Abstract
Diabetic nephropathy (DN) is often accompanied by mesangial cell proliferation and fibrosis. Tanshinone IIA (Tan-IIA) is the main fat-soluble component of Salvia miltiorrhiza. N6-Methyladenosine (m6A) modification is a widely studied epigenetic mechanism. This study aimed to investigate the role of Tan-IIA in DN and the underlying mechanism. Cell viability and proliferation were assessed via cell counting kit-8 and ethynyldeoxyuridine assays. Protein levels of fibrosis-related indicators were detected by Western blot. Reverse transcription-quantitative polymerase chain reaction was used to detect the levels of m6A-related enzymes. The interaction betweenWT1 associated protein (WTAP) and prolactin receptor (PRLR) was examined through RNA immunoprecipitation and dual-luciferase reporter assays. The animal DN models was established. Biochemical measurements of rat serum were performed using commercial kits. Hematoxylin&eosin and Masson trichrome staining were used for histopathological analysis. Results showed that Tan IIA treatment inhibited the cell proliferation and fibrosis of human renal mesangial cells (HRMCs). Besides, Tan IIA treatment regulated WTAP-mediated m6A modification. Overexpression of WTAP upregulated the cell proliferation and fibrosis of HRMCs. Mechanically, WTAP enhanced the stability of PRLR mRNA via m6A methylation. Subsequent rescue investigations revealed that overexpression of PRLR increased the cell proliferation and fibrosis of HRMCs. In the in vivo study, Tan IIA treatment reversed the renal injury in rats and decreased the protein levels of WTAP, PLRP, and fibrosis-related indicators in kidney tissues. Tan IIA suppressed the proliferation and fibrosis of HRMCs in DN though WTAP-mediated m6A methylation of PRLR.
Similar content being viewed by others
Introduction
Diabetic nephropathy (DN) is a serious complication of diabetes that is characterized by damage to small blood vessels in the kidneys. This results in compromised kidney function and, ultimately, kidney failure1. It is approximated that DN will affect up to 40% of individuals with diabetes, positioning it as a prominent contributor to end-stage kidney disease on a global scale1,2. The progression of DN is intricately linked to inadequate management of blood sugar levels, hypertension, and genetic predisposition3. Prolonged elevated glucose level in the bloodstream leads to the impairment of kidneys’ filtration function, resulting in the leakage of proteins into the urine and the buildup of waste products in the blood. It is crucial to identify and address DN at an early stage in order to prevent its progression to kidney failure. This necessitates consistent monitoring of blood sugar levels, blood pressure, and kidney function, along with adopting a healthy lifestyle3,4. Treatment options for DN include medications to control blood sugar and blood pressure and, in severe cases, dialysis or kidney transplantation5,6. Research into new therapies for DN is ongoing, focusing on the underlying mechanisms of diabetic kidney damage.
Salvia miltiorrhiza, commonly referred to as Danshen in Chinese, is a traditional Chinese herbal medicine made from the dried roots and rhizomes of the Salvia miltiorrhiza Bge7. It has a variety of functions, including activating blood circulation, removing blood stasis, relieving pain, eliminating irritability, and cooling blood8. Various pharmacological studies have found that Salvia miltiorrhiza has protective effects on cardiovascular diseases and diabetes9,10. Tanshinone IIA (Tan-IIA) is a diterpenoid compound found in abundance in the root of Salvia miltiorrhiza11. Due to its potential therapeutic properties and ability to exert various biological effects, Tan-IIA has been the subject of many studies. At present, Tan-IIA shows great potential in the treatment of a variety of diseases such as cardiovascular diseases, neurodegenerative diseases, diabetic complications, and cancers12,13,14,15. The potential protective role of Tan-IIA in DN has been investigated before16, but the specific mechanism remains unclear.
N6-methyladenosine (m6A) represents a dynamic and reversible methylation alteration occurring at the N6 position of adenosine17. This chemical modification is widely observed in mRNA post-transcriptional modification and is considered the predominant epigenetic modification, involving processes including methylation, demethylation, and recognition17. M6A modification is regulated by “writers”, “erasers”, and “readers”. The methyltransferase complex, known as “writers”, is responsible for catalyzing m6A. The demethylase, or “erasers”, removes m6A. RNA reader proteins identify m6A, attach to the RNA, and carry out specific functions. Crosslink among “writers”, “erasers”, and “readers”, is involved in the pathogenesis and progression of various diseases. Methyltransferase-like protein (METTL)3, METTL14, and Wilms tumor 1-associating protein (WTAP) are three m6A methyltransferases18. Multiple studies have demonstrated that m6A modification affects the progression of diabetes and its complications19,20. Whereas, the role of Tan-IIA in m6A modification in DN has not been researched.
This study aimed to investigate the role of Tan-IIA in DN and the associated mechanism, with the goal of identifying a possible therapeutic approach for DN.
Methods and materials
Cell culture and treatment
Human renal mesangial cells (HRMCs) obtained from Pricella Life Technology Co., LTD (Wuhan, China) were cultured in commercial complete medium (Pricella) containing 10% fetal bovin serum (FBS; Gibco) and 1% penicillin/streptomycin (Pricella). The atmosphere required for cell culture was 37 °C and 5% CO2. HRMCs were cultured with normal (5.5 mmol/L) or high (25 mmol/L) concentrations of D-glucose (MedChem Express, Monmouth Junction, NJ, USA) in serum-free medium. Besides, HRMCs were treated with different concentrations (5, 10, 20, and 40 μg/mL) of Tan IIA.
