Abstract
Flowering is a key developmental event for plants, marking the transition from the vegetative phase to the reproductive phase. Artificial induction of flowering holds great impact on agriculture by accelerating fruit and seed setting. However, to date, only a limited number of externally applicable methods have been established for flowering induction. In this study, we focused on FLOWERING LOCUS T (FT), a pivotal flowering inducer conserved in many flowering plants. We developed a non-transgenic flowering induction system in the model plant Arabidopsis, employing the exogenous introduction of FT mRNA through a functional peptide-mediated delivery. Utilizing the functional peptide BP100-KH9 as a nanocarrier for nucleic acid delivery into plant cells, we confirmed the production of FT protein from the introduced FT mRNA, which was fused with a reporter gene Citrine. The introduction of FT mRNA was also evidenced by the induction of expression of floral genes in treated seedlings, leading to the optimization of the timing for mRNA delivery. Measurement of flowering demonstrated accelerated flowering resulting from the introduction of FT mRNA as indicated by a shortened flowering time and a reduced number of leaves at flowering. This proof-of-concept study proposes an innovative non-transgenic flowering induction method using mRNA delivery.
Similar content being viewed by others
Introduction
Flowering is a crucial developmental stage for plants, signifying the shift from vegetative growth to reproductive growth, which enables the production of the next generations and introduces genetic diversity. Flowering is regulated precisely at the molecular level, with studies in Arabidopsis significantly advancing our understanding of its mechanisms. The process integrates information about temperature and day length through various pathways: the thermosensory and vernalization/autonomous pathways, and the photoperiod pathway1. FLOWERING LOCUS T (FT), which encodes a small globular protein similar to the mammalian Raf kinase inhibitor protein2 is known as a florigen and plays a critical role in the photoperiodic pathway in the long-day plant Arabidopsis thaliana2,3. FT expression is induced by the zinc-finger protein CONSTANS (CO), a transcription factor that acts under long day condition4,5, in coordination with the developmental growth of plants. FT is normally expressed in the phloem companion cells of leaves6 and its protein is transported to the shoot apical meristem (SAM) via phloem sieve elements7,8,−9. In the SAM, FT binds to the transcription factor FD to directly or indirectly induce floral genes such as APETALA1 (AP1), LEAFY (LFY), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), SEPALLATA3 (SEP3)10,11,12,13,14,15,−16.
Inducing flowering has significant potential for applications in breeding and agriculture by promoting faster seed and fruit setting. Genetic engineering approaches, such as overexpressing FT or other flowering-promoting genes, can effectively induce flowering by either enhancing these genes or suppressing their regulators. For example, overexpression of FT under a strong constitutive promoter significantly promotes early flowering and reduces the number of rosette leaves in Arabidopsis Col-0 17. Flowering induction by FT overexpression has also been observed in other plant species, including the annual monocot rice18 and the perennial dicot tree poplar19. Other than genetic engineering, limited number of phytohormones and compounds are known to induce flowering. Phytohormones such as salicylic acid and cytokinin have been reported to induce flowering in Arabidopsis through external application20,21. While endogenous molecules like reactive oxygen species (ROS) and sugars are involved in flowering induction, their external application has not yet been demonstrated to induce flowering in Arabidopsis22.
Recent advances in molecular-level understanding of plant mechanisms have stimulated their application for plant modification. Genetic engineering is the major and forceful method for the modification and has been utilized for many years. Agrobacterium-mediated transformation23 and biolistic transformation24 are well-established genetic engineering techniques used for plant modification. In addition to these traditional methods, emerging methods utilizing nanomaterials (such as peptides, carbon materials, and metals) are now being employed for plant modification25. Functional peptides are short peptides (~ 50 amino acids) with specific in vivo functions. Cell-penetrating peptides (CPPs) are a subset of functional peptides that can traverse cell membranes26,27. BP100 is a widely used CPP for various plant species, except for rice leaf28. When combined with a tandemly fused polycationic peptide KH9, which efficiently binds to nucleic acids through electrostatic interactions, this fusion peptide can deliver a range of biomacromolecules, including DNA and mRNA, to produce proteins in plant cells29,30,−31. Thus, this peptide-mediated method allows for non-transgenic protein production in plants via mRNA delivery. In this study, we applied the peptide-mediated mRNA delivery method to accelerate flowering in Arabidopsis as a model system. We optimized the conditions for delivering FT mRNA into Arabidopsis seedlings using the fusion peptide BP100-KH9. This delivery method led to upregulation of floral genes and earlier flowering in the treated plants.
