Abstract
Semantic segmentation involves an imminent part in the investigation of medical images, particularly in the domain of microvascular decompression, where publicly available datasets are scarce, and expert annotation is demanding. In response to this challenge, this study presents a meticulously curated dataset comprising 2003 RGB microvascular decompression images, each intricately paired with annotated masks. Extensive data preprocessing and augmentation strategies were employed to fortify the training dataset, enhancing the robustness of proposed deep learning model. Numerous up-to-date semantic segmentation approaches, including DeepLabv3+, U-Net, DilatedFastFCN with JPU, DANet, and a custom Vanilla architecture, were trained and evaluated using diverse performance metrics. Among these models, DeepLabv3 + emerged as a strong contender, notably excelling in F1 score. Innovatively, ensemble techniques, such as stacking and bagging, were introduced to further elevate segmentation performance. Bagging, notably with the Naïve Bayes approach, exhibited significant improvements, underscoring the potential of ensemble methods in medical image segmentation. The proposed EnsembleEdgeFusion technique exhibited superior loss reduction during training compared to DeepLabv3 + and achieved maximum Mean Intersection over Union (MIoU) scores of 77.73%, surpassing other models. Category-wise analysis affirmed its superiority in accurately delineating various categories within the test dataset.
Similar content being viewed by others
Introduction
Currently, deep neural networks have made significant inroads in the realm of biomedical image analysis1,2,3,4. They have been applied extensively in roles such as image classification, identification, segmentation, and brain research5,6,7,8,9, delivering impressive outcomes. Among these applications, image semantic segmentation stands out as a pivotal area in digitalimage processing and system vision study. One of the effective methods related to segmenting semantically is dealt by Csurkaet al.10, involves categorizing every class pixels in the selected image according to label of the class, yielding predictions rich in “semantic” information11. This technique finds widespread utility in diverse fields, spanning virtual reality, industry, civil engineering, medicine, and beyond, where it has demonstrated remarkable efficacy. In the domain of medicine, cerebrovascular images are typically captured through techniques such as ComputedTomography Angiography (CTA), DigitalSubtraction angiography (DSA), and MagneticResonance angiography (MRA). These images have historically been processed using traditional algorithms and more recently, deeplearning methods. But there hasn’t been much research done on analyzing true-color vascular decompress images. The advantage of true-color medical images lies in their enhanced comparability with traditional medical images, facilitating a seamless transition. Therefore, a key frontier in the continued development of smart medical treatments is the accurate segmentation of brain arteries and spinal nerves using these vascular decompress images. The overarching objective is multifaceted. It encompasses reducing the cognitive burden on surgeons, enhancing surgical speed, minimizing adverse surgical events and complications, enabling general practitioners to attain a level of expertise akin to specialists, and empowering experts to conduct their procedures with greater efficiency. The goal of this research project is to close the gap among modern technology and the medical profession, ushering in a time when healthcare delivery will be more efficient and effective.
Traditional methods for blood-vessel segmentation have encompassed a range of techniques, including filter by matching12,13,14, multiscale techniques15,16,17,18,19,20, morphological strategies21, active contour models22,23,24,25,26,27, growing of region28,29,30,31, level-set methods32,33, and merging of region34. Deeplearning-based advances in the area of semantic categorization of cerebral pictures, however, have called for particular methods for data gathering and labeling. The capture and tagging of these essential photos is the first stage in research that uses neural learning to semantically separate images of the vascular system. Given the unique characteristics of cerebral pictures, specific machinery is frequently used to facilitate their acquisition. The subsequent step entails meticulous manual annotation of the acquired data.
Imagery segmentation with semantics using region categorization (ISSbRC) and pixel categorization (ISSbPC) are two main categories for deeplearning-based imagery semantics segmentation techniques. While useful in some situations, ISSbRC has some drawbacks, including lesser segment reliability, shorter segment rates, and poorer computational effectiveness. These challenges have led to the development of ISSbPC techniques.
In this study, the main focus is to enhance the semantic segmentation of microvascular decompression (MVD) images using a novel ensemble learning approach. MVD imaging, a crucial part of neurosurgery, is hindered by limited publicly available datasets and the labor-intensive task of manual annotation. This work addresses these challenges by creating a specialized dataset of 2003 RGB MVD images with annotated masks and employing extensive data preprocessing and augmentation strategies. The research introduces EnsembleEdgeFusion, an innovative technique that combines the strengths of multiple semantic segmentation models (DeepLabv3+, U-Net, DANet, and DilatedFastFCN) to improve segmentation accuracy, boundary delineation, and feature extraction. The proposed method significantly enhances the robustness of medical image segmentation, offering a promising solution to assist in surgical planning, reduce cognitive load on surgeons, and enable more efficient medical diagnostics. Furthermore, this study aims to bridge the gap between modern computational techniques and practical medical applications, thereby contributing to the advancement of smart healthcare solutions.
Literature review
One pivotal milestone in this journey was the introduction of completely connected convolutions (FastFCN) by Long et al.35. This invention used completely supervised instruction for imagery and semantic segmentation and was made to work with photos of various sizes. FastFCN builds upon the Visual Geometry Group VGG-16 architecture36. The proposed part substitutes the fully connected part with convolutions in conventional neural networks (CNN)37 and employs a skip-layer mechanism to integrate feature-maps produced by the intermediaries in the network. This introduction of skip based connections facilitates the fusion of deep contextual features with fine-grained spatial details. Bilinear interpolation is then employed for upsampling, enabling pixel-wise classification and the transformation of coarse segmentation results into refined outputs.
Recognizing that pooling operations diminish resolution of feature maps, Ronneberger et al.38 suggested U-Net technique. Encoding-Decoding based approach is adopted in U-Net, wherein downsampling occurs during encoding phase to progressively reduce the feature map’s resolution. Conversely, during the decoding phase, upsampling is applied to progressively recover details and resolution of the picture. This approach has proven highly effective in semantic segmentation tasks, particularly in domain of medicine, where fine-grained anatomical particulars are critical. These developments signify significant strides in realm of deeplearning-based semantic separation, offering a pathway to more accurate and efficient cerebrovascular image analysis.
Another notable modelling is SegNet39, which operates by classifying pixels based on some probabilities. SegNet’s encoding component utilizes complete convolutions in network that undergoes downsampling and is then deciphered using various processes, including convolutional pooling. Decoder, on the other hand, employs deconvolution to upsample its input based on the encoder’s transition indices. Deconvolution plays a crucial role in restoring intricate info and maintaining consistent spatial dimensions. This encoding-decoding architecture effectively circumvents the issue of reduced feature map resolution following pooling operations, ensuring the preservation of spatial dimensions and pixel location info within image.
In prior research efforts, NasrEsfahani et al.40. introduced common Convolution NN model for coronary angiogram vessel categorization wherein the obtained outcomes are not that much worthy. Phellan et al.41 suggested a complete deeper neurals for analyzing MRA images by referring the earlier suggestions related to common CNN in cerebral vasculature categorization. However, due to limitations stemming from a small sample size and the shallowness of the networking model, evaluation was constrained. Mo et al.42 introduced a FCN with multiple leveling architecture with deeper supervision. The introduced model demonstrated proficient segmentation of larger vessels but struggled for smaller/microlevel vessels. Jiang et al.43 introduced the concept of transfer learning within FCNs to enhance vascular structure segmentation but faced challenges while categorizing finer regions of vascular. Approximated CNN is introduced by Noh et al.44 that preserves receptiveness thereby augmenting depth of the network and delivering impressive efficacy in segmenting blood vessels. However, the removal of downsampling layers negatively impacted certain datasets. Livne et al.45 adopted an encoder-decoder U-Net architecture to categorize cerebral blood vessels in MRI. This architecture was successful in collecting context data and propagating it to greater-resolution levels, but it had difficulties processing intricate details like tiny blood vessels or aggregating more features.
Chen et al.46 introduced DeepLab as an advancement over FCN, addressing the shortcomings of 3-D inconsistency and vague segmentation. To create coarse-categorization map, this model utilized a completely linked conditionally randomizedField (FCCRF). Expanding the open area of map features using boundary optimization approaches and atrous convolution eventually enhanced semantic segmentation’s performance. Building upon DeepLab, DeepLabv247 introduced the ASPP (Atrous Spatial Pyramid Pooling) module, which effectively integrated multiple scale parameters, expanded receptiveness, and heightened accuracy of the segments.
Expanding on the ideas from DeepLab and DeepLab version2, DeepLab version348 was conceived. It enhanced the ASPP part by incorporating BatchNormalization (BN) and eliminating FCCRF. However, DeepLabv3’s use of pooling operations led to a loss of detailed target boundary information, and the computational load of dilated convolution was comparatively high. Subsequently, DeepLabv3 + was developed, surpassing the performance of DeepLabv1, v2, and v3. It achieved this by employing deeper divisible convolutions in depth-wise. DeepLabv3 served as the encoder, requiring the addition of a decoder to reinstate target border particulars.
