Abstract
Kyphosis is a prevalent spinal condition where the spine curves in the sagittal plane, resulting in spine deformities. Curvature estimation provides a powerful index to assess the deformation severity of scoliosis. In current clinical diagnosis, the standard curvature estimation method for quantitatively assessing the curvature is performed by measuring the vertebral angle, which is the angle between two lines, drawn perpendicular to the upper and lower endplates of the involved vertebra. However, manual Cobb angle measurement requires considerable time and effort, along with associated problems such as interobserver and intraobserver variations. Hence, in this study, we propose UNet-based Attention Network for Thoracolumbar Vertebral Compression Fracture Angle (UANV), a vertebra angle measuring model using lateral spinal X-ray based on a deep convolutional neural network (CNN). Specifically, we considered the detailed shape of each vertebral body with an attention mechanism and then recorded each edge of each vertebra to calculate vertebrae angles.
Similar content being viewed by others
Introduction
The spine is one of the most crucial parts of the human body. It provides humans with many significant functions such as carrying the weight of the body and protecting the spinal cord and nerves within. Spinal deformities, such as scoliosis and kyphosis, are prevalent conditions that significantly affect patients’ quality of life by causing pain, reducing mobility, and resulting in severe complications if remain untreated1,2,3. The vertebral angle, a crucial metric in diagnosing and assessing the progression of these deformities, quantifies the degree of vertebral deformities on X-ray images. The traditional vertebral angle measurement involves manually drawing lines along the upper and lower endplates and calculating the intersection angle of these lines. Despite its clinical importance, manual angle measurement is labor-intensive, time-consuming, and prone to interobserver variability, thereby requiring the development of automated and accurate methods4,5,6. Automating the Cobb angle measurement requires precise vertebral segmentation from X-ray images, a challenging task due to spinal anatomy complexity and image quality variability. Recent advances in deep learning have revolutionized medical image analysis, with convolutional neural networks (CNNs) playing a pivotal role7. Among various architectures, the UNet and feature pyramid network (FPN) have appeared as prominent models for biomedical image segmentation. The UNet architecture, which was originally designed for biomedical image segmentation, has gained widespread adoption due to its encoder–decoder structure with skip connections. The encoder captures the contextual features through a series of down-sampling operations, whereas the decoder reconstructs the segmentation map with upsampling operations, which ensures high-resolution outputs by concatenating features from corresponding encoder layers. This architecture is superior in capturing both global and local features, making it highly effective for segmenting vertebrae in X-ray images. To further improve the performance of segmentation models, various advanced architectures such as Attention UNet8 and Residual UNet9, have been proposed. The Attention UNet integrates attention mechanisms within the UNet architecture to improve focus on the most relevant features. This enables the network to prioritize important regions in the image, causing more accurate segmentation results. The attention gates selectively highlight salient features while suppressing irrelevant information, thereby refining the segmentation output. Further, the Residual UNet incorporates residual connections into the UNet architecture to facilitate the flow of gradients during training and mitigate the vanishing gradient problem. By adding shortcuts between layers, Residual UNet ensures that the network learns deeper representations without performance degradation. This results in improved segmentation accuracy, particularly for complex structures such as the vertebrae.
In this study, we propose a novel approach that combines a UNet-based architecture with an integrated attention mechanism to automate vertebral segmentation from lateral X-ray images and calculate the vertebral angle. Our model uses the strengths of UNet for detailed segmentation and incorporates attention gates to improve focus on relevant features. The segmented vertebrae are converted into polygons to extract precise vertex coordinates, which are then utilized to compute the vertebral angle. This method aims to provide an accurate, efficient, and reliable solution for clinical assessments of spinal deformities. The main contributions of this research are summarized as follows:
-
Established an X-ray-based spine fracture dataset of 1,349 images for segmentation.
-
Proposed an improved UNet-based model for automating vertebral segmentation from lateral X-ray images and calculating the vertebral angle.
-
Investigated the efficiency of using an attention mechanism for vertebrae segmentation.