Cell transfection
Short hairpin (sh) negative control (NC), shWTAP, negative control pcDNA 3.1, pcDNA 3.1-WTAP overexpression, and pcDNA 3.1-PRLR overexpression vectors were synthesized by NOVIN Biotechnology Co. Ltd., (Beijing, China). HRMCs cultured in 6-well plates were performed cell transfection using Lipo8000™ transfection reagent (Beyotime Biotechnology Co., LTD, Shanghai, China) when the cell confluence reached 80%.
Cell counting kit-8 (CCK-8) assay
HRMCs were seeded into 96-well plates at a density of 1 × 103 cells per well in 100 µL of complete medium and incubated at 37 °C for 24 h. Then, 10 µL of CCK8 reagent (Beyotime) was added to each well, and the cells were further incubated at 37 °C for 2 h. Finally, the optical density value (OD450) was measured using a microplate reader.
Ethynyldeoxyuridine (EdU) analysis
EdU assay was performed to analyze cell proliferation. HRMCs (5 × 103 cells/well) were cultured in 96-well plates. Then, 10 μL of EdU labeling media was added to the 96-well plates and then incubated at 37 °C under 5% CO2 for 2 h. Then, the cells were treated with 4% paraformaldehyde (Beyotime) and 0.5% Triton X-100 (Beyotime) and stained with the anti-EdU working solution and Hoechst 33342 (Beyotime). Next, the cells were visualized using a fluorescence microscope (Olympus). Finally, Image J 1.8.0 was used to quantify the EdU positive cells.
Western blot
Total RNA in rat kidney tissues or HRMCs was isolated using Trizol reagent (Beyotime). Bradford assay protein quantitative kit (Thermo Fisher) was used for protein quantification. Next, 30 μg of the proteins was accomplished with 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) for separation. Then, the proteins were transferred to the polyvinylidene fluoride (PVDF) membrane (Jiancheng Biotechnology Co., LTD, Nanjing, China). After blocking for 10 min, the membranes were incubated overnight with primary antibodies at 4 °C. Next, the membrane was washed by Tris-buffered saline Tween (TBST; Thermo Fisher) thrice, and incubated with the secondary antibody for 1 h at room temperature. Finally, the membranes were washed with TBST and proteins were visualized by electrochemiluminescence. Intensity of the bands was analyzed with ImageJ software. The used antibodies are obtained from Thermo Fisher and listed here: WTAP, rabbit-anti-rat, PA5-144681, 1/1000; PRLR, rabbit-anti-rat, PA5-87960, 1/1000; TGF β-1, rabbit-anti-human, MA5-15065, 1/1000; TGF β-1, rabbit-anti-rat, MA5-44667, 1/1000; fibronectin, rabbit-anti-human/rat, PA5-29578, 1/1000; collagen IV, rabbit-anti-human/rat, PA5-104508, 1/1000; β-actin, rabbit-anti-human/rat, PA1-183, 1/1000; goat anti-rabbit IgG secondary antibody, 31460, 1/1000.
M6A dot blot assay
RNA extracted from HRMCs underwent a heat treatment at 95 °C for 3 min. The concentration of RNA was determined using a Nanodrop 2000 (Thermo Fisher). Subsequently, 600 ng of total RNA were separately applied to a nitrocellulose filter membrane and dried at 37 °C for 30 min. Next, the membrane was washed with TBST for 5 min. Then, the sample was blocked with 5% bovine serum albumin for 1 h after removing the TBST, and incubated with m6A antibody (68055-1-Ig; 1/2000; Proteintech Biotechnology Co. LTD, Wuhan, China) overnight at 4 °C. The membrane was then washed thrice with TBST. Finally, the samples were treated with secondary antibody (RGAM001; 1/1000; Proteintech) for 1 h, and the signals from the dot blot were visualized using an ECL Western blotting detection kit (Thermo Fisher).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from HRMCs using Trizol reagent. Subsequently, the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotechnology Co. LTD, Nanjing, China) was employed for reverse transcription to generate cDNA. The AceQ qPCR SYBR Green Master Mix (Vazyme) was utilized for qPCR amplification, following the suggested reaction conditions. The primers utilized in this study were synthesized by Genecfps Biotechnology Co., LTD (Wuxi, China) and are listed in Table 1.
Molecular docking
Molecular docking was used to analyze the action mode of Tan IIA and the target protein WTAP. The Tan IIA molecular structure of this docking was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The WTAP protein structure was obtained from the RCSB Protein Data Bank database (https://www.rcsb.org/). The molecular docking was performed by the Glide module in Schrodinger Maestro software (Version 11.9, https://www.schrodinger.com/products/maestro). Finally, the mode of action of the compound and the target protein was analyzed, and the interaction between the compound and the protein residue was obtained.
Methylated-RNA immunoprecipitation (Me-RIP)
Total RNA was extracted from HRMCs. Afterwards, the RNA samples were fragmented into fragments that were 100 nucleotides long using the RNA fragmentation reagent (Thermo Fisher). Following this, fragmented mRNAs (400 ng) were incubated with anti-m6A (68055-1-Ig; Proteintech Biotechnology Co. LTD, Wuhan, China) antibody at room temperature for 1 h. The mixtures were then subjected to immunoprecipitation by incubating with prewashed protein A magnetic beads (Thermo Fisher) at 4 °C for 5 h, resulting in the formation of complexes consisting of magnetic bead-antibody-RNA. Subsequently, the complexes were digested using proteinase K (Thermo Fisher) digestion buffer at 55 °C for 1 h to release the RNA bound to the antibody from the complex. Finally, the RNAs were isolated for RT-qPCR analysis.