Results and discussion
Formation of FT mRNA-peptide complexes
We first prepared A. thaliana FT mRNA by in vitro transcription and obtained high quality FT mRNA without any prominent by-product (Fig. S1). To analyze the complexation of the FT mRNA and the peptide BP100-KH9, which is critical for delivering nucleic acids into cells through nanocarrier-mediated delivery methods, we performed gel shift and hydrodynamic diameter analyses at various N/P ratios (the ratio of the number of positive charges in peptides to the number of negative charge in nucleic acids). The gel shift analysis showed formation of RNA-peptide complexes, evidenced by decreased RNA band intensity and retarded mobility in the gel (Fig. 1A). Complexation between FT mRNA and peptide was observed at every N/P ratio, with binding efficiency reaching 100% at an N/P ratio 2 or higher. Notably, the mobility of the complexes was highly retarded at N/P ratios 2 and 4, with some complexes retained in the well (Fig. 1A), suggesting the formation of higher-order mRNA-peptide complexes at higher N/P ratios. Next, we evaluated the in vitro protection of mRNA against RNase through complex formation. Following RNase digestion of the complexes formed between FT mRNA and BP100-KH9 at various N/P ratios, we observed reduced mRNA degradation at all tested N/P ratios (Fig. 1B). This protection of FT mRNA by binding with the peptides via the cationic KH9 region supports the notion that the peptide acts not only as a carrier but also provides protection from RNase during delivery into plant cells.
Complexation of FT mRNA with the peptide BP100-KH9. (A) Gel shift assay for mRNA-peptide complexes. FT mRNA complexed with the peptide BP100-KH9 at N/P ratio = 0.25, 0.5, 1, 2, and 4 were analyzed by agarose gel electrophoresis. Binding efficiency of the RNA at each N/P ratio was calculated from the gel image. (B) Protection of mRNA from RNase by peptides. FT mRNA was complexed with BP100-KH9 at N/P ratio = 0.25, 0.5, 1, 2, and 4, and then treated with RNase A. After dissociation from BP100-KH9, FT mRNA was analyzed by agarose gel electrophoresis. Original gel images are shown in Fig. S6.
The hydrodynamic diameter of FT mRNA without peptide showed a broad size distribution with a peak at around 500 nm. The size distribution of the complexed molecules shifted with the addition of the peptide; an additional peak with smaller size around 50 nm at N/P ratio of 0.25 and peaks with larger sizes at other N/P ratios. The size distribution of the complexes at an N/P ratio 2 was bimodal, with a main peak larger than 1000 nm, supporting the formation of higher order complex at this N/P ratios (Fig. S1). Despite the higher order complex formation at an N/P ratio 4 as confirmed by the gel shift assay, the complexes at this ratio exhibited a main peak around 200 nm in hydrodynamic diameter. This may be because the predicted larger complex were removed by filtration before the analysis. These properties of FT mRNA-peptide complexes collectively suggest the formation of smaller complex at lower N/P ratios, with larger complexes forming at higher N/P ratios. Considering that the maximum size of molecules to pass through the cell wall is approximately 50 nm32, the physicochemical properties of the FT mRNA-peptide complexes suggest that mRNA delivery should be performed at lower N/P ratios. Additionally, it is advisable to use the lowest possible peptide concentration (N/P ratio), as higher concentration of CPPs may cause stress to plants due to cytotoxicity28.