Methods
Dataset
Korea University Hospital provided data used in this study. This study was approved by the Institutional Review Board of Korea University Anam Hospital (IRB No. 2023AN0268). Spine X-rays of adult patients aged > 18 years with or without compression fractures were collected based on admission dates from January 2018 to December 2019 from Korea University Anam Hospital. Exclusion criteria included imaging findings of previous spine surgery including instrumentation, vertebroplasty, radiographic evidence of tumor or infection around the spine, and poor image quality, including an inappropriately positioned lateral view. All the images were desensitized before being used (e.g., removal of name, age, examination date, sex). This study enrolled 1,349 patients based on image and disease incidence findings. These 1,349 images are then randomly split into train, validation, and test with an 8:1:1 ratio. The polygon is utilized to show the shape and location of the lumbar vertebral body. Neurosurgeons performed data annotation, an important part of object detection because an accurate decision on medical image data is required. Data annotation was performed by four trained researchers under the supervision of four neurosurgeons. A minimum of six points, including the four corners, were drawn along each single vertebral body using an annotation tool (Aview, Coreline Soft, Korea). Annotation was conducted using a consensus approach.
First, we applied contrasted limited adaptive histogram equalization10 to increase the contrast of images. Each image is then resized to I(x) ∈ R800 × 1333 pixels and used as input for the proposed method. We applied random flip and rotation for data augmentation. Significant differences in brightness, contrast, resolution, and size of X-ray images significantly affect the performance of the model; thus, we applied a few pre-processing methods: data cleaning, image normalization, and data augmentation. Although we implemented data pre-processing steps such as contrast adjustment and normalization, these steps currently require semi-manual intervention. Full automation of pre-processing remains a goal, and future work will aim to develop end-to-end automated pipelines to further minimize operator involvement.
Base model
The proposed UNet11 is the base model for vertebral body segmentation, and it attached an attention mechanism to address detailed characteristics of vertebral bodies. Specifically, the model operates in two primary stages: one is a vertebral segmentation using a segmentation module based on UNet with an attention network and the other is the vertebral angle calculation from segmented regions. Figure 1 illustrates the overall architecture. The traditional UNet model is used for the segmentation task due to its efficacy in biomedical image segmentation. The architecture consists of an encoder–decoder structure with skip connections to preserve spatial information. The encoder path captures the context of the input image through a series of convolutional and down-sampling operations, whereas the decoder path reconstructs the image to produce a detailed segmentation map. However, to improve model performance, we introduce a locational attention module to integrate the information about each vertebral body location.
The encoder consists of several convolutional layers, each followed by a ReLU activation function and a max-pooling operation. The left side of Fig. 1 represents the encoder path of the UNet, consisting of multiple convolutional layers followed by max-pooling operations (indicated by red arrows). Each layer extracts features at different abstraction levels, which progressively reduce the spatial dimensions while increasing the depth, thereby capturing higher-level features of the input X-ray images.
The bottleneck layer at the deepest part of the network performs convolutions that further refine the feature representation. The decoder part, presented on the right side of Fig. 1, is where upsampling operations (indicated by green arrows) are applied to increase the spatial dimensions, which reconstructs the segmented output. Further, the decoder path includes convolutional layers that combine features from corresponding encoder layers through skip connections (indicated by dashed lines). This structure ensures the recovery of the fine details lost during down-sampling.
Attention module
Attention modules are integrated at various network stages, positioned between the encoder and decoder paths. These modules improve the features by focusing on relevant areas and suppressing irrelevant ones. Each attention module processes low- and high-level features. Low-level features from the encoder are processed through a 3 × 3 convolutional layer, whereas high-level features from the decoder undergo global pooling followed by a 1 × 1 convolutional layer. These low- and high-level features are then combined using element-wise multiplication for attention coefficient computation. The attention-modulated features are subsequently upsampled and added to the original low-level features, which improves the feature map.