RNA immunoprecipitation (RIP) assay
A commercial RIP kit (Geneseed, Biotechnology Co. LTD, Guangzhou, China) was utilized to examine the interaction between WTAP and prolactin receptor (PRLR) in HRMCs. HRMCs were lysed in 400 μL complete RIP lysis buffer at 4 °C for 30 min. After centrifugation at 12,000 rpm for 10 min, the cell supernatant was collected. The collected supernatant (100 μL) was labeled as the Input group, whereas 900 μL of the supernatant was subjected to pre-treatment with protein A + G beads at 4 °C for 10 min. Subsequently, protein A + G beads (200 μL) were combined with IgG (30000-0-AP, Proteintech) or WTAP (60188-1-Ig, Proteintech) antibodies at 4 °C for 2 h. The antibody-bead complex was then incubated with the supernatant at 4 °C for 12 h. Following the washing steps, RNA extraction was performed, and the level of PRLR expression was measured using qPCR.
Prediction of m6A sites of PRLR
The sequence-based RNA adenosine methylation site predictor database (http://www.cuilab.cn/sramp) was used to predict m6A sites of PRLR.
Dual-luciferase reporter assay
Dual-luciferase reporter assay was performed to further clarify WTAP regulated PRLR through which site. The plasmids included PRLR-wild-type, PRLR-mutant (Mut)1 (1#), PRLR-Mut2 (2#), and PRLR-Mut3 (3#) were constructed from Genescript Biotechnology Co., LTD, (Nanjing, China). Wild-type PRLR sequences with possible m6A sites were amplified and inserted into pGL3 vectors (Promega, Madison, WI, USA). Their Mut sequences were cloned into pGL3 vectors. HRMCs were co-transfected with the WT or Mut plasmids together with empty or WTAP inhibition vectors using transfection reagent. The luciferase activity was assessed 48 h later using the dual-luciferase reporter assay system (Promega).
RNA stability assessment
RNA stability assessment was performed to verify the stability of PRLR after WTAP deficiency in HRMCs. HRMCs were treated with actinomycin D (MedChem Express; 0.5 µg/mL), then existing PRLR expression at different time points (1, 4, 8, and 12 h) was analyzed by RT-qPCR.
Animal study
A total of 22 male Sprague–Dawley (SD) rats aged 6–8 weeks (160 ± 10 g) were obtained from Aniphe Biolaboratory Inc. (Nanjing, China) and placed in cages with a temperature of 24℃, a 12-h light/dark cycle, and unlimited access to food and water. Following a week of adaptive feeding, six rats were selected as the normal controls (NC group) and were provided with a standard diet. The remaining 16 rats were fed a high-fat diet (HFD) for a duration of 6 weeks. The HFD consisted of 78.7% standard diet, 10% glucose, 10% animal fat, 1% total cholesterol, and 0.3% sodium cholate. After that, rats fed with HFD were given an intraperitoneal injection of streptozocin (STZ; 35 mg/kg; Yeason Biotechnology Co Ltd., Shanghai, China) for three consecutive days to induce diabetes. Simultaneously, the rats in the NC group were received an injection of 10 mM citrate buffer with pH = 4.5 at a volume of 2 mL/kg. After 72 h, the rats with plasma glucose level exceeding 16.7 mmol/L were categorized as diabetic, and in this process, four STZ-injected rats that did not meet the criteria for diabetes were excluded. After two weeks, the urine microalbumin (UMA) level in the rats was measured, and rats with 24 h UMA levels above 30 mg were considered to have DN21. DN rats (n = 12) were randomly divided into DN group (n = 6) and Tan IIA-treatment group (DN + Tan IIA group, n = 6). The DN + Tan IIA group was applied daily with Tan IIA (dissolved in dimethyl sulfoxide; 4 mg/kg/day22) by oral gavage for 42 d. While the NC and the DN group rats were given an oral gavage of 0.9% saline (2 mL/kg) as control.
After 42-day treatments, all rats were anesthetized with intraperitoneal injection of sodium pentobarbital (60 mg/kg) and weighted. Fasting blood-glucose (FBG) was tested using the blood drawn from tail vein by a glucose analyzer (Roche, Germany). Blood was extracted from the carotid arteries, and the serum was obtained from the blood samples through centrifugation at 1500 g for 15 min at 4 °C. Serum samples were kept at -20 °C for measurement of serum creatinine (Scr) and blood urea nitrogen (BUN). Besides, 24 h urine samples were collected for the measurement of UMA. The kidneys were immediately removed and the left one was weighted. The ratio between kidney weight (KW) and body weight (BW) was calculated (KW/BW). One kidney was fixed with 4% paraformaldehyde for further histopathological analysis and the other one was frozen in -80℃ for further Western blot analysis. Finally, the rats were anesthetized using isoflurane gas and euthanized by cervical dislocation.
Biochemical measurements
Serum Scr and BUN and uric UMA levels were detected using commercial enzyme-linked immunosorbent assay (ELISA) kits (Baililai Biotechnology Co., LTD, Shanghai, China). All operations were carried out according to the manufacturer’s protocols. Finally, the OD value of each well was detected by a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA). The obtained results were normalized against total protein concentration in each sample for intersample comparison.
Histopathological analysis
The left kidneys isolated from rats were fixed using 4% paraformaldehyde solution for 24 h, and embedded in paraffin. Then, the embedded tissues were sliced into 4 μm sections followed by staining with hematoxylin&eosin (H&E) and Masson trichrome staining. Finally, the sections were observed by a biopathology microscope (Olympus, Tokyo, Japan).