Production of FT-Citrine from the delivered mRNA
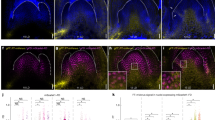
To obtain evidence of protein production from exogenously delivered mRNA, we tested FT-Citrine mRNA delivery mediated by the carrier peptide BP100-KH9. The FT-Citrine mRNA, synthesized by in vitro transcription, was complexed with BP100-KH9 and introduced into Arabidopsis leaves via infiltration using a needleless syringe33. Confocal laser-scanning microscopy (CLSM) analysis of the infiltrated leaves showed Citrine fluorescence, with FT-Citrine distributed in both the cytoplasm and nucleus of the leaf cells (Fig. 2A). The fluorescence was observed in stomata, likely through the permeation pathway used by non-viral carriers for nucleic acid cargo30 and this biased incorporation complicates the accurate measurement of FT-Citrine mRNA delivery efficiency in leaves. To validate the results, FT-Citrine was expressed under the constitutive promoter CaMV 35 S from exogenously introduced DNA using biolistics. The produced FT-Citrine was localized to both cytoplasm and nucleus, consistent with the localization observed from the introduced mRNA (Fig. 2B). While the fluorescence patterns resembled the characteristic autofluorescence typically observed in the cytosol and the chloroplasts of guard cells34, the presence of fluorescence within the nuclei suggests that the signal was derived from FT-Citrine. Furthermore, these observations align with the localization of FT-GFP in both the nucleus and cytoplasm when expressed endogenously in Arabidopsis shoot apex and leaf cells11. Collectively, the observed fluorescence and intracellular localization of FT-Citrine validate the production and correct localization of FT-Citrine protein from the exogenously introduced mRNA in Arabidopsis leaf cells. However, FT-Citrine protein was not detected by Western blotting, likely due to its low abundance. Since FT protein fused to GFP is reported to exhibit reduced activity in Arabidopsis8, we employed FT without any reporter protein in subsequent analyses.
Production and localization of FT-Citrine protein in Arabidopsis leaf cells. (A) Production of FT-Citrine protein from FT-Citrine mRNA introduced by the peptide-mediated method. Chlorophyll fluorescence shows leakage to YFP fluorescence. (B) Production of FT-Citrine protein from plasmid DNA introduced by biolistic delivery method. Confocal laser-scanning microscopy (CLSM) of the cells represents fluorescence of Citrine (YFP) and chlorophyll (chl). N and C denote nucleus and cytosol, respectively. Bars, 20 mm.
Induction of floral gene expression by the introduction of FT mRNA
Since FT transcript levels dramatically increase during seedling development in Arabidopsis under long day conditions17, seedling stage is thought to be more effective for flowering induction by FT mRNA delivery. Peptide-mediated mRNA delivery has only been applied to mature leaves so far31. Thus, we first tested the delivery of FT mRNA into Arabidopsis seedlings using the peptide-mediated method and analyzed the retention of the delivered FT mRNA. As shown in Fig. S2, the relative amount of FT mRNA in the seedlings treated with FT mRNA and peptide showed a marked increase one day after the introduction (DAI) compared to the controls. The FT mRNA level in the treated seedlings was then decreased to almost basal level by 3 DAI. These results suggest that the externally introduced FT mRNA persists in Arabidopsis seedlings for at least two days after introduction. However, the assessment does not reflect the amount of functional FT mRNA, as the assay quantifies total FT mRNA in the seedlings, including that in intercellular spaces.
In Arabidopsis, FT expression remains in a low level until 9 days after germination (DAG), then abruptly increases around 11 DAG35. We therefore assessed the effect of FT mRNA delivery at various growth stages up to 11 DAG in Arabidopsis seedling by analyzing expression of floral genes, which should be upregulated in response to the internalization of functional FT mRNA into plant cells. This analysis aim to determine the optimal timing for FT mRNA delivery for effective flowering induction and to demonstrate the functionality of externally introduced FT mRNA in Arabidopsis seedlings.