Angle calculation module
Upon obtaining the segmentation results, a postprocessing algorithm is used to convert these results into polygon representations. The conversion process begins by extracting the contours of the segmented regions, which effectively translates the pixel-based segmentation output into vector-based polygons. Each polygon is then simplified to a quadrilateral by determining its four corner points: the top–left (VT L), top–right (VT R), bottom–right (VB L), and bottom–left (VB R). The coordinates of these vertices are denoted as follows:
After establishing the quadrilateral approximation of each polygon, the coordinates of these corner points are used to calculate the top and bottom edge angles of the quadrilateral. The angle θtop of the top edge, formed by the line connecting VT L and VT R is presented as:
Similarly, the angle θbottom of the bottom edge, formed by the line connecting VB L and VB R is expressed as:
The angle θ between the top and bottom edges is then computed as the difference between these two angles as follows:
This angle is a quantitative measure of the relative orientation between the top and bottom edges of the polygon. This postprocessing step is crucial for applications that require precise geometric and positional information from segmentation outputs. This method facilitates the accurate interpretation and analysis of segmented regions by converting segmentation results into polygonal representations and calculating the angles between their edges.
Experiment methods
The environment of this study is the server equipped with a GPU with an operating system Ubuntu version 10.02, and we used Python with Pytorch as a deep learning framework. We first split the data to train, validation, and test with an 8:1:1 ratio, then combined the image file name, size, image ID, and annotation information of each train, validation, and test dataset. Finally, we train and test the model using train, validation, and test datasets, with an optimizer and a loss function. The optimizer utilized beta 0.9, beta2 0.999, and epsilon 1e −8 with an initial learning rate of 0.005 for 50 epochs. We selected the final optimal model based on validation set performance. We validated the efficiency of the proposed network by comparing it with common detection methods such as UNet, UNeXt12, and FPN. We used BCE loss and Dice loss during training as well as computed Dice loss for each anatomical landmark of the image for segmentation. The training of our model is conducted by minimizing the loss function using the Adaptive Moment Estimation (Adam) optimizer, with parameters beta1 = 0.9, beta2 = 0.999, epsilon = 1e-8, and an initial learning rate of 0.005. Training was performed for 50 epochs. To prevent overfitting, we used early stopping based on validation performance and regularization techniques. Further training with a larger and more diverse dataset is planned to enhance generalizability.
Results
In this experiment, we input lumbar spine X-ray images into the model. The proposed model primarily uses CNNs to extract crucial features, generate vertebral edges from the resulting feature maps, and perform vertebral angle measurements. Considering the variability in the original X-ray image dimensions, all images were resized to a standardized input size of 800 × 1333. Multiple image sizes were assessed to optimize performance, and this size was selected for its superior results.
Table 1 compares the performance of the proposed method with several well-known segmentation methods, namely UNet, UNeXt, and FPN, on the provided dataset. The evaluation metrics used include mean squared error (MSE) and dice coefficient (Dice), which measure the accuracy of segmentation and model loss during the training, validation, and test phases. Table 1 shows that the proposed method consistently outperforms the competing models across all phases. It achieves the lowest train loss during the training phase, with an MSE of 0.03275 and a Dice score of 0.8895, indicating high efficiency in learning from the training data. This is further reflected in its superior Dice score, indicating a better overlap between predicted and actual segmentation masks compared to the other models. The validation performance emphasizes the robustness of the proposed method, with a validation loss with MSE of 0.03201 and a Dice score of 0.8899. This indicates the model’s ability to maintain high accuracy when applied to unseen validation data, thereby effectively minimizing the risk of overfitting. The proposed model continues to excel, recording the lowest, when assessing test data. Test loss revealed an MSE of 0.02321 and a Dice score of 0.8865, emphasizing its strong generalization ability. UNeXt also performs well, with a test loss demonstrating an MSE of 0.02762 and a Dice score of 0.8800, indicating its efficacy in generalizing to new data, although it remains slightly behind the proposed method. In contrast, the feature pyramid network model exhibits the highest losses across all metrics with a train loss of 0.03625, validation loss of 0.03361, and test loss of 0.03502, indicating that it struggles with learning and generalizing. UNet performs reasonably well across the board, with lower train, validation, and test losses t than those of the FPN but still do not match the efficiency of the proposed method or UNeXt.