In the DeepLabv3 + architecture, Xception49, one of the lightweight technique was initially used for extracting parameters, followed by ASPP component to acquire parameter info in multiple scales. The gotten multiscale feature info underwent processing through 1 × 1 convolutions. It was combined with the 1 × 1 convolution-processed characteristics of the core network following four passes of upward sampling. Subsequently, fine-tuning was carried out using 3 × 3 convolution, followed by four additional rounds of upsampling to yield final output. DeepLabv3 + demonstrated robust performance on datasets commonly utilized in semantic segmentation tasks, achieving results of 89% and 82% on the PASCAL VOC2012 and Cityscapes datasets, respectively50. However, when applied to the nuanced domain of microvascular decompression images, it encountered challenges, including target pixel blending and the presence of other deficiencies like imprecise target boundaries, blurred contours, and insufficient feature information.
Transformer-oriented approaches have made notable progress in the field of polyp division. CaraNet51 utilizes axial and reverse attentiveness techniques to examine peripheral regions. DuAT52 utilizes a spatial aggregate module that operates from global to small scale, as well as a selective border aggregation component, to accurately identify objects with different dimensions. PVTCASCADE53 relies on a hierarchical visual converter for integrating features at different scales, makes use of an attention gates to fuse upper and lower characteristics through bypassing relationships, and relates a convolutional attention section to reduce surroundings data and enhance detached connections. FCBFormer54 utilizes capabilities of transformer to accurately outline distinctive features of polyps and utilizes a parallel thorough convolution branch for providing detailed information at local scale.
The attention system has the ability to alleviate problem of polyp division. XBoundFormer55 utilizes boundaries attention to confine inflection points inside the border, considers boundary information as vector that embeds for boundary, and combines this vector with desired features using multiple heads of attention. H2 Former56 employs convolution algorithms with different scales to extract multiscale characteristics, leverages adaptive attention channeling to determine significance of these multiple-scale characteristics for enhancing local component representation, and incorporates the transformer to establish global context dependency. FRCNNAACIF57 integrates attention perception unit into every layer of underlying system, eliminating impurities by utilizing local crosschannel facts, and enhancing integration of contextual data with ROI attentiveness. The efficient attentiveness process enhances salient elements and reduces the impact of irrelevant ones.
Recent research has illuminated a compelling Medical Image Classification (MIC) trend: the most effective and accurate MIC pipelines frequently employ ensemble learning strategies56,58,59,60,61,62,63,64,65,66,67,68,69. Finding a hypothesis in computational learning that improves the precision of predictions is the primary goal. However, because pursuing an ideal hypothesis is intrinsically tricky, an approach combining various hypotheses to produce a better predictor that comes close to an ideal hypothesis has developed. These theories are learned neural network simulations applied to deep convolutional neural networks (CNNs). In order to improve prediction efficiency, these algorithms are combined in a learning ensemble. Ensemble learning techniques are incorporated into the DL workflow as part of deep ensemble learning.
The use of deep ensemble learning has been demonstrated to be effective in improving the performance and robustness of various MIC pipelines61,62,63,64,65,66,67,68,69,70,71. Empirical evidence indicates that ensemble learning-based workflow is used to outperform single-model approaches. This is rooted in the assumption that combining diverse models allows them to focus on different features, effectively compensating for each other’s limitations71,72,73,74. Nevertheless, the extent and specific ensemble learning approaches that are valuable in medical visual classification using deep learning remain open questions. While the notion of ensemble learning is not new, previous research has yet to thoroughly examine the influence of ensemble learning methodologies on deep learning-powered medical picture categorization. Several authors have offered extensive reviews of ensemble learning in general, such as Ganaiea et al.67, who explored the field of deep ensemble-based learning. A survey on bioinformatics applications using deep ensemble-based learning strategies is done in74.
Meanwhile, a general analysis of deep ensemble-based learning is done in71,75,76. This project aims to provide a repeatable analytical channel to analyze the efficacy of ensemble-based learning with CNN in healthcare visual segmentation. Our goal in evaluating several ensemble approaches to learning is to contrast how they perform to a benchmark process. This investigation will aid in identifying possible performance improvements attainable via ensemble learning methodologies in deep learning-driven healthcare image categorization. A novel hybrid approach combines U-Net and SegNet with a logistic regression classifier to improve segmentation accuracy for retinal blood vessels. The model addresses challenges like size variation and contrast in retinal images, achieving a segmentation accuracy of 97.02%77,78. An ensemble method combining EfficientNetB0, EfficientNetB2, and ResNet101 using transfer learning and beta normalization achieved outstanding accuracies of 97.88% and 97.47% on two GI datasets. The model outperforms individual base models and uses Grad-CAM for interpretability79. A lightweight model using ConvLSTM layers, ConvNext Blocks, and Knowledge Distillation achieved an accuracy of 99.38% with low computational cost and disk space usage. It offers an efficient solution for GI disease detection in clinical settings80. A hybrid ensemble of ResNet34, Inception V3, and VGG16 for retinal blood vessel segmentation on the DRIVE and HRF datasets showed significant improvements in accuracy, precision, and recall, with ResNet34 + U-Net achieving an accuracy of 99.6% and AUC of 0.999 for detecting diabetic retinopathy (DR)81. Table 1 details the latest existing works with their limitations and the need for proposed work.
To comprehensively address these limitations, we introduce an innovative ensemble algorithm coined ‘EnsembleEdgeFusion.’ This approach amalgamates cutting-edge algorithms tailored for medical image segmentation, including DeepLabv3+, U-Net, DANet, and FastFCN. The objective is to influence exclusive strengths of individual algorithm to collectively rectify identified shortcomings.
Our ensemble technique, ‘EnsembleEdgeFusion,’ targets issues like target pixel mixing, striving to deliver refined target boundary delineations by harnessing the precision of U-Net and FastFCN. Moreover, the incorporation of DANet enhances context awareness and enables the effective capture of intricate features, mitigating the problem of insufficient feature information. This ensemble framework further addresses the occurrence of blurry contours, attributing this improvement to the robust boundary-preserving capabilities of DeepLabv3+.
By synergistically harnessing the capabilities of DeepLabv3+, U-Net, DANet, and FastFCN, our ensemble approach seeks to provide a comprehensive solution to the identified limitations. The collaborative strength of these algorithms aspires to yield semantic segmentation outcomes for microvascular decompression images that surpass those obtained through individual methodologies. The major highlights of the paper are listed below:
-
A specialized dataset for microvascular decompression images is created, addressing the shortage of openly accessible medical image datasets.
-
The proposed ensemble technique, EnsembleEdgeFusion, provides a robust approach for selecting the best model for microvascular decompression image segmentation.
-
Targeting issues such as target pixel blending, imprecise boundaries, blurred contours, and insufficient feature information.
-
Aiming to enhance surgical precision, reduce cognitive burden on surgeons, and empower general practitioners in medical diagnostics.
By enhancing the latest developments in semantic categorization of vascular decompress visuals, the present study aims to greatly advance the area of smart medical therapy.
Materials and methods
DeepLabv3 + was employed to conduct semantic segmentation on vascular decompression visulas, utilizing dedicated microvascular-decompression-image data for model training. Results of the tests, however, revealed mediocre results in the semantic division of these photos. The approach encountered challenges related to the blending of target pixels, along with several other deficiencies. These shortcomings encompassed issues such as the imprecise demarcation of target boundaries, the presence of blurry contours, and a dearth of comprehensive feature information.
To address these limitations comprehensively, a novel ensemble algorithm named ‘EnsembleEdgeFusion’ was devised, amalgamating diverse conventional algorithms tailored for medicinal image categorization. The aim was to leverage distinctive strengths of each algorithm to collectively rectify the inadequacies that had been observed. This ensemble technique harnessed the capabilities of DeepLabv3+, U-Net, DANet, and FastFCN, among others.
Through the collaborative efforts of these algorithms, a holistic resolution was sought. The proposed ensemble technique ‘EnsembleEdgeFusion’ aimed to ameliorate the issue of target pixel mixing, offering refined delineations of target boundaries by capitalizing on the precision of U-Net and FastFCN. Furthermore, the incorporation of DANet brought forth heightened context awareness and the effective capture of intricate features, mitigating the problem of insufficient feature information. The ensemble framework additionally countered the occurrence of blurry contours, attributing this enhancement to the robust boundary-preserving capabilities of DeepLabv3+.
By synergistically combining the competencies of DeepLabv3+, U-Net, DANet, and FastFCN, the ensemble approach sought to provide a comprehensive remedy to the identified shortcomings. The collaborative strength of these algorithms aspired to yield semantic segmentation outcomes for microvascular decompression images that surpass those obtained through individual methodologies.