Statistical analysis
Statistical analyses were performed using SPSS 21.0 software (IBM, USA) and GraphPad Prism software (v8.0.1, GraphPad Software Inc., San Diego, CA, USA). Data are expressed as mean ± standard deviation (SD). For comparisons among multiple groups, one-way analysis of variance (ANOVA) was used, followed by Tukey’s post hoc test for pairwise comparisons. The rationale for using one-way ANOVA is that it is appropriate for comparing the means of three or more independent groups, which aligns with our experimental design (e.g., NC group, DN group, and DN + Tan IIA group). For comparisons between two groups, an unpaired Student’s t-test was applied, as it is suitable for assessing the significance of differences between two independent groups. The normality of data distribution was assessed using the Shapiro–Wilk test, and homogeneity of variance was confirmed using Levene’s test. A p value < 0.05 was considered statistically significant. All experiments were performed with appropriate biological replicates (n = 6 for animal experiments and n = 3 for cell experiments) to ensure the reliability and reproducibility of the results.
Results
Tan IIA treatment inhibited the cell proliferation and fibrosis of HRMCs
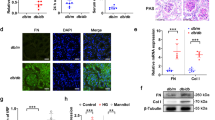
Tan IIA is a compound extracted from Salvia miltiorrhiza, shows promise in the management of various medical conditions23. This study aimed to examine that whether Tan IIA has a protective effect on DN. An in vitro DN model was established. Cell viability findings demonstrated that high glucose (HG) treatment increased cell viability compared with the control group, and the results were reversed with the increasing concentration of Tan IIA (5, 10, and 20 μg/mL), while the concentration of 40 μg/mL could not further inhibit cell viability (Fig. 1A). Tan IIA at the concentration of 20 μg/mL was chosen for followed experiments. The results from EdU analysis revealed that the treatment of HG led to an augmentation in the number of EdU positive cells. Conversely, the administration of Tan IIA hindered this increase, suggesting that Tan IIA effectively suppressed the proliferation of HRMCs (Fig. 1B,C). In addition, data from fibrosis-related indicators using Western blot indicated upregulation of TGF-β1, fibronectin, and collagen IV in the HG group compared with the control group, and these effects were restored after Tan IIA administration (Fig. 1D–G). These results suggested that Tan IIA treatment inhibited the proliferation and fibrosis of HRMCs induced by HG.
Tan IIA treatment inhibited the cell proliferation and fibrosis of HRMCs. HRMCs were cultured in commercial complete medium containing 10% fetal bovin serum and 1% penicillin/streptomycin. The atmosphere required for cell culture was 37 °C and 5% CO2. HRMCs were cultured with normal (5.5 mmol/L) or high (25 mmol/L) concentrations of D-glucose in serum-free medium. Besides, HRMCs were treated with different concentrations (5, 10, 20, and 40 μg/mL) of Tan IIA. (A) Cell viability in each group was detected by CCK-8 assay (n = 3); (B) EdU assay was performed to analyze cell proliferation (n = 3); (C) Quantification of EdU positive cells in each group (n = 3); (D) Western blot analysis of TGF-β1, fibronectin, and collagen IV protein levels in each group (n = 3); Quantification of (D) TGF-β1, (E) fibronectin, and (F) collagen IV protein levels in each group (n = 3). Data are expressed as the mean ± SD. Tan IIA, Tanshinone IIA; HRMCs, human renal mesangial cells; CCK-8, cell counting-kit 8; EdU, ethynyldeoxyuridine; TGF-β1, transforming growth factor-beta1.
Tan IIA treatment regulated WTAP-mediated m6A modification
M6A modification has been shown to regulate kidney injury in DN24. A prior investigation has demonstrated Tan IIA alleviates cardiac hypertrophy through m6A modification of galectin-325, yet the potential of Tan IIA to regulate m6A modification in DN remains unexplored. In this study, it was discovered that HG-treated HRMCs exhibited an increase in m6A level in comparison to the control group. However, the outcome was reversed following Tan IIA treatment (Fig. 2A). Besides, RT-qPCR findings revealed that the mRNA level of WTAP was reduced after Tan IIA treatment in comparison to the HG group. However, there were no alterations observed in the levels of METTL3, METTL14, RBM15, VIRMA, ZC3H13, FTO, and ALKBH5 between the two groups (Fig. 2B). Molecular docking is widely used to assay potential bioactivity of compounds. The results of molecular docking showed that Tan IIA and the target protein had good binding effect and high matching degree. The complex formed by the docking compound and the protein was visualized by Pymol2.1 software, and the binding mode of the compound and the protein was obtained. According to the binding mode, the amino acid residues of the compound and the protein pocket could be clearly seen. For example, Tan IIA compounds are highly hydrophobic and can form strong hydrophobic interactions with active site amino acids (PRO-115, LEU-989, HIS990, LEU-97, LYS-98, etc.). It plays an important role in stabilizing molecules in protein cavities. These results showed that Tan IIA and WTAP had high matching degree and formed stable complexes (Fig. 2C). Therefore, WTAP was a possible potential target of Tan IIA.
Tan IIA treatment regulated WTAP-mediated m6A modification. (A) Dot blot assay was performed to assess the total m6A level in each group (n = 3); (B) RT-qPCR was performed to analyze the mRNA levels of METTL3, METTL14, RBM15, VIRMA, WTAP, ZC3H13, FTO, and ALKBH5 in each group (n = 3); (C) The 3D structure of the binding complex of WTAP with Tan IIA and the detail binding mode of the complex. The backbone of the protein was rendered in tube. Compound is rendered by green and the OGT is shown in blue sticks. Yellow and ray dash represents hydrogen bond. Data are expressed as the mean ± SD. Tan IIA, Tanshinone IIA; WTAP, Wilms tumor 1-associating protein; m6A, N6-methyladenosine; RT-qPCR, reverse transcription-polymerase chain reaction; METTL, methyltransferase; RBM15, RNA binding motif protein 15; VIRMA, vir-like m(6)A methyltransferase-associated protein; ZC3H13, zinc finger CCCH-type containing 13; FTO, alpha‐ketoglutarate dependent dioxygenase; ALKBH5, AlkB homolog 5.