Overall, the results reveal the superior performance of the proposed method in terms of training efficiency and generalization to new data, making it a promising approach for medical image segmentation tasks, particularly in analyzing lumbar spine X-rays.
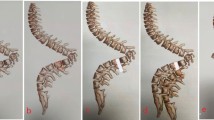
The image presented in Fig. 2 illustrates the outcomes of a multi-step process involved in spinal vertebrae segmentation and analysis. The left panel exhibits the segmentation result, where individual vertebral bodies have been successfully isolated from the background. This segmentation is crucial for subsequent analysis steps because it provides a clear delineation of each vertebra, ensuring that each segment can be independently analyzed without interference from adjacent structures. The segmented vertebrae appear to be accurately identified, with each vertebra distinguishable from the others.
The image in the middle presents the edge detection results, highlighting the boundaries of the segmented vertebrae. Edge detection is a vital step in improving the segmentation by pinpointing the precise borders of each vertebra. This process helps in enhancing the segmentation accuracy by ensuring that the edges are clearly defined. The colored dots in the middle panel indicate key points along these edges, likely representing crucial landmarks used for further analysis.
The image on the right shows the angle calculation results, where each colored dot corresponds to a different vertebral body. These colors are utilized to uniquely determine each vertebra, thereby facilitating a detailed analysis of their individual orientations and spatial associations. The angle calculation is crucial for assessing spine alignment and curvature, which is important in diagnosing and monitoring spinal conditions. The use of different colors for each vertebral body enables easy visual differentiation and helps in tracking changes or abnormalities in vertebral alignment over time. Overall, the image illustrates a comprehensive workflow from segmentation to edge detection and angle calculation, with each step building on the previous to provide a detailed and accurate analysis of the spinal structure.
Comparison of each vertebra
In this study, we analyzed the performance of our model for various vertebral bodies (L5, L4, L3, etc.) in spinal segmentation and edge detection. We assessed using two key performance metrics: the average DICE coefficient for segmentation and the average MSE for edge detection. Table 2 shows the Average DICE coefficient, which measures the overlap between predicted and actual segmentation regions, revealing that the L4 vertebral body achieved the highest segmentation accuracy, with a DICE coefficient of 0.9151. The T12 vertebral body followed closely, with a DICE coefficient of 0.9174, indicating similarly high segmentation accuracy.
In contrast, the L5 vertebral body recorded the lowest DICE coefficient of 0.8326, indicating that it had the least accurate segmentation among the vertebrae analyzed. However, the T12 vertebral body demonstrated the best performance, with the lowest error rate of 0.0413, when considering the Average MSE Loss, which assesses the squared differences between predicted and true values. Despite the lower DICE coefficient, L5 demonstrated a relatively low MSE loss of 0.0424, indicating that the model’s predictions for L5 were still quite precise whereas its segmentation overlap accuracy may be lower.
Overall, the results indicate that L4 and T12 vertebrae were segmented with the highest accuracy and minimal error, respectively, whereas the L5 and T11 vertebrae may benefit from additional focus to improve segmentation performance.
Discussion
In this work, we present a novel approach that integrates a UNet-based architecture with an attention mechanism to automate vertebra segmentation from lateral X-ray images and vertebral angle computation. This method overcomes the limitations of manual measurement techniques using advanced segmentation to provide a more reliable tool for diagnosing and monitoring spinal deformities. CNNs, which are fundamental algorithms in computer vision, have seen increasing use in spinal image analyses. Li et al.13 applied deep learning to determine osteoporotic lumbar fractures in spinal X-rays. Similarly, Galbusera et al.14 developed a fully automated approach for measuring spinal parameters using CNNs. Further, Cho et al.15 demonstrated the successful automated measurement of lumbar lordosis to segment vertebral bodies with the UNet model applied to spinal X-rays. The UNet, a popularly adopted CNN architecture in medical imaging, is designed for effective binary semantic segmentation, particularly with small biomedical image datasets16. However, the UNet has limitations when detecting and performing segmentation of fractured and adjacent normal vertebrae.