DeepLabv3+
DeepLabv3 + stands as an advanced deep learning algorithm tailored to achieve precise and intricate semantic segmentation within images, especially in complex scenarios where accuracy is paramount. Emerging as a natural extension of the DeepLab lineage, DeepLabv3 + introduces a fusion of architectural advancements that markedly refine segmentation performance. Central to this approach are atrous (or dilated) convolutions, which play a pivotal role in capturing contextual information across varying scales within the input images. This innovative technique permits the network to analyze different levels of detail while minimizing the computational load.
A decoder and an encoder are both included in the DeepLabv3 + design, which has a double layout. With the input image, the encoding process helps to identify the most important features, which are then painstakingly refined by the decoder in order to provide the finished segmentation mapping. The AtrousSpatial PyramidPooling (ASPP) section is at the core of DeepLabv3+’s power. This module adopts parallel atrous convolutions with diverse dilation rates, intelligently aggregating contextual insights at varying scales and facilitating the recognition of objects spanning different dimensions. DeepLabv3 + seamlessly harmonizes high-resolution features from encoder with context-rich information garnered through the ASPP module. This fusion empowers the algorithm to meticulously delineate object boundaries and elevate the overall segmentation accuracy. Notably, the decoder stage plays a pivotal role in this augmentation. By elevating low-resolution features through upsampling and synergizing them with their high-resolution counterparts, the decoder preserves intricate nuances within the segmented outputs.
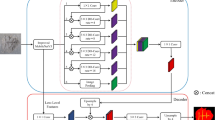
Moreover, the incorporation of skip connections in DeepLabv3 + enables the harmonization of features from disparate scales within the decoder. This, in turn, empowers the model to encapsulate both local intricacies and broader contextual understanding, thereby accentuating its capacity to produce meticulous segmentation outcomes. DeepLabv3+’s forte lies in semantic segmentation tasks, where every image pixel is meticulously categorized, proving invaluable across domains such as medical imaging, autonomous driving, satellite image analysis, and beyond. Being able to continuously show outstanding results among standard datasets, DeepLabv3 + underscores its prowess in generating high-quality segmentation outputs. With its adeptness at addressing intricate details and context, it emerges as a sophisticated solution for critical task of semantic image segmentation. The process of segmentation using DeepLabv3 + algorithm for microvascular decompression images are demonstrated in Fig. 1.
Imageries Using DeepLabv3+1.
In the DeepLabv3 + framework, which includes both the encoder and decoder parts, the task of clearly identifying the boundaries of the target objects in microvascular decompression images is challenging1. The decoder’s direct upsampling process, which enlarges the image four times, results in the loss of some important feature details. To address this, we included the ASPP component in the decoding. This component handles the lowest-level features in the decoding steps, but the highest-level feature map, that has been amplified by four in the encoding process, is additionally engaged. By combining these two sets of information, we were able to enhance the completeness of the segmentation’s boundary details and achieve a clearer representation of the semantic information1.
UNet
The UNet al.gorithm is used to segment microvascular decompression images, where the goal is to distinguish different structures2. UNet is a specialized model for this task. It works by taking an input image and producing a segmented image that highlights specific regions of interest. One special thing about UNet is its U-shaped architecture. It has an encoder part that captures important features from the input image, and a decoder part that refines the features and generates the final segmented image. In microvascular decompression images, UNet is particularly useful because it can handle complex structures and details that are important for accurate segmentation. It focuses on both local and global features, ensuring that the boundaries of structures are well-defined and the overall segmentation is accurate. UNet’s architecture allows it to capture intricate patterns in the images, which is crucial for identifying and differentiating various components in microvascular decompression images. UNet’s success in this context is due to its ability to handle medical images effectively. It can distinguish subtle differences in tissues and structures, which is crucial for tasks like identifying blood vessels and nerves in microvascular decompression images. Its encoder-decoder architecture ensures that both fine details and broader context are considered, leading to accurate and meaningful segmentations. UNet al.gorithm for microvascular decompression images are illustrated in Fig. 2.
DANet (deformable attention network)
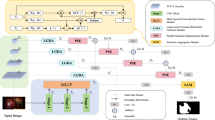
Rajamani et al.3, combined ideas from UNet and CCNet to create the Deformable Attention Net (DANet) for segmenting microvascular decompression images. The DANet architecture as proposed in3 is utilized in the research study for segmenting microvascular decompression images and the same is outlined in Fig. 3. A modified U-Net structure is utilized to process our 256 × 256 images. The structure includes three blocks for downsampling and three for upsampling. Each block has Batch Normalization, 2D Convolution with a 3 × 3 kernel, and ReLU activation. The last block has a 1 × 1 convolution. Downsampling is done using max pooling, and ConvTranspose2 d is used for upsampling. As the image gets progressed across the network, the numbers of features also get changes. The ending layer of the U-Net exactly match with the number of segmentation class labels.
The key innovation is addition of the Deformable Attention Module, combining the principles of CCNet and UNet. The local features from the U-Net’s downsampling blocks are given to the attention module, which is placed in the bottleneck for efficient processing. Unlike traditional criss-cross attention4, our method smartly captures only the essential contextual information, enhancing segmentation accuracy. In DANet3, a dynamic and learnable pattern is created by using the Deformable Attention Module. The entire pattern is tuned to extract the noteworthy non-local info from the input image. The results from the Deformable Attention Module are then combined with the original features and passed through the upsampling path of the U-Net. Our approach uses deformable criss-cross attention to efficiently gather non-local information. The pattern is adjusted dynamically using learnable offsets, ensuring that the attention mechanism focuses only on the most important details. This method of attention sampling is differentiable, allowing for seamless training. This dynamic deformable attention mechanism significantly improves segmentation results, especially for complex microvascular decompression images.
DilatedFastFCN with JPU
Initially, in5 a popular technique called DilatedFCN is introduced for semantic image segmentation. Then, this approach is enhanced by introducing a unique Joint Pyramid Upsampling (JPU) module. This module improves the performance of DilatedFCNs while keeping computations manageable. DilatedFCN starts by turning a CNN designed for image classification into a FullyConvolutionalNetwork (FCN). It replaces certain layers to produce labeled maps from images. One challenge is that the final feature map’s low resolution can lead to inaccuracies. To address this, DeepLab removes some downsampling and uses dilated convolutions. This is called DilatedFCN, and it preserves more detail.
The method in5 improves DilatedFCNs by introducing the Joint Pyramid Upsampling (JPU) module. This module focuses on approximating the final feature map of DilatedFCN without overwhelming computations. The proposed approach maintains the backbone of the original FCN, using three feature maps. Then JPUs’ are introduced to refine predictions. JPU, the core of the method, is designed to generate a feature map similar to DilatedFCN’s final one. This process is formulated as joint upsampling, where details from high-resolution visuals guide generation of high-resolution target visual. To achieve this, convolutional operations are utilized. By integrating JPU, the issues of maintaining high-resolution features without overwhelming computations in the microvascular decompression image segmentation context is addressed.
Joint upsampling
Joint upsampling involves enhancing a lower-resolution image using a higher-resolution reference. To explain further, imagine having a fuzzy image (low-res target) and a clear, detailed image (high-res guidance). The aim is to improve the fuzzy image by borrowing details and structure from the detailed one. This is like refining a rough sketch using a more polished version as a guide. The process works like this: For the fuzzy image (low-res target) created from a basic transformation of the detailed image (low-res guidance), we want to figure out a simpler transformation (ˆf) that gets similar results. This way, we can achieve high quality without the complex calculations of the initial transformation (f). For instance, if f involves multiple steps (like a multi-layer perceptron), we try to find a shortcut (ˆf) that still does a great job. When we apply this simplified transformation to the detailed image (high-res guidance), we get a high-resolution image that’s as good as if we used the full transformation. In technical terms, given the two versions of the lower-resolution image (xl,yl) and higher-resolution image (xh), joint upsampling is defined as creating a new high-resolution image (yh) using a simpler transformation (ˆf) that approximates the more complex transformation (f) while minimizing difference among original low-res image (yl) and the transformed type (h(xl)) within a set of possible transformations (H). This difference is measured using a predefined metric as denoted in Eq. (1).
where h is a transformation possible functions’ set and ||·|| is the distance metric that is pre-defined.
Dilated convolution
Dilated convolution was introduced in the DeepLab method6 as a technique to capture detailed information in high-resolution feature maps while preserving a wide field of view. Imagine a simplified example in one dimension (1D), with a dilation rate of 2. In this case, the process can be broken down into three main stages: First, the input features (fin) are divided into two sets based on whether their indices are even or odd. Then, a convolution is applied to both sets using the same convolution layer. This produces two sets of processed features (\(\:{f}_{out}^{0}\) and \(\:{f}_{out}^{1}\)). Finally, these processed sets are combined in an interleaved manner to form the final output feature map (fout).