Overexpression of WTAP upregulated the cell proliferation and fibrosis of HRMCs
In order to explore the significance of WTAP in HG-treated HRMCs, both empty and WTAP overexpression vectors were introduced into HRMCs. The findings indicated a rise in WTAP level following WTAP overexpression (Fig. 3A). Furthermore, overexpression of WTAP resulted in increases in cell viability, EdU positive cells, as well as the protein levels of fibrosis-related indicators (TGF-β1, fibronectin, and collagen IV) in HRMCs when compared with the vector group (Figs. 3B–H).
Overexpression of WTAP upregulated the cell proliferation and fibrosis of HRMCs. Negative control pcDNA 3.1 and pcDNA 3.1-WTAP overexpression vectors were transfected into HRMCs using Lipo8000™ transfection reagent. (A) RT-qPCR was used to assess the expression of WTAP when WTAP was overexpressed in HRMCs (n = 3); (B) Cell viability in each group was detected by CCK-8 assay (n = 3); (C) EdU assay was performed to analyze cell proliferation (n = 3); (D) Quantification of EdU positive cells in each group (n = 3); (E) Western blot analysis of TGF-β1, fibronectin, and collagen IV protein levels in each group (n = 3); Quantification of (F) TGF-β1, (G) fibronectin, and (H) collagen IV protein levels in each group (n = 3). Data are expressed as the mean ± SD. HRMCs, human renal mesangial cells; RT-qPCR, reverse transcription-polymerase chain reaction; WTAP, Wilms tumor 1-associating protein; CCK-8, cell counting-kit 8; EdU, ethynyldeoxyuridine; TGF-β1, transforming growth factor-beta1.
WTAP enhanced the stability of PRLR mRNA via m6A methylation
After transfecting shWTAP vector into HRMCs, the expression of WTAP and PRLR were decreased (Fig. 4A,B). Additionally, MeRIP assay indicated that inhibition of WTAP showed decreased m6A level of PRLR in HRMCs (Fig. 4C). RIP assay suggested that WTAP bound with the mRNA of PRLR in HRMCs (Fig. 4D). The potential m6A methylation sites (sites 4727, 6446, and 6496) of PRLR mRNA was predicted using the sequence-based RNA adenosine methylation site predictor database (Fig. 4E,F). Dual-luciferase reporter assays showed that WTAP was specifically bound to PRLR at site 2# (6446) rather than site 1# (4727) or site 3# (6496) in HRMCs (Fig. 4G–I). Moreover, RNA stability results displayed that WTAP inhibition resulted an accelerated degradation of PRLR mRNA in HRMCs, suggesting that WTAP weakened the stability of PRLR mRNA via m6A methylation in HRMCs (Fig. 4J).
WTAP enhanced the stability of PRLR mRNA via m6A demethylation. Short hairpin negative control (shNC) and shWTAP vectors were transfected into HRMCs using Lipo8000™ transfection reagent. (A) RT-qPCR was used to assess the expression of WTAP when WTAP was inhibited in HRMCs (n = 3); (B) RT-qPCR was used to assess the expression of PRLR when PRLR was suppressed in HRMCs (n = 3); (C) The m6A level of PRLR after WTAP inhibited was detected using MeRIP-qPCR assay (n = 3); (D) RIP assay was conducted to examine the interaction between WTAP and PRLR in HRMCs cells (n = 3); (E) Sequence-based RNA adenosine methylation site predictor database was used to predict m6A sites of PRLR; (F) The three predictive sites of PRLR; Dual-luciferase reporter assay was performed to evaluate the binding of WTAP and PRLR at sites (G) 1#, (H) 2#, and (I) 3# in HRMCs; (J) RNA stability assay was used to detect the existing PRLR expression when actinomycin D treated at different time points (1, 4, 8, and 12 h) in HRMCs (n = 3). Data are expressed as the mean ± SD. WTAP, Wilms tumor 1-associating protein; PRLR, prolactin receptor; m6A, N6-methyladenosine; RT-qPCR, reverse transcription-polymerase chain reaction; HRMCs, human renal mesangial cells; RIP, RNA immunoprecipitation; MeRIP, Methylated-RIP.
Overexpression of PRLR increased the cell proliferation and fibrosis of HRMCs
In final rescue studies, we introduced PRLR overexpression and empty vectors into HRMCs, leading to an observed upregulation of PRLR expression in comparison to the vector group (Fig. 5A). Additionally, in comparison to the vector group, overexpression of PRLR reversed the decreased cell viability, EdU positive cells, as well as the protein levels of fibrosis-related indicators (TGF-β1, fibronectin, and collagen IV) in HRMCs (Fig. 5B–H).
Overexpression of PRLR increased the cell proliferation and fibrosis of HRMCs. Negative control pcDNA 3.1, pcDNA 3.1-PRLP overexpression, shNC, and shWTAP vectors were transfected into HRMCs using Lipo8000™ transfection reagent. (A) RT-qPCR was used to assess the expression of PRLR when PRLR was overexpressed in HRMCs (n = 3); (B) Cell viability in each group was detected by CCK-8 assay (n = 3); (C) EdU assay was performed to analyze cell proliferation (n = 3); (D) Quantification of EdU positive cells in each group (n = 3); (E) Western blot analysis of TGF-β1, fibronectin, and collagen IV protein levels in each group (n = 3); Quantification of (F) TGF-β1, (G) fibronectin, and (H) collagen IV protein levels in each group (n = 3). Data are expressed as the mean ± SD. PRLR, prolactin receptor; RT-qPCR, reverse transcription-polymerase chain reaction; HRMCs, human renal mesangial cells; CCK-8, cell counting-kit 8; EdU, ethynyldeoxyuridine; TGF-β1, transforming growth factor-beta1.