To overcome some limitations, we customized the UNet to enable accurate segmentation. The UNet framework facilitates detailed segmentation, whereas the attention gates improve focus on critical features. The segmented vertebrae are then converted into polygons to extract precise vertex coordinates for accurate angle calculation. Our approach aims to achieve high segmentation accuracy and dependable vertebral angle measurements by combining these elements, thereby improving diagnostic precision and patient care outcomes. The highest accuracy of our architecture was the results indicating L4 (Dice: 0.9151 and MSE: 0.0837) and T12 vertebrae (Dice: 0.9174 and MSE: 0.0413). The proposed architecture exhibited better performance than other algorithms. The MSE and dice coefficient of our results in the test dataset were 0.02321 and 0.8865, respectively.
However, some limitations remained in this study that need to be improved. First, the size and diversity of our dataset is limited. A greater volume of data is expected to increase the performance of deep learning models because the dataset with 1,349 patients is quite small for deep learning. Hence, our data collection strategy requires the inclusion of diverse views of X-ray images alongside the acquisition of more data. Furthermore, investigating the performance of the model across diverse clinical contexts and patient populations, including varying spinal pathologies, will be crucial for examining its overall applicability and potential for broader clinical implementation. Our model can process external images; however, we cannot guarantee optimal performance across all populations. Since the current dataset consists of Korean adult patients, fine-tuning may be required for images from different ethnic groups, pediatric populations, or different imaging systems to maintain high accuracy.
Specifically, we considered the detailed shape of each vertebral body with an attention mechanism and then took an edge of each vertebra to calculate vertebral angles. In particular, our algorithms support the automatic Cobb angle measurement without considerable time and effort in clinical field. In this study, deep learning algorithms were developed to measure a vertebra angle using lateral spinal X-ray. In clinical practice, high accuracy is crucial because current manual measurements often suffer from operator variability and human error. Automated, reliable measurements can significantly reduce these issues and support more consistent clinical decision-making.
As clinical practice continues to evolve toward personalized medicine, the demand for patient-specific instrumentation is growing. Consequently, engineers are also increasingly required to develop computational models tailored to individual patients, which necessitates automated and precise measurement tools17,18. We believe that our tool directly addresses this need and will serve as a valuable asset in advancing biomechanical engineering research. The ultimate goal of our model is to develop a generalized system that can be widely applied beyond our institution, potentially for multi-institutional clinical use.
Data availability
The code used in this study is available at https://github.com/YurimALee/UANV. Datasets in the study are available from the corresponding author upon reasonable request with our IRB allowance.
Change history
22 November 2025
The original online version of this Article was revised: The original version of this Article contained an error in Reference 17, which was incorrectly given as: Prasannaah Hadagali, J. R., Peters, S. & Balasubramanian Morphing the feature-based multi-blocks of normative/healthy vertebral geometries to scoliosis vertebral geometries: development of personalized finite element models. Comput. Methods Biomech. Biomed. Eng. 21 (2018). The correct reference is: Hadagali, P., Peters, J.R., & Balasubramanian, S. Morphing the feature-based multi-blocks of normative/healthy vertebral geometries to scoliosis vertebral geometries: development of personalized finite element models. Comput. Methods Biomech. Biomed. Eng. 21 (2018).
References
Mohamadi, A., Googanian, A., Ahmadi, A. & Kamali, A. Comparison of surgical or nonsurgical treatment outcomes in patients with thoracolumbar fracture with score 4 of TLICS: A randomized, single-blind, and single-central clinical trial. Medicine 97, e9842 (2018).
Shen, J., Xu, L., Zhang, B. & Hu, Z. Risk factors for the failure of spinal burst fractures treated conservatively according to the thoracolumbar injury classification and severity score (TLICS): A retrospective cohort trial. PLoS ONE. 10, e0135735 (2015).
Alimohammadi, E. et al. Predictors of the failure of Conservative treatment in patients with a thoracolumbar burst fracture. J. Orthop. Surg. Res. 15, 514 (2020).
Sadiqi, S. et al. Measurement of kyphosis and vertebral body height loss in traumatic spine fractures: An international study. Eur. Spine J. 26, 1483–1491 (2017).