Stride convolution, on the other hand, is a technique designed to decrease the spatial resolution of input features. In a simplified scenario, the input features (fin) undergo two primary steps: First, a regular convolution is applied to the input to generate an intermediate feature set (fm). Next, only the elements with even indices are retained, effectively reducing intensity of feature map, resulting in output feature map (fout).
Reformulating the concept for joint upsampling
The distinctions between our approach’s framework and the DilatedFCN technique become evident in the final two convolution stages. To illustrate, consider the 4 th convolution stage (Conv4). In the case of DilatedFCN, the input feature map is initially treated by a regular convolution layer, which is then followed by a sequence of dilated convolutions (d = 2). In contrast, our method initially processes the input feature map using a stride convolution (s = 2), and subsequently employs multiple regular convolutions to produce the final output. If the feature_map of the input i.e., x is given, the corresponding output feature map yd in Dilated FCN is obtained as given below5:
whereas in the proposed method, ys, the feature_map of the output is generated as follows5:
Cr, Cs and Cd shows a regulated or dilated convolution respectively whereas \(\:{C}_{r}^{n}\) is regular convolution layers and S, R and M illustrates the split operation, merge operation and reduce operation respectively5. When the x and ysvalues are provided, the feature_map y which approximates the value of yd is obtained by5:
Which is equivalent to Eq. (1). Similar type of conclusions is obtained for fifth stage of convolution also.
In the JPU module, processing input feature maps with convolution blocks is initiated first. This generates intermediate maps and reduces dimensionality. These maps are then upsampled and combined. The multiple convolutional operations are employed in parallel to capture various information from the maps. These operations enable us to capture both the relation between different maps and the transformation needed to achieve the desired high-resolution outcome. Finally, a convolution block further refines the generated features to create the ultimate prediction5. The Dilated FastFCN architecture as proposed in5 is utilized in the research study for segmenting microvascular decompression images and the same is outlined in Fig. 4.
Custom vanilla architecture
The custom Vanilla architecture was designed as a simple baseline model to compare the performance of more advanced segmentation models. It consists of four convolutional layers with 3 × 3 filters and ReLU activations, followed by max-pooling layers for downsampling. A final fully connected layer outputs the segmentation map, with a softmax activation function used to produce class probabilities.
The motivation behind this architecture was to establish a reference point for evaluating the impact of advanced segmentation models such as DeepLabv3+, U-Net, DANet, and DilatedFastFCN with JPU. The Vanilla model also served as a simple, interpretable model to observe the basic performance of deep learning for segmentation tasks in MVD images, providing insight into how more complex architectures improve upon this baseline.
Proposed ensembleedgefusion method
In the realm of computer vision tasks, particularly image segmentation, the supremacy of deep convolutional neural networks (CNNs) in terms of accuracy and robustness is widely acknowledged7,8,9,10. Instead of fixating on a single model architecture, our approach prioritized training a diverse set of deep learning architectures to ensure the reliability of our results. The ensemble of architectures selected for our study includes DeepLabv3+, U-Net, DANet, DilatedFastFCN, and a custom Vanilla architecture, which serves as a comparative benchmark.
The Vanilla architecture comprises four convolutional layers, each of which in-lined by maxpooling layers. All architectures, including the Vanilla one, employ a consistent classification head consisting of global avg pooling, denselayers, dropoutlayer, and activation using softmax layer for generating probabilities of final class. To harness the benefits of transfer learning, we initiated the training process by pretraining all models on the microvascular decompression image dataset. During this initial phase, the layers of the architecture were initially frozen, except for the classification head. Subsequently, these layers were unfrozen for finetuning.
The frozen transferlearning stage was carried out over ten epochs, utilizing Adamoptimization algorithm with a preliminary learningrate of 1E04. In contrast, finetuning phase, which encompassed both transfer learning and additional fine-tuning, concluded after a maximum training duration of 1000 epochs. During this phase, a dynamic learning rate strategy, based on the Adam optimization approach, was implemented. The learning rate commenced at 1-E05 and gradually decreased to a minimum of 1-E07. If there was no improvement in the monitored validation loss after 8 epochs, the learning rate decreased by a factor of 0.1. The training utilized the weighted Focal loss introduced by Lin et al.11 as the loss function.
The weighted Focal loss (FL) is defined in Eq. (5), where pt represents original groundtruth probabilities class t, γ is user-defined specific parameter (that is equal to 2.0 in our study), and αt represents corresponding weight of the class t. These weights of the class were determined using the distribution of the class among the model training samples. Additionally, we incorporated early stop and a checkpoint strategy for model during the fine-tuning phases. Training was terminated if there was no improvement after 15 epochs, and the best-performing model was saved, guided by monitoring the validation loss. Throughout the analysis, a batch size of 28 was used, and computations were carried out in parallel on a workstation equipped with 4x NVIDIA Titan RTX GPUs, each with 24GB VRAM, and an Intel Xeon Gold 5220R CPU boasting 96 cores and 384GB RAM.
Traditionally, deep ensemble learning referred to combining predictions from various deep convolutional neural network models12. However, recent advancements have reshaped the understanding of ensemble learning in the realm of deep learning. This modern perspective involves merging information, usually predictions, for a single inference, which can initiate from numerous models or even a singular one. In our examination, the performance implications were explored for various ensemblelearning techniques, namely Augmenting, Bagging, and Stacking. Notably, the Boosting concept was omitted from our study, a departure from its conventional use in ensemble learning. The reason for this choice was its impracticality in image classification tasks involving deepconvolutional neuralnetworks due to the significant increase in time of training12,13. To provide a visual representation and overview of these four techniques, we created an illustrative diagram presented in Fig. 5. In our comparative study, we established Baseline models for each selected architecture. These Baseline models acted as benchmarks, enabling us to identify potential trends in performance enhancement or degradation resulting from the application of ensemble learning techniques.
Augmenting
The Augmenting technique, often known as test-time data augmentation, involves applying reasonable image alterations before making inferences14,15,16,17,18. Its purpose is to counter potential issues like overfitting or overly rigid pattern learning by generating multiple images of the same sample. These varied images are then utilized to create multiple predictions14,15,16. In our specific study, we extended Baseline models and introduced random rotations and reflecting along all axes during the implication stage. For each individual example, we generated 15 randomly altered images, and predictions made were amalgamated using unweighted Meanpooling function.
Stacking
Unlike approaches relying on a single algorithm, combining various deepconvolutional neuralnetwork architectures, also known as inhomogeneous ensemblelearning, has shown significant advantages in enhancing overall performance19,20,21,22,23. This form of ensemble learning is intricate and applicable to a wide array of computervision tasks19,20,21,22. The essence of Stacking technique lies in utilizing the diverse and autonomous models by introducing an additional ML algorithm that operates on the predictions generated by these models. In our study, we utilized Baseline models, which comprised lot of architectures, working as an ensemble for the Stacking technique. Here, diverse poolingfunctions were applied directly on top of all these distinct architectures to harness their collective predictive power.
Bagging
In the realm of homogeneous model ensembles, multiple models are utilized, all of which share the same algorithm, hyperparameters, or architecture20,24. One prominent technique within this approach is Bagging, a popular method in ensemble learning, which aims to enhance training dataset sampling. Unlike the conventional single training/validation split, which produces just one model, Bagging entails training numerous models on randomly chosen data subsets. Essentially, a k-fold crossvalidation technique is given to dataset, ensuing in k distinct mockups25. In the given study, 5-fold cross-validation strategy is employed for Bagging. This approach yielded 5 models for individual architecture. The resultant predictions generated by ensemble models for a sample wes then amalgamated using various pooling functions.
Pooling strategies
To synthesize the diverse predictions generated by our ensemble, we delved into a variety of methodologies and algorithms. Each prediction yielded a probability distribution across unknown sample classes, normalized using softmax. Within the realm of Bagging and Stacking techniques, we meticulously evaluated several pooling functions, including the Best Model approach, Global Argmax, MajorityVote (both Soft and Hard versions), Decision Tree, Unweighted and WeightedMean, NaïveBayes, Gaussian Process classifier, Logistic Regression, SVM, and kNN26. In the context of the Augmenting technique, we opted for an exclusive use of the Unweighted Mean as the designated pooling function. This comprehensive exploration allowed us to merge the ensemble predictions effectively, ensuring a unified outcome that encompasses the strengths of various models and algorithms.