Tan IIA treatment reversed the renal injury induced by DN
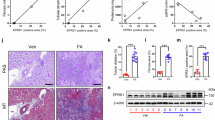
A diabetic rat model was established. Results demonstrated that DN group rats showed increased KW/BW and UMA, FBG, and BUN levels compared with NC group, suggesting that the kidneys in DN group rats were hypertrophic and injured. However, Tan IIA treatment reversed the impaired kidney function in DN group, implying that Tan IIA has a protective effect on DN (Fig. 6A–C,E). Besides, the Scr level among three groups showed no significant differences (Fig. 6D). H&E and Masson trichrome staining were performed to examine the kidney pathologic changes. H&E results indicated that deposited mesangial matrix, mesangial expansion and the fractional mesangial area were significantly higher in the DN group compared with the NC group, while they were significantly improved by Tan IIA treatment. Besides, Masson trichrome staining results revealed that lots of blue stained tissues was observed in DN group rats relative to CN group rats, suggesting accelerated renal fibrosis. Besides, compared with the DN group, renal fibrosis was mitigated after Tan IIA treatment (Fig. 6F). These results suggested that Tan IIA treatment reversed the renal injury induced by DN. In addition, Western blot results indicated that compared with the NC group, DN group rats showed increased WTAP, PRLR, TGF-β1, fibronectin, and collagen IV protein levels in kidneys, and the results were reversed after Tan IIA treatment (Fig. 6G).
Tan IIA treatment reversed the renal injury induced by DN. Creating a rat model mimicking diabetes in humans through a combination of HFD and STZ treatment. Twenty-two male Sprague–Dawley rats (6 weeks old) were randomly assigned to either negative controls (NC; n = 6) or HFD (n = 16) groups for 6 weeks at the start of the experiment. After that, the HFD group rats received intraperitoneal injections of STZ (35 mg/kg), while the NC group rats were injected with vehicle citrate buffer. Rats (n = 12) were randomly divided into DN group (n = 6) and Tan IIA-treatment group (DN + Tan IIA group, n = 6) after excluding four STZ-injected rats that did not meet the criteria for diabetes. (A) KW/BW, (B) UMA, (C) FBG, (D) Scr, and (E) BUN levels of the each group rats were shown (n = 6); F, Representative images of H&E and Masson trichrome staining of the each group kidney sections (magnification: × 200; n = 6); (G) Western blot was used to analyze the protein levels of WTAP, PRLR, TGF-β1, fibronectin, and collagen IV in each group rat kidneys (n = 6). Data are expressed as the mean ± SD. Tan IIA, Tanshinone IIA; DN, diabetic nephropathy; HFD, high-fat diet; STZ, streptozocin; KW/BW, kidney weight to body weight ratio; UMA, urinary microalbumin; FBG, fasting blood glucose; Scr, serum creatinine; BUN, blood urea nitrogen; H&E, hematoxylin&eosin; WTAP, Wilms tumor 1-associating protein; PRLR, prolactin receptor; TGF-β1, transforming growth factor-beta1.
Discussion
In many clinical practices, traditional strategies such as renin–angiotensin–aldosterone system blocking, blood glucose level control, and weight loss have failed to achieve satisfactory results in the treatment of DN26. Nowadays, more and more studies emphasize the significance of the application of traditional Chinese medicine (TCM) in the treatment of DN26. Tan IIA, a bioactive compound extracted from Salvia miltiorrhiza, has been identified as playing a role in the advancement of kidney diseases, including DN23. The proliferation of mesangial cells and the development of fibrosis are key indicators of kidney damage, which can result in various lesions like glomerulosclerosis21. Hence, the prevention or delay of DN involves the implementation of strategies to inhibit the proliferation and fibrosis of mesangial cells.
The early detection of kidney disease relies on UMA detection, which is known to be the most sensitive and reliable diagnostic index4. Additionally, the damage degree of glomerular filtration function can be reflected by serum Scr and BUN levels26. In this study, Tan IIA inhibited renal injury and fibrosis in DN rats, which were consistent with previous studies27,28. In addition, Tan IIA also shows protective roles in other diabetes complications. Wu et al., indicate that Tan IIA improved the pathological morphology of cardiomyocytes and reduced cell mortality in diabetic cardiomyopathy15. Besides, Tan IIA improves retinal structure and inhibited oxidative stress in mice with diabetic retinopathy29. In in vitro study, we found that Tan IIA reversed the increased cell proliferation and fibrosis of HG-treated HRMCs. Similarly, Tan IIA inhibits the expression of fibrosis-related proteins in renal tissue of DN rats30. Moreover, another study has found that Tan IIA improves diabetes-induced fibrosis area in HK-2 cells28. Moreover, other TCM compound or active ingredients also inhibit excessive mesangial cell proliferation and renal fibrosis caused by DN31,32.
