene expression by dietary polyphenolic compounds. Free Radic Biol. Med. 42 (3), 315–325 (2007).
Ikarashi, N., Ogawa, S., Hirobe, R. & etal High-dose green tea polyphenol intake decreases CYP3A expression in a liver-specific manner with increases in blood substrate drug concentrations. Eur. J. Pharm. Sci. 89, 137–145 (2016).
Ikarashi, N. et al. Epigallocatechin gallate induces a hepatospecific decrease in the CYP3A expression level by altering intestinal flora. Eur. J. Pharm. Sci. 100, 211–218 (2017).
Li, X. et al. -)-Epigallocatechin-3-gallate (EGCG) inhibits starch digestion and improves glucose homeostasis through direct or indirect activation of PXR/CAR-mediated phase II metabolism in diabetic mice. Food Funct. 9 (9), 4651–4663 (2018).
Paine, M. F. et al. First-pass metabolism of Midazolam by the human intestine. Clin. Pharmacol. Ther. 60 (1), 14–24 (1996).
Qin, X. & Wang, X. Role of vitamin D receptor in the regulation of CYP3A gene expression. Acta Pharm. Sin B. 9 (6), 1087–1098 (2019).
Lu, J. et al. CRISPR knockout rat cytochrome P450 3A1/2 model for advancing drug metabolism and pharmacokinetics research. Sci. Rep. 7, 42922 (2017).
Kusaba, J., Kajikawa, N., Kawasaki, H., Kurosaki, Y. & Aiba, T. Comparative study on altered hepatic metabolism of CYP3A substrates in rats with glycerol-induced acute renal failure. Biopharm. Drug Dispos. 33 (1), 22–29 (2012).
Sun, Z. et al. BDE47 induces rat CYP3A1 by targeting the transcriptional regulation of miR-23b. Sci. Rep. 6, 31958 (2016).
Sevrioukova, I. F. & Poulos, T. L. Interaction of human cytochrome P4503A4 with Ritonavir analogs. Arch. Biochem. Biophys. 520 (2), 108–116 (2012).
Basheer, L., Schultz, K. & Kerem, Z. Inhibition of cytochrome P450 3A by acetoxylated analogues of Resveratrol in in vitro and in Silico models. Sci. Rep. 6, 31557 (2016).
Imaoka, S. et al. Multiple forms of human P450 expressed in Saccharomyces cerevisiae. Systematic characterization and comparison with those of the rat. Biochem. Pharmacol. 51 (8), 1041–1050 (1996).
Funding
This work is supported in part by Natural Science Foundation of Hainan Province (821RC1067 to PL), Natural Science Foundation of China (82360056 to WH), Project supported by the Education Department of Hainan Province (Hnky2023ZD-10 to WH), Hainan Medical University Talent Research Launch Fund (2022026 to WH).
Author information
Authors and Affiliations
Contributions
Wei Huang and Hyo In Kim wrote the draft of the manuscript. Jinbong Park and Peiqiong Li corrected the draft. Wei Huang and Peiqiong Li supervised the experiments. Wei Huang, Hyo In Kim, Feng Cao, and Shuang Li performed the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, W., Kim, H.I., Cao, F. et al. Effects of mallotus furetianus extract on CYP3A activities in rats. Sci Rep 15, 18530 (2025). https://doi.org/10.1038/s41598-025-02193-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-02193-7