The Best Model approach involves choosing the model that has F1 score, on the ‘ensembletrain’ sampling set. DecisionTrees were trained using the Giniimpurity criterion to measure informationgain27. The Gaussian Process classifier used the Laplace approximation with a ‘one vs rest’ multi strategy. The GlobalArgmax technique involves identifying class with probability among all predictions and setting probabilities of classes to zero. For LogisticRegression we used ‘newton_cg’ solver with L2 regularization and a multinomial multiclass strategy32. The MajorityVote Soft variant combines class likelihoods for each class. Softmax is utilized then for normalization across each classes. On hand in MajorityVote variant fundamental class voting is used where each prediction chooses the class with the highest probability as its vote. Unweighted Mean calculates an average of class probabilities from predictions while Weighted Mean incorporates weighted averaging based on each models F1 score achieved on the ‘ensembletrain’ set. Our implementation of Naïve Bayes follows Rennie et al.s Complement variant82. The Support Vector Machine classifier adheres, to LIBSVMs implementation33. We used a neighbor count of five for the k Nearest Neighbors classifier. The algorithm below illustrates the process of our proposed model.
Hyperparameter optimization and optimization strategies
While hyperparameter tuning was not performed in this study, several standard optimization strategies were employed to enhance the performance of the models. Transfer Learning was the first strategy used, where all models were pretrained on large-scale datasets such as ImageNet. This approach allowed the models to learn general features from the pretraining phase, which were then fine-tuned on the MVD dataset. Fine-tuning on the MVD dataset enabled the models to adapt to the specific task of semantic segmentation for microvascular decompression images.
For model training, the Adam optimizer was selected, with an initial learning rate set to 1E-04. The learning rate was dynamically adjusted during training based on the performance on the validation set. This adaptive approach helped the models converge efficiently while avoiding issues like getting stuck in local minima. A dynamic learning rate scheduling strategy was implemented, where the learning rate was reduced by a factor of 0.1 if the validation loss did not show improvement for 8 consecutive epochs. This strategy helped the models continue learning more effectively when convergence slowed down.
Early stopping was another important strategy used to prevent overfitting. Training was halted if the validation loss did not improve for 15 consecutive epochs, ensuring that the models did not continue to train unnecessarily and avoid overfitting on the training data. Finally, to address the issue of class imbalance in the dataset, the Weighted Focal Loss function was utilized. The weights for the loss function were adjusted based on the class distribution in the dataset, enabling the model to focus more on the harder-to-classify classes, thereby improving segmentation accuracy. These optimization strategies were applied to ensure effective model convergence, reduce overfitting, and enhance the performance of the models on the MVD dataset.
Results and discussion
Dataset
The 2003 microvascular decompression (MVD) images used in this study were curated from multiple renowned medical imaging centers and hospitals, ensuring a diverse and representative sample of MVD cases. These images, primarily depicting MVD procedures for conditions such as hemifacial spasm (HFS) and trigeminal neuralgia (TN), were collected using standard medical imaging equipment. The inclusion criteria focused on images that clearly depicted anatomical features of the vascular system, including cranial nerves, arteries, and other neurovascular structures, with sufficient quality for accurate segmentation. Images with poor resolution or significant motion artifacts were excluded from the dataset. Using the Labelme tool, experts outlined key anatomical structures such as cranial nerves (e.g., CNV, CNVII, CNIX, CNX) and arteries (e.g., AICA, PICA). The annotations were meticulously cross-checked and validated to ensure accuracy and consistency. Each image was paired with a segmentation mask, formatted similarly to the Pascal VOC 2012 dataset, enabling seamless integration with semantic segmentation models. Sample original images used for this study is illustrated in Fig. 6.
In this study, we provide a self-annotated dataset containing 2003 RGB images of microvascular decompression, each supplied with meticulously labeled masks designed to segment microvascular decompression images. The dataset consists of images with varying dimensions, including 758 × 586 and 1920 × 1080. It encompasses nine distinct categories, with the background category denoted as 0, resulting in a total of ten categories. Table 2 illustrates the different types of categories and their associated colors. The table provides information on each category, such as “CNV” (CarnialNerve-5) representing the trigeminal nerve, “CNVII” indicating the nerve of the face, “CNIX” representing the glossopharyngeal nerve, “P.I.C.A” represents PartialIntracranialArtery, “A.I.C.A” represents AnteriorInferior CerebellarArtery and “Pet.V” represents Petitville and so on.
For the experimental dataset, we harnessed 2003 images. Within this dataset, 1822 images comprised the training part, and 177 images comprised the testing part, and both were selected randomly. Due to variations in image sizes, we standardized them to a size of 512 × 512 for the sake of training and consistency.
Data preprocessing and augmentation
The dataset underwent several preprocessing and augmentation steps to improve model performance and generalization.
Preprocessing
-
Normalization was applied to scale pixel values to the range [0, 1], ensuring that each image contributed equally during training.
-
Resizing was performed to standardize image dimensions to 512 × 512 pixels.
-
Mean subtraction was applied by subtracting the mean pixel value of the entire dataset from each image to center the data around zero, improving training stability.
Data augmentation
-
Random Horizontal Flipping was applied to introduce variance in the orientation of the vascular structures, helping the model to generalize better.
-
Random Rotation (within the range of − 20° to + 20°) was used to simulate different perspectives of the structures and improve rotational invariance.
-
Random Cropping of 256 × 256 pixel patches allowed the model to learn from various regions of the images, promoting better generalization.
-
Gaussian Blurring was randomly applied to reduce the impact of high-frequency noise, helping the model focus on the most relevant features.
-
Elastic Deformations introduced random distortions to the images, enhancing the model’s ability to recognize varying shapes and sizes of vascular structures.
-
Brightness and Contrast Adjustments were randomly applied to simulate lighting variations, improving the model’s robustness to different imaging conditions.
These preprocessing and augmentation techniques collectively contributed to reducing overfitting, enhancing the model’s ability to generalize, and ultimately improving segmentation accuracy, especially for complex structures in MVD images. The most beneficial augmentation techniques for segmentation accuracy were random rotation and elastic deformations, as they allowed the model to handle variations in the orientation and shape of vascular structures more effectively.
Techniques such as flipping horizontally in random, cropping in random, Gaussian blurring in random, and normalization are noteworthy and the sample of preprocessing is depicted in Fig. 7. These preprocessing techniques are completely necessary to progress efficacy, simplification of model proposed and augmenting the robustness.
Network training
We conducted the network training in a controlled experimental environment consisting of an Intel(R)Core™ i7-9700 K CPU@3.60 GHz, Ubuntu 18.04 64-bit operating system, 32GB of RAM, NVIDIA GEFORCE RTX 2080 Ti, CUDA 10.1, CuDNN 7.6.0, and Python 3.7. We employed the following training parameters, as summarized in Table 3.
In Table 3, “Number of Clones” denotes GPUs’ count utilized during training; “Iterations” signifies total number of training iterations; “Atrous Rate” specifies dilatedconvolution rate applied within ASPP module during training; “Output Stride” refers to output stride of encoder architecture; “Decoder Output Stride” indicates outputstride of decoder structure; “Crop Size” represents image dimensions, and “Batch Size” reflects quantity of images processed in each batch.
The proposed methodology entails an extensive training process, taking approximately 2 h for every 10,000 iterations. This rigorous training regimen was conducted in a consistent experimental environment, which included a setup featuring an Intel Core i7-9700 K CPU, Ubuntu 18.04 as the operating system, 32GB of RAM, an NVIDIA GeForce RTX 2080 Ti GPU, CUDA 10.1, CuDNN 7.6.0, and Python 3.7.
In this controlled environment, we conducted comprehensive training sessions for a selection of state-of-the-art semantic segmentation models, including DeepLabv3+, U-Net, DilatedFastFCN with JPU, DANet, and a custom Vanilla architecture. These models were trained using a dedicated microvascular decompression image dataset.
The Baseline models include DeepLabv3+, Unet, Dilated FastFCN, DANet and Custom Vanilla architecture without utilizing any ensemble techniques. The various performance metrics utilized for comparing the outcomes are accuracy, F1 score, Specificity and Sensitivity and the same is represented in Table 4. According to the F1 score, the best architecture is DeepLabv3+. By utilizing the Augmenting technique of ensemble method in comparison with baseline models clearly revealed no drastic changes with respect to baseline approaches. Similar to the baseline, the F1 score of the DeepLabv3 + model achieved the best compared to the others.