In diabetic kidney diseases, multiple m6A modifications have been identified. For instance, Zhang et al. indicate that WTAP-mediated m6A modification of tripartite motif-containing (TRIM)22 promotes DN by inducing mitochondrial dysfunction33. In addition, Fu et al.34 demonstrate that silence the expression of WTAP inhibits the migration and proliferation of renal tubular epithelial cells. In the present study, we for the first time found that Tan IIA treatment regulated WTAP-mediated m6A modification and overexpression of WTAP upregulated the cell proliferation and fibrosis of HRMCs. These results suggested that Tan IIA inhibited the cell proliferation and fibrosis of HRMCs though WTAP-mediated m6A modification. Tan IIA-regulated m6A modification has been found in a study of cardiac hypertrophy before25. Besides, a recent study indicates that Tan IIA restrains hepatocellular carcinoma cell viability, proliferation, invasion, and stemness by regulating METTL3-mediated m6A modification of tribbles pseudokinase-3 (TRIB3) mRNA35. At present, there is a lack of epigenetic mechanism studies on the regulation of DN by Tan IIA. The regulation of Tan IIA on DN mainly focuses on related signaling pathways, such as phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt)/nuclear factor-kappaB (NF-κB) and vitamin D receptor (VDR)/Wnt/β-catenin pathways27,30. In further mechanism studies, it was discovered that PRLR serves as a downstream objective of WTAP in HRMCs. PRLR, a type-1 cytokine receptor, is overexpressed in a number of diseases36. In our study, we found that WTAP enhanced the stability of PRLR mRNA via m6A methylation and overexpression of PRLR increased the cell proliferation and fibrosis of HRMCs. Similarly, a previous in vivo study demonstrates that DN is associated with increases in the renal expression of PRLR37. Besides, knockdown of PRLR ameliorates high glucose-induced fibrosis and enhances autophagy in rat kidney cells38. Interestingly, abnormal expression of PRLR is also implicated in other diabetic complications, such as diabetes-related cognitive impairment. Jiang et al.39 reveal that knockout of PRLR in microglia leads to hippocampal synaptic loss and cognitive impairment in HFD-fed diabetic mice.
To summarize, our findings suggested that Tan IIA inhibited the cell proliferation and fibrosis of HRMCs though WTAP-mediated m6A modification of PRLR. This study could provide a reference for developing a new drug therapy for DN.
While this study provides valuable insights into the protective role of Tan-IIA in DN through WTAP-mediated m6A modification of PRLR, several limitations should be acknowledged. First, the sample size used in the animal experiments was relatively small (n = 6 per group), which may limit the generalizability of the findings. A larger sample size would enhance the statistical power and reliability of the results. Second, the study duration was limited to 42 days of Tan-IIA treatment, which may not fully capture the long-term effects of the intervention. Extending the treatment period could provide a more comprehensive understanding of Tan-IIA’s therapeutic potential in DN. Third, the study utilized a single animal model (Sprague–Dawley rats) and a single cell (HRMCs). While these models are widely used in DN research, the inclusion of additional animal models or cell types could strengthen the validity of the findings. Finally, the study lacked clinical validation, as all experiments were conducted in preclinical models. Future studies should include clinical trials to confirm the translational relevance of these findings in human patients with DN.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Alicic, R. Z., Rooney, M. T. & Tuttle, K. R. Diabetic kidney disease: Challenges, progress, and possibilities. Clin. J. Am. Soc. Nephrol. 12, 2032–2045 (2017).
Gupta, S., Dominguez, M. & Golestaneh, L. Diabetic kidney disease: An update. Med. Clin. North. Am. 107, 689–705 (2023).
Gross, J. L. et al. Diabetic nephropathy: Diagnosis, prevention, and treatment. Diabetes Care 28, 164–176 (2005).
Samsu, N. Diabetic nephropathy: challenges in pathogenesis, diagnosis, and treatment. Biomed Res. Int. 2021, 1497449 (2021).
Zou, M. et al. Safety and efficacy of hemodialysis and peritoneal dialysis in treating end-stage diabetic nephropathy: A meta-analysis of randomized controlled trials. Int. Urol. Nephrol. 54, 2901–2909 (2022).
Lee, K. W. et al. Recoverability of diabetic nephropathy of donor kidney after kidney transplantation. Transpl. Int. 35, 10714 (2022).
Li, M. H., Chen, J. M., Peng, Y., Wu, Q. & Xiao, P. G. Investigation of danshen and related medicinal plants in China. J. Ethnopharmacol. 120, 419–426 (2008).
Hao, D. C., Ge, G. B. & Xiao, P. G. Anticancer drug targets of salvia phytometabolites: Chemistry biology and omics. Curr. Drug Targets. 19, 1–20 (2018).
Jia, Q. et al. Salvia miltiorrhiza in diabetes: A review of its pharmacology, phytochemistry, and safety. Phytomedicine 58, 152871 (2019).
Wang, L. et al. Salvia miltiorrhiza: A potential red light to the development of cardiovascular diseases. Curr. Pharm. Des. 23, 1077–1097 (2017).
Guo, R. et al. Pharmacological activity and mechanism of tanshinone iia in related diseases. Drug Des. Dev. Ther. 14, 4735–4748 (2020).
Gao, S. et al. Cardiovascular actions and therapeutic potential of tanshinone iia. Atherosclerosis 220, 3–10 (2012).
Subedi, L. & Gaire, B. P. Tanshinone Iia: A phytochemical as a promising drug candidate for neurodegenerative diseases. Pharmacol. Res. 169, 105661 (2021).
Alam, S., Samanta, A., Uddin, F., Ali, S. & Hoque, M. Tanshinone Iia targeting cell signaling pathways: A plausible paradigm for cancer therapy. Pharmacol. Rep. 75, 907–922 (2023).
Wu, S. et al. Tanshinone Iia ameliorates experimental diabetic cardiomyopathy by inhibiting endoplasmic reticulum stress in cardiomyocytes via Sirt1. Phytother. Res. 37, 3543–3558 (2023).
Zhang, L. et al. Exploring the mechanisms underlying the therapeutic effect of salvia miltiorrhiza in diabetic nephropathy using network pharmacology and molecular docking. Biosci. Rep. 41, BSR20203520 (2021).