Using the peptide BP100-KH9, we introduced FT mRNA into Arabidopsis seedlings at 7, 9, and 11 DAG and analyzed the expression of the floral genes AP1, LFY, SEP3, and SOC1, which are directly or indirectly induced by FT (Fig. 3A), two days after the introduction. Expression analysis showed a stepwise increase in all the tested floral gene transcripts according to the growth of seedlings treated with water or peptide only (Fig. 3B), suggesting a minimal effect of the delivery method on seedling growth. The introduction of FT mRNA mediated by the peptide significantly induced the expression of the floral genes, with the most pronounced induction observed at 7 DAG and 9 DAG, compared to the controls (Fig. 3B). While the addition of only peptide occasionally induced the expression especially in AP1, likely be due to stress-induced flowering36 caused by cytotoxicity of CPPs28, the induction caused by the introduction of FT mRNA with peptide exceeded that induced by peptide, demonstrating that the induction of floral gene expression is primarily attributed to FT mRNA delivery. Furthermore, these results suggest that our FT mRNA delivery system is more effective when introduced into 7 to 9 DAG seedlings for inducing floral genes. This effectiveness can be explained by two reasons. First, the ratio of delivered FT mRNA to endogenous FT mRNA was higher in younger seedlings due to the abrupt increase in endogenous FT transcript levels after 9 DAG. Second, the cell wall network becomes denser with the increase of cell wall stiffness as seedlings grow37, which impedes the mRNA-peptide nanocomplex from passing through cell wall pores. Either or both of these factors likely contribute to the effective introduction of FT mRNA into 7–9 DAG Arabidopsis seedlings. Collectively, the induction of floral genes by FT mRNA delivery suggests that FT protein was produced and functioned to induce flowering in younger seedlings at 7 to 9 DAG. These results, on the other hand, raise the question of whether delivering FT mRNA into seedlings younger than 7 DAG or using multiple introductions would be more effective for flowering induction. However, delivery to very young seedlings or multiple infiltrations would be unsuitable, since the delivery method utilizes pressure and compression of seedlings in the mRNA/peptide solution, which substantially delays growth.
Expression analysis of floral genes after peptide-mediated delivery of FT mRNA in Arabidopsis seedlings. (A) Transcriptional regulation of floral genes by FT. FT protein associated with FD induces transcription of floral genes (AP1, LFY, SEP3, SOC1) directly or indirectly. (B) Transcript abundance of floral genes was assessed by quantitative reverse transcription PCR and normalized by that of an internal control PEX4. FT mRNA was delivered into 7, 9, or 11 days after germination Arabidopsis seedlings by the peptide BP100-KH9, and total RNA was extracted from the shoot apex of the seedlings two days after the introduction. Seedlings were treated only with the peptide BP100-KH9 or water as controls. Data are shown as means with standard deviation of three biological replicates (n = 3). The lowercase letters indicate statistically significant differences (Tukey’s multiple comparison test, P < 0.05).
Acceleration of flowering by FT mRNA delivery
Using the optimized conditions for the FT mRNA to peptide ratio and the timing for delivery, we delivered FT mRNA into Arabidopsis seedlings to test the acceleration of flowering. 8 DAG seedlings were subjected to the peptide-mediated FT mRNA delivery and then cultivated under long-day condition, along with FT mRNA only and peptide only controls. Since flowering is affected by light intensity and temperature, we measured flowering time by cultivating plants on an agar medium, ensuring a homogenous condition for all plants being compared. As a result of measurement of the flowering date, plants treated with either FT mRNA or peptide flowered at a median of 26 DAG (Fig. 4A and C), which nearly agrees with that of Arabidopsis Col-0 in a pot38, confirming that plants flowered normally under these conditions. In comparison, plants treated with the FT mRNA-peptide complex flowered at a median of 25 DAG, which is statistically earlier than any of the controls (Fig. 4A). We also investigated the effect of FT mRNA delivery on the leaf number at bolting, a common metric for assessing the impact on flowering2. As shown in Fig. 4B, the results largely agreed with the flowering time of the plants; plants treated with the FT mRNA-peptide complex flowered with significantly fewer leaves compared to any of the controls. We validated the flowering induction with FT mRNA delivery by testing peptide-mediated mRNA delivery of FT-Citrine, which has limited ability to induce flowering39, and observed a significant reduction in flowering acceleration with FT-Citrine mRNA delivery (Fig. S4). These findings collectively suggest that the introduction of FT mRNA mediated by the peptide BP100-KH9 accelerates flowering in the Arabidopsis wild-type plants, even under the long-day condition.