Ruiz Santiago, F. et al. Classifying thoracolumbar fractures: role of quantitative imaging. Quant. Imaging Med. Surg. 6, 772–784 (2016).
Street, J. et al. Intraobserver and interobserver reliabilty of measures of kyphosis in thoracolumbar fractures. Spine J. 9, 464–469 (2009).
Qu, B. et al. Current development and prospects of deep learning in spine image analysis: A literature review. Quant. Imaging Med. Surg. 12, 3454–3479 (2022).
Oktay, O. et al. Attention u-net: learning where to look for the pancreas. ArXiv Preprint arXiv:180403999 (2018).
Diakogiannis, F. I., Waldner, F., Caccetta, P. & Wu, C. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm Remote Sens. 162, 94–114 (2020).
Yadav, G., Maheshwari, S. & Agarwal, A. Contrast limited adaptive histogram equalization based enhancement for real time video system in international conference on advances in computing, communications and informatics (ICACCI) 2392–2397 (IEEE, 2014). (2014).
Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical. in Medical image computing and computer-assisted intervention–MICCAI 2015:18th international conference, Munich, Germany, October 5–9, proceedings, part III 18 234–241 (Springer, 2015).
Valanarasu, J. M. J., Patel, V. M. & Unext Mlp-based rapid medical image segmentation network. in International conference on medical image computing and computer-assisted intervention 23–33 Springer, (2022).
Li, Y. C. et al. Can a deep-learning model for the automated detection of vertebral fractures approach the performance level of human subspecialists? Clin. Orthop. Relat. Res. 479, 1598–1612 (2021).
Galbusera, F. et al. Fully automated radiological analysis of spinal disorders and deformities: A deep learning approach. Eur. Spine J. 28, 951–960 (2019).
Cho, B. H. et al. Automated measurement of lumbar lordosis on radiographs using machine learning and computer vision. Global Spine J. 10, 611–618 (2020).
Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation in medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference; Oct 5–9; Munich, Germany. Proceedings, Part III 182015 234–241 (Springer International Publishing, 2015).
Hadagali, P., Peters, J.R., & Balasubramanian, S. Morphing the feature-based multi-blocks of normative/healthy vertebral geometries to scoliosis vertebral geometries: development of personalized finite element models. Comput. Methods Biomech. Biomed. Eng. 21 (2018).
Joeri Kok. et al. Automatic generation of subject-specific finite element models of the spine from magnetic resonance images. Front. Bioeng. Biotechnol. 11 (2023).
Acknowledgements
We would like to thank the Advanced Medical Imaging Institute in the Department of Radiology, Korea University Anam Hospital in the Republic of Korea, and the researchers for providing software, datasets, and various forms of technical support. JHK, YUK, ISY, and YAH assisted with data organization and case-level annotation.
Funding
This work was supported by the Ministry of Education of the Republic of Korea, the National Research Foundation of Korea (RS-2022-NR073261, RS-2023-00239603, RS-2023-00262309). This study was supported by grants from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare (KR) [HR22C1302]. This work was supported by the Soonchunhyang University Research Fund.
Author information
Authors and Affiliations
Contributions
Y.L. and Y.C. wrote the main manuscript. Y.L., J.K., and Y.C. performed the experiments and prepared the figures. J.K., K.L., S.A., KSA, and JWH prepared the dataset and confirmed the datasets. Y.L. and Y.C. revised the main manuscript. All authors reviewed the manuscript. All authors were involved in writing the paper and approved the final submitted and published versions.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The study protocol was approved by the institutional review board for human investigations at the Korea University Anam Hospital, and the requirement for informed consent was waived owing to the retrospective design of our study. Total datasets were deidentified to preserve patient privacy. All processes were performed according to relevant regulations and guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lee, Y., Kim, J., Lee, KC. et al. UANV: UNet-based attention network for thoracolumbar vertebral compression fracture angle measurement. Sci Rep 15, 19952 (2025). https://doi.org/10.1038/s41598-025-03514-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03514-6