Zhang, N., Ding, C., Zuo, Y., Peng, Y. & Zuo, L. N6-methyladenosine and neurological diseases. Mol. Neurobiol. 59, 1925–1937 (2022).
Zhang, X. & Jia, G. F. Rna epigenetic modification: N6-methyladenosine. Yi Chuan 38, 275–288 (2016).
Meng, L. et al. Mettl14 Suppresses pyroptosis and diabetic cardiomyopathy by downregulating tincr lncrna. Cell Death Dis. 13, 38 (2022).
Wang, Y. et al. The M6a methylation profiles of immune cells in type 1 diabetes mellitus. Front. Immunol. 13, 1030728 (2022).
Han, F. et al. Triptolide suppresses glomerular mesangial cell proliferation in diabetic nephropathy is associated with inhibition of Pdk1/Akt/Mtor pathway. Int. J. Biol. Sci. 13, 1266–1275 (2017).
Chen, K., Yu, B. & Liao, J. Lncrna Sox2Ot alleviates mesangial cell proliferation and fibrosis in diabetic nephropathy via Akt/Mtor-mediated autophagy. Mol. Med. 27, 71 (2021).
Xu, S. et al. Tanshinone Iia ameliorates streptozotocin-induced diabetic nephropathy, partly by attenuating perk pathway-induced fibrosis. Drug Des. Dev. Ther. 14, 5773–5782 (2020).
Lan, J., Xu, B., Shi, X., Pan, Q. & Tao, Q. Wtap-mediated N(6)-methyladenosine modification of Nlrp3 Mrna in kidney injury of diabetic nephropathy. Cell Mol. Biol. Lett. 27, 51 (2022).
Zhang, M. et al. Tanshinone Iia alleviates cardiac hypertrophy through M6a modification of galectin-3. Bioengineered 13, 4260–4270 (2022).
Tang, G. et al. Clinical efficacies, underlying mechanisms and molecular targets of Chinese medicines for diabetic nephropathy treatment and management. Acta Pharm. Sin. B. 11, 2749–2767 (2021).
Xu, Z. et al. Salvianolic acid B and tanshinone iia synergistically improve early diabetic nephropathy through regulating Pi3K/Akt/Nf-kappab signaling pathway. J. Ethnopharmacol. 319, 117356 (2024).
Zhang, L. & Yang, F. Tanshinone iia improves diabetes-induced renal fibrosis by regulating the Mir-34-5P/Notch1 axis. Food Sci. Nutr. 10, 4019–4040 (2022).
Zeng, X. et al. Study on the antioxidant effect of tanshinone iia on diabetic retinopathy and its mechanism based on integrated pharmacology. Evid.-Based Complement Altern. Med. 202, 9990937 (2022).
Zeng, J. Y. et al. Tanshinone Iia is superior to paricalcitol in ameliorating tubulointerstitial fibrosis through regulation of vdr/wnt/beta-catenin pathway in rats with diabetic nephropathy. Naunyn. Schmiedebergs. Arch. Pharmacol. 397, 3959–3977 (2024).
Mao, Q. et al. Astragaloside IV inhibits excessive mesangial cell proliferation and renal fibrosis caused by diabetic nephropathy via modulation of the Tgf-Beta1/Smad/Mir-192 signaling pathway. Exp. Ther. Med. 18, 3053–3061 (2019).
Shen, Y. L. et al. Erhuang formula improves renal fibrosis in diabetic nephropathy rats by inhibiting Cxcl6/Jak/Stat3 signaling pathway. Front. Pharmacol. 10, 1596 (2019).
Zhang, Z. et al. Wtap-mediated M(6)a modification of Trim22 promotes diabetic nephropathy by inducing mitochondrial dysfunction via ubiquitination of Opa1. Redox Rep. 29, 2404794 (2024).
Fu, K. et al. Wtap and Mettl14 regulate the m6a modification of Dkk3 in renal tubular epithelial cells of diabetic nephropathy. Biochem. Biophys. Res. Commun. 738, 150524 (2024).
Jiang, Y. et al. Tanshinone iia restrains hepatocellular carcinoma progression by regulating Mettl3-mediated M6a modification of Trib3 Mrna. Turk. J. Gastroenterol. (2025).
Gharbaran, R., Onwumere, O., Codrington, N., Somenarain, L. & Redenti, S. Immunohistochemical localization of prolactin receptor (Prlr) to Hodgkin’s and reed-sternberg cells of Hodgkin’s lymphoma. Acta Histochem. 123, 151657 (2021).
Al-Trad, B. Prolactin receptor Mrna expression in experimental diabetic nephropathy: Relationship with urinary albumin excretion. Neuro Endocrinol. Lett. 36, 552–556 (2015).
Chen, Z. et al. Astragalus polysaccharide promotes autophagy and alleviates diabetic nephropathy by targeting the Lncrna Gm41268/Prlr pathway. Ren. Fail. 45, 2284211 (2023).
Jiang, J. et al. Prolactin deficiency drives diabetes-associated cognitive dysfunction by inducing microglia-mediated synaptic loss. J. Neuroinflamm. 21, 295 (2024).
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript. H P drafted the work and revised it critically for important intellectual content and was responsible for the acquisition, analysis and interpretation of data for the work; X Z made substantial contributions to the conception or design of the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Xiangyang Hospital of Traditional Chinese Medicine. All animal experiments should comply with the ARRIVE guidelines. All methods were carried out in accordance with relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peng, H., Zhang, X. Tanshinone IIA suppresses the proliferation and fibrosis of mesangial cell in diabetic nephropathy though WTAP-mediated m6A methylation. Sci Rep 15, 21261 (2025). https://doi.org/10.1038/s41598-025-03738-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03738-6