For stacking approach, lot of pooling functions were utilized for integrating all baseline predictions and the resultant F1 score estimation. From the results illustrated in Table 5, the Naïve Bayes scored the best compared to the others. Through the utilization of a 5-fold cross-validation approach, new models were trained to explore the impacts of Bagging on predictive capabilities. Diverse pooling strategies was utilized to accumulate the prophecies obtained from the five models. Among these experiments, the DeepLab v3 + model exhibited the highest F1-scores and was consequently chosen for further analysis and to exemplify the Bagging technique’s outcomes. Upon evaluating the amalgamated predictions from the above models, averaged F1-score outcomes were obtained. In comparison to the Baseline, it became evident that Bagging had a noteworthy adverse effect on performance. Notably, in contrast to earlier ensemblelearning methods, ‘Best Model’ pooling functions did not always indicate Baseline model with greatest validation rating, but rather the model with the highest performance in the 5-fold crossvalidation. It was discovered that these functions produced scores that were strongly clustered when evaluating the order of importance of pooling functions for the DeepLabv3 + cross-validation with five folds. With the exception of Decision Trees, all pooled functions in the data set given earned an F1-score of 0.92. On the other hand, all pooled functions worked for F1-scores of 0.94%, with the exception of BestModel, the DecisionTree, GlobalArgmax, and NaveBayes. Thus the proposed EnsembleEdgeFusion technique operates in choosing the best model for segmenting the microvascular decompression images.
Subsequently, we evaluated the performance of these models by comparing them to the test set, using our Ensemble Edge Fusion approach. Figures 8 and 9 visually illustrate progression of the average losscurve during training and validation phases for both the enhanced network model and DeepLabv3+.
The loss curve plots reveal that during the initial training stages, there is a rapid reduction in loss. However, with the gradual uplift in training iterations, the loss gradually stabilizes, indicating that the models are converging towards a solution. Remarkably, our Ensemble Edge Fusion method exhibits superior performance in terms of loss reduction compared to DeepLabv3+. This enhanced loss reduction signifies the efficacy of our approach in achieving more accurate and robust semantic segmentation results for microvascular decompression images when compared to DeepLabv3 + and other individual models.
The assessment of results involved subjecting the test set to the semantic-segmentation models, including DeepLabv3+, U-Net, DilatedFastFCN with JPU, DANet, a custom Vanilla architecture, all of which were trained using our Ensemble Edge Fusion approach. This comparative analysis aimed to discern the performance disparities among these models.
In Fig. 10, a visual representation showcases the results in a top-to-bottom sequence. The sequence includes the original image, followed by the outcomes generated by DeepLabv3+, U-Net, DilatedFastFCN with JPU, DANet, and custom Vanilla architecture. Subsequently, our proposed method’s results are presented, along with the ground truth images for reference. This comprehensive evaluation provides a holistic view of the models’ capabilities in semantic segmentation. It enables a direct visual comparison of the segmentation quality achieved by each model, including our Ensemble Edge Fusion approach, against the ground truth images.
As evident from the outcomes depicted in Fig. 10, a detailed examination of the first column reveals that DeepLabv3+, U-Net, DANet, DilatedFastFCN with JPU, and the custom Vanilla architecture encounter challenges in accurately delineating the segmentation boundary of “df10”. In these cases, the object contours lack precision and clarity. Moreover, discernible multipixel mixing issues are apparent in the results obtained using PSPNet and DANet methods. Notably, in the second column, when segmenting “df5”, deficiencies in boundary-segmentation are observed across Deeplabv3+, U-Net, DANet, DilatedFastFCN with JPU, and the custom Vanilla architecture. These shortcomings manifest as evident gaps in the segmentation of the target contour, resulting in incomplete representations. Expanding the analysis to include the additional object “df7”, we note that the U-Net method exhibits multipixel mixing issues. In the case of DeepLabv3+, misclassifications occur concerning the “pv” and “pica” categories. In contrast, the proposed Ensemble Edge Fusion segmentation method offers a more comprehensive and informative segmentation. However, it’s worth noting that even though it provides richer feature information, it still faces challenges in segmenting “df10” and “pv” accurately. The segmentation outcome for “df10” in the first column notably differs from the actual scenario. Nevertheless, when compared to the other methods, the results produced by our proposed approach align more closely with the ground truth. Our method excels in capturing additional feature information and achieving segmentation results that closely approximate real-world conditions.
In the examination and comparative analysis of the test dataset, we employed the Mean IntersectionoverUnion (MIoU) metric represented in Eq. (6) to evaluate the performance of various network models, including DeepLabv3+, U-Net, DANet, DilatedFastFCN with JPU, the custom Vanilla architecture, and our proposed method. MIoU is a pivotal indicator used to gauge accuracy of image based segmentation. It remains computed by cumulatively calculating Intersection over Union (IoU) values for each sort and subsequently averaging them. IoU quantifies degree of overlap among predicted segmentation area and ground truth. This ratio represents the joint of two areas divided by their union, with an ideal situation resulting in a ratio of one. A higher MIoU value signifies more precise segmentation outcomes and superior network model performance. The MIoU calculation is outlined as follows71:
where.
-
k signifies number of categories. If background is comprised, there are k + 1 classes71.
-
i denotes true category, while j signifies predicted category71.
-
pii signifies total number of pixels correctly classified as group i71.
-
pij signifies total number of pixels where category i is predicted as category j, and pji represents the converse case71.
-
pij and pji signify pixels that are misclassified71.
We applied this MIoU calculation to assess the segmentation accuracy of the deep learning models and our proposed EnsembleEdgeFusion network after training on the test set. During testing, both networks used an output stride of 16. The fallouts are presented in the Table 6 below. This table affords a quantitative comparison of segmentation performance between the deep learning models and our proposed EnsembleEdgeFusion method, offering insights into their respective capabilities in accurately delineating different categories within the test dataset.
In Table 6, “Train OS” refers to output stride used in the training phase, while “Eval OS” represents output stride utilized in the validation phase. An output stride of 16 was chosen for both training and evaluation. We compared these settings with established segmentation models such as DeepLabv3+, UNet, DilatedFastFCN with JPU, DANet, and a custom Vanilla Architecture. Following the training of segmentation models with designated trainingset, testset to estimate Mean Intersection over Union (MIoU) value is employed for each trained model. The Ensemble Edge Fusion method achieved a Mean Intersection over Union (MIoU) score of 77.73%, outperforming several widely used models in the field of semantic segmentation. For comparison, DeepLabv3 + achieved an MIoU of 73.56%, U-Net achieved 72.66%, DilatedFastFCN with JPU achieved 74.02%, and DANet achieved 74.89%. These results highlight the effectiveness of combining multiple models through the Ensemble Edge Fusion approach, which significantly enhances segmentation accuracy. When compared to other state-of-the-art methods in the field, our method demonstrates a competitive advantage, particularly in the task of microvascular decompression image segmentation. While other methods may perform similarly on different datasets, the performance of Ensemble Edge Fusion on this specific task shows that the proposed ensemble approach is highly effective for improving segmentation accuracy in medical imaging. The resulting precision values for semantic-segmentation are summarized in the Table 7 below:
From Table 5 it is analyzed that the proposed methods achieve good outcomes in segmentation compared to other models. The graphical representation of the pre-class outcomes of the proposed EnsembleEdge Fusion methods compared to the other deep learning architectures considered in the model like DeepLabv3+, UNet, DilatedFastFCN with JPU, DANet, and a custom Vanilla Architecture are illustrated in the below Fig. 11.
Figure 12 presents instances of unsuccessful outcomes from semantic-segmentation network technique. The uppermost record in figure displays groundtruth images, while row beneath exhibits experimental outcomes generated by suggested method. These results highlight certain faults in cerebral vessels segmentation and cranial nerves segmentation. Specifically, in first image, “pica” remains unsegmented. The second image exhibits multipixel mixing issues, and the third image displays problems related to both incorrect segmentation and multipixel mixing.
In terms of computational efficiency and training complexity, bagging is generally more efficient than stacking. Bagging involves training multiple models independently on different subsets of the data, which is less resource-intensive than stacking, which requires additional training for a meta-learner. However, stacking provides improved performance by combining predictions from diverse models, albeit at the cost of increased training time and computational resources.
Ablation study
For the Ablation Study, we evaluated the contribution of each individual component in the Ensemble Edge Fusion approach, including the performance of each base model (DeepLabv3+, U-Net, DANet, DilatedFastFCN with JPU) and the impact of ensemble techniques (stacking and bagging). We performed a series of experiments using the following configurations:
-
1.
Single Model Performance Evaluated the performance of each model independently (DeepLabv3+, U-Net, DANet, and DilatedFastFCN with JPU).
-
2.
Ensemble Techniques Evaluated the performance of stacking and bagging applied to the models independently.
-
3.
Ensemble Edge Fusion Evaluated the full Ensemble Edge Fusion approach combining both stacking and bagging.
Each experiment was run using the same 2003 RGB MVD images, and performance was measured using the Mean Intersection over Union (MIoU) metric. The ablation study outcomes are mentioned in Table 8.
The Ablation Study results provide valuable insights into the contribution of each component in the Ensemble Edge Fusion approach. DANet and DilatedFastFCN with JPU performed better than DeepLabv3 + and U-Net, with DANet achieving the highest single-model performance (74.89% MIoU). This suggests that models with advanced attention mechanisms, like DANet, are effective for segmenting complex structures in MVD images, though even the best individual models could not match the performance of ensemble methods.
When comparing stacking and bagging, stacking provided the highest improvement in performance, yielding a 75.42% MIoU. By combining multiple models through a meta-learner, stacking capitalizes on their individual strengths, leading to better segmentation. In contrast, bagging resulted in a 75.12% MIoU, stabilizing predictions by training multiple models on different subsets of the data but falling slightly short of stacking’s performance.
The Ensemble Edge Fusion approach, which combines both stacking and bagging, achieved the highest MIoU of 77.73%, demonstrating the synergy between the two ensemble techniques. This improvement of 2.31% over stacking alone indicates that using both strategies together enhances model robustness and performance. These results highlight the effectiveness of ensemble learning in medical image segmentation. Combining diverse models through stacking and bagging provides a substantial boost in accuracy. Future research could explore additional ensemble methods, such as boosting, to further improve segmentation performance and robustness.
Conclusion and future insights
In this study, the intricate task of semantic segmentation in microvascular decompression images has been addressed, a domain challenged by the scarcity of publicly available medical image datasets and the need for expert annotation. A self-curated dataset of 2003 RGB microvascular decompression images, meticulously paired with annotated masks, was introduced. Through rigorous data preprocessing and augmentation, the training dataset’s robustness was significantly improved. Extensive experimentation involved training various state-of-the-art semantic segmentation models, including DeepLabv3+, U-Net, DilatedFastFCN with JPU, DANet, and a custom Vanilla architecture. These models underwent evaluation using a range of performance metrics, demonstrating competitive results in accuracy, F1 score, sensitivity, and specificity. Notably, the DeepLabv3 + model displayed exceptional F1 score performance. The introduction of ensemble techniques, such as stacking and bagging, further improved segmentation performance. Bagging, particularly with the Naïve Bayes approach, yielded significant enhancements, emphasizing the potential of ensemble methods in medical image segmentation. The proposed EnsembleEdgeFusion technique exhibited superior loss reduction during training compared to DeepLabv3 + and achieved the highest Mean Intersection over Union (MIoU) score of 77.73%, surpassing other models. Detailed category-wise analysis confirmed its superiority in accurately delineating various categories within the test dataset.
This research in semantic segmentation of microvascular decompression images paves the way for upcoming exploration, including integrating data from various modalities, developing real-time segmentation techniques, investigating transfer learning possibilities, and extending the work to 3D semantic segmentation for volumetric medical data, enabling more comprehensive analysis83.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Plis, S. M. et al. Deep learning for neuroimaging: A validation study. Front. NeuroSci. 8, 229 (2014).
Li, Q. et al. Medical image classification with convolutional neural network. In 2014 13th international conference on control automation robotics & vision (ICARCV) 844–848 (IEEE, 2014).
Ypsilantis, P. P. et al. Predicting response to neoadjuvant chemotherapy with PET imaging using convolutional neural networks. PloS one 10(9) (2015).
Do, D. et al. Using deep neural networks and biological subwords to detect protein S-sulfenylation sites. Brief. Bioinform. 22(3) (2021).
Turaga, S. C. et al. Convolutional networks can learn to generate affinity graphs for image segmentation. Neural Comput. 22(2), 511–538 (2010).
Roth, H. R. et al. Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation. In MICCAI 2015: 18th International Conference, Munich, Germany, Proceedings 556–564 (Springer International Publishing, 2015).
Roth, H. R. et al. Evrim Turkbey, and Ronald M. Summers.: A new 2.5 D representation for lymph node detection using random sets of deep convolutional neural network observations. In MICCAI 2014: 17th International Conference, Boston, MA, USA, Proceedings 520–527 (Springer International Publishing, 2014).
Le, N. Q. et al. A computational framework based on ensemble deep neural networks for essential genes identification. Int. J. Mol. Sci. 21(23), (2020).
Koyamada, S. et al. Deep learning of fMRI big data: A novel approach to subject-transfer decoding. arXiv preprint arXiv,1502.00093, (2015).
Csurka, G. & Perronnin, F. An efficient approach to semantic segmentation. Int. J. Comput. Vis. 95, 198–212 (2011).
Guo, Y., Liu, Y., Georgiou, T. & Lew., M. S. A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr. 7, 87–93 (2018).
Odstrcilik, J. et al. Retinal vessel segmentation by improved matched filtering: Evaluation on a new high-resolution fundus image database. IET Image Proc. 7(4), 373–383 (2013).
Chakraborti, T. et al. A Self-adaptive matched filter for retinal blood vessel detection. Mach. Vis. Appl. 26, 55–68 (2015).
Frangi, A. F. et al. Multiscale vessel enhancement filtering. In MICCAI’98: First International Conference Cambridge, 1998 Proceedings 1 130–137 (Springer Berlin Heidelberg, 1998).
Nguyen, U. T. V., Alauddin Bhuiyan, Laurence, A. F., Park & Ramamohanarao, K. An effective retinal blood vessel segmentation method using multi-scale line detection. Pattern Recogn. 46(3), 703–715 (2013).
Saffarzadeh, V., Mohammadi, A., Osareh & Shadgar, B. Vessel segmentation in retinal images using multi-scale line operator and K-means clustering. J. Med. Signals Sens. 4(2) (2014).
Zhang, L., Fisher, M. & Wang, W. Retinal vessel segmentation using multi-scale textons derived from keypoints. Comput. Med. Imaging Graph. 45, 47–56 (2015).
Carballal, A. et al. Automatic multiscale vascular image segmentation algorithm for coronary angiography. Biomed. Signal Process. Control. 46, 1–9 (2018).
Khawaja, A. et al. A multi-scale directional line detector for retinal vessel segmentation. Sensors 19(22) (2019).
Sun, K., Chen, Z. & Jiang, S. Morphological multiscale enhancement, fuzzy filter and watershed for vascular tree extraction in angiogram. J. Med. Syst. 35, 811–824 (2011).
Kass, M., Witkin, A. T. & Tetzopoulos, D. Active contour models. International Journal of Computer Vision. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 1 (1998).
Zhao, Y., Rada, L., Chen, K. & Harding, S. P. Automated vessel segmentation using infinite perimeter active contour model with hybrid region information with application to retinal images. IEEE Trans. Med. Imaging 34(9), 1797–1807 (2015).
Zhao, Y. et al. Saliency driven vasculature segmentation with Infinite perimeter active contour model. Neurocomputing 259, 201–209 (2017).
Nirmala Devi, S. Comparison of active contour models for image segmentation in X-ray coronary angiogram images. J. Med. Eng. Technol. 32(5), 408–418 (2008).
Tagizaheh, M., Sadri, S. & Doosthoseini, A. M. Segmentation of coronary vessels by combining the detection of centerlines and active contour model. In 2011 7th Iranian Conference on Machine Vision and Image Processing 1–4 (2011).
Wang, J. et al. An active contour model based on adaptive threshold for extraction of cerebral vascular structures. Comput. Math. Methods Med. 2016(1), 6472397 (2016).
Brieva, J. et al. A Level Set Method for Vessel Segmentation in Coronary Angiography. In 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference 6348–6351 (2006).
Roychowdhury, S. et al. Iterative vessel segmentation of fundus images. IEEE Trans. Biomed. Eng. 62(7), 1738–1749 (2015).
Lara, D. S. D. et al. A semi-automatic method for segmentation of the coronary artery tree from angiography. In 2009 XXII Brazilian Symposium on Computer Graphics and Image Processing 194–201 (2009).
Shoujun, Z., Jian, Y. & Yongtian, W. Automatic segmentation of coronary angiograms based on fuzzy inferring and probabilistic tracking. Biomed. Eng. Online 9(1), 1–21 (2010).
Wan, T. et al. Automated coronary artery tree segmentation in x-ray angiography using improved hessian based enhancement and statistical region merging. Comput. Methods Programs Biomed. 157, 179–190 (2018).
Sum, K. W. & Paul, Y. S. C. Vessel extraction under non-uniform illumination: A level set approach. IEEE Trans. Biomed. Eng. 55(1), 358–360 (2007).
Lázár, I. & András, H. Segmentation of retinal vessels by means of directional response vector similarity and region growing. Comput. Biol. Med. 66, 209–221 (2015).
Long, J. et al. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 3431–3440 (2015).
Simonyan, K., & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv 1409.1556 (2014).
LeCun, Y. et al. Gradient-based learning applied to document recognition. Proc. IEEE 86(11), 2278–2324 (1998).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In MICCAI 2015: 18th International Conference, Germany, Proceedings, Part III Vol. 18 234–241. (Springer International Publishing, 2015).
Badrinarayanan, V. & Kendall, A. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017).
Nasr-Esfahani, E. et al. Vessel extraction in x-ray angiograms using deep learning. In 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 643–646 (2016).
Phellan, R. et al. Vascular segmentation in TOF MRA images of the brain using a deep convolutional neural network. In Intravascular Imaging and Computer Assisted Stenting, and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis: 6th Joint International Workshops, CVII-STENT 2017 and Second International Workshop, LABELS 2017, Held in Conjunction with MICCAI 2017, Canada, 2017, Proceedings 2 39–46 (Springer International Publishing, 2017).
Mo, J. Multi-level deep supervised networks for retinal vessel segmentation. Int. J. Comput. Assist. Radiol. Surg. 12, 2181–2193 (2017).
Jiang, Z., Zhang, H., Wang, Y. & Ko, S. B. Retinal blood vessel segmentation using fully convolutional network with transfer learning. Comput. Med. Imaging Graph. 68, 1–15 (2018).
Noh, K. J., Park, S. J. & Lee, S. Scale-space approximated convolutional neural networks for retinal vessel segmentation. Comput. Methods Programs Biomed. 178, 237–246 (2019).
Livne, M. et al. A U-Net deep learning framework for high performance vessel segmentation in patients with cerebrovascular disease. Front. Neurosci. 13(97), (2019).
Chen, L. C., Zhu, Y., Papandreou, G., Schroff, F. & Adam, H. Encoder-Decoder with atrous separable convolution for semantic image segmentation. In Proceedings of ECCV 801–818 (Munich, Germany, 2018).
HuBMAP. Kaggle.com; Kaggle. (2023). https://www.kaggle.com/code/mersico/hubmap-eda-pycocotools-submission.
Alexander Kirillov. (n.d.). Google.com.: From (2023). https://scholar.google.com/citations?user=bHn29ScAAAAJ.
Chen, L. C. et al. DeepLab: Semantic Image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. Arxiv.org. (2023).
Lin, T. Y. et al. Feature Pyramid Networks for Object Detection. In arXiv [cs.CV]. (2017).
Chen, L. C., Papandreou, G., Kokkinos, I., Murphy, K. & Yuille, A. L. Semantic image segmentation with deep convolutional Nets and fully connected Crfs. ArXiv:1412.7062 (2014).
Wang, G. A perspective on deep imaging. IEEE Access 4, 8914–8924 (2016).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017).
Shen, D. et al. Suk Deep learning in medical image. Anal. Annual Rev. Biomedical Eng. 19, 221–248 (2017).
Lee, K. et al. Superhuman accuracy on the SNEMI3D connectomics challenge. arXiv preprint arXiv:1706.00120 (2017).
Puttagunta, M. & Ravi., S. Medical image analysis based on deep learning approach. Multimed. Tools Appl. 80, 24365–24398 (2021).
Yang, Y., Hu, Y., Zhang, X. & Wang, S. Two-stage selective ensemble of CNN via deep tree training for medical image classification. IEEE Trans. Cybern. 52(9), 9194–9207 (2021).
Xue, D. et al. An application of transfer learning and ensemble learning techniques for cervical histopathology image classification. IEEE Access 8 (2020).
Logan, R. et al. Deep convolutional neural networks with ensemble learning and generative adversarial networks for alzheimer’s disease image data classification. Front. Aging Neurosci. (2021).
Rajaraman, S. et al. iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access 8, 115041–115050 (2020).
Mohammed, M., Mwambi, H., Mboya, I. B., Elbashir, M. K. & Omolo, B. A stacking ensemble deep learning approach to cancer type classification based on TCGA data. Sci. Rep. 11(1) (2021).
Hameed, Z. et al. Breast cancer histopathology image classification using an ensemble of deep learning models. Sensors 20(16) (2020).
Das, A. et al. Design of deep ensemble classifier with fuzzy decision method for biomedical image classification. Appl. Soft Comput. 115 (2022).
Zhang, J., Wang, Y., Li, G. & Yuantian Sun, and Strength of ensemble learning in multiclass classification of rockburst intensity. Int. J. Numer. Anal. Meth. Geomech. 44(13), 1833–1853 (2020).
Ju, C., Bibaut, A. & Mark van der Laan. The relative performance of ensemble methods with deep convolutional neural networks for image classification. J. Appl. Stat. 45(15), 2800–2818 (2018).
Sułot, D. et al. Glaucoma classification based on scanning laser ophthalmoscopic images using a deep learning ensemble method. PloS One 16(6) (2021).
Kuncheva, L. I. Measures of diversity in classifier ensembles and their relationship with the ensemble accuracy. Mach. Learn. 51, 181–207 (2003).
Ganaie, M. A. et al. Ensemble deep learning: A review. Eng. Appl. Artif. Intell. 115 (2022).
Cao, Y., Geddes, T. A., Yang, J. Y. H. & Yang, P. Ensemble deep learning in bioinformatics. Nat. Mach. Intell. 2(9), 500–508, (2020).
Sagi, O. & Rokach, L. Ensemble learning: A survey. Wiley Interdiscip. Rev: Data Min. Knowl. Discov. 8(4) (2018).
Kandel, I. Comparing stacking ensemble techniques to improve musculoskeletal fracture image classification. J. Imaging 7(6) (2021).
Berman, M. e al. The Lovász-softmax loss: A tractable surrogate for the optimization of the intersection-over-union measure in neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 4413–4421, (2018).
Liang, C., Yang, L., Zhang, B., Li, R. & Shiwen Guo. 3D multimodal image fusion based on MRI in the preoperative evaluation of microvascular decompression: A meta analysis. Exp. Ther. Med. 25 (4), 1–8 (2023).
Shono, N., Kin, T. & Saito, N. Preoperative 3D microvascular decompression simulation. No Shinkei Geka. Neurol. Surg. 52(1), 163–176 (2024).
Zhang, Y. et al. Predictive value of preoperative magnetic resonance imaging structural and diffusion indices for the results of trigeminal neuralgia microvascular decompression surgery. Neuroradiology 1–7 (2023).
Liu, F. et al. Thalamus and Facial Nerve Nuclei Glucose Metabolism in Patients with Hemifacial Spasm Before and After Microvascular Decompression Surgery. pp. 2428–2428, (2022).
Wang, J. et al. Application of neuronavigation in microvascular decompression: Optimizing craniotomy and 3D reconstruction of neurovascular compression. J. Craniofac. Surg. 34(7) (2023).
Jeon, C., Kim, M., Lee, H. S., Kong, D. S. & Park, K. Outcomes after microvascular decompression for hemifacial spasm without definite radiological neurovascular compression at the root exit zone. Life 13, 10 (2023).
Dodda, A. et al. Ensemble Approach for blood vessel segmentation in retinal images: Combining UNet and SegNet models.
Waheed, Z., Gui, J., Amjad, K., Waheed, I. & Asif, S. An ensemble approach of deep CNN models with Beta normalization aggregation for Gastrointestinal disease detection. Biomed. Signal Process. Control 105, 107567 (2025).
Waheed, Z. et al. (2025). A novel lightweight deep learning based approaches for the automatic diagnosis of gastrointestinal disease using image processing and knowledge distillation techniques. Comput. Methods Programs Biomed. 260, 108579.
Vij, R. & Arora, S. A hybrid evolutionary weighted ensemble of deep transfer learning models for retinal vessel segmentation and diabetic retinopathy detection. Comput. Electr. Eng. 115, 109107 (2024).
Hong, Q. et al. 3D vasculature segmentation using localized hybrid level-set method. Biomed. Eng. 13(1), 1–15 (2014).
Chen, L. C., Papandreou, G., Kokkinos, I., Murphy, K. & Yuille, A. L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected Crfs. IEEE Trans. Pattern Anal. Mach. Intell. 40, 834–848 (2017).
Author information
Authors and Affiliations
Contributions
B.D. conceptualised and designed the study, prepared the dataset, implemented the proposed methodologies, and draughted the manuscript. B.D. & M.V. contributed to the preprocessing and augmentation strategies, as well as the training and evaluation of semantic segmentation models. P.S. & D.V. conducted experiments on ensemble techniques, including stacking and bagging, and analysed the results. B.D. & D.V. provided support for the implementation of the EnsembleEdgeFusion technique and contributed to the interpretation of results and manuscript refinement. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dhiyanesh, B., Vijayalakshmi, M., Saranya, P. et al. EnsembleEdgeFusion: advancing semantic segmentation in microvascular decompression imaging with innovative ensemble techniques. Sci Rep 15, 17892 (2025). https://doi.org/10.1038/s41598-025-02470-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-02470-5
Keywords
This article is cited by
-
Enhancing privacy in IoT-based healthcare using provable partitioned secure blockchain principle and encryption
Scientific Reports (2025)
-
Feature fusion context attention gate UNet for detection of polycystic ovary syndrome
Scientific Reports (2025)