Flowering induction of Arabidopsis plants by FT mRNA delivery. (A) and (B) Box plot showing flowering date (A) and leaf number at flowering (B) of plants treated with peptide-mediated delivery of FT mRNA, FT mRNA with 5’Cap and polyA (FT-C), and the controls treated with FT mRNA (RNA control) or peptide only (Peptide control). The asterisks indicate statistically significant differences (P < 0.05, Tukey’s multiple comparison test, excluding outliers). (C) Flowering rate of plants treated with FT mRNA and/or peptides shown by days after germination.
Since both 5’ cap and 3’ polyA modifications of mRNA enhance expression of externally introduced mRNA in plant cells by increasing stability and translation efficiency40, we tested these modifications on FT mRNA. The modified FT mRNA formed a complex with the peptide BP100-KH9 similarly to the unmodified FT mRNA (Fig. S3). As shown in Fig. 4, plants treated with the modified FT mRNA flowered earlier than the controls, with statistical significance in both flowering date and leaf number. However, there was no significant difference in the flowering status between plants treated with modified or unmodified FT mRNA (Fig. 4), suggesting that the flowering acceleration by the delivery of modified FT mRNA was comparable to that of the unmodified version. Since our carrier peptide binds to mRNA to compact the complex and thus protect it from RNase (Fig. 1B), this likely reduced the effect of mRNA modification on stability. Moreover, the complexation of mRNA with the peptide by N/P ratio results in the introduction of a reduced number of mRNA copies into cells for the modified mRNA due to its higher molecular weight. Considering these factors, the results suggest a minor effect of mRNA modification on our peptide-mediated FT mRNA delivery for flowering acceleration.
Given that the uptake of nucleic acids mediated by the BP100-KH9 peptide is mainly via leaf hydathodes in Arabidopsis leaves30, FT protein, which is thought to be the predominant mobile form8 translated from the exogenously delivered FT mRNA in leaves, would move to the SAM via the phloem and subsequently induce the floral genes in our system. Additionally, the delivered FT mRNA might be directly transported to the SAM, leveraging its potential mobility to induce floral genes41, though induction of flowering via direct transport of FT mRNA to the SAM is still debated42. Direct delivery of FT mRNA to the SAM should accelerate flowering more efficiently. However, the SAM region is likely protected against externally introduced biomolecules, as demonstrated against the viruses43. Thus, we believe that our system demonstrated in this study offers the only feasible method for flowering acceleration utilizing FT mRNA delivery to date.
Conclusion
In this study, we established a method to accelerate flowering in a model Arabidopsis by utilizing FT mRNA delivery mediated by the carrier peptide BP100-KH9. The Arabidopsis ecotype used in the study was the early flowering ecotype Col-044, and the conditions were long-day, which typically make flowering acceleration challenging. Despite these conditions, we observed a shortened flowering time, confirmed both by the flowering time and the number of leaves at flowering, validating the effectiveness of the method. Considering the short half-life of the delivered FT mRNA, the acceleration of flowering is likely due to the advanced progression of the flowering stage triggered by a transient increase in FT protein levels, rather than a sustained elevation. We chose FT mRNA for external flowering induction, because FT is functionally conserved in many crop plants45. Indeed, genetic engineering with FT overexpression has been shown to induce flowering in various plant species18,19,46,47,−48, suggesting the applicability of the method established in this study. The method also offers promise for biosafety when used for crop plants, as it employs a biocompatible peptide carrier, and mRNA delivery is already utilized for vaccination in human49. The method established in this study serves as a proof of concept for external FT mRNA delivery for flowering acceleration and mRNA delivery for plant modification.
Materials and methods
Plant materials and cultivation
Arabidopsis thaliana ecotype Col-0 was used in the study. Arabidopsis seeds were sterilized with 70% ethanol for 1 min and subsequent 2.5% hypochlorite for 5 min. After vernalization at 4 °C storage for 3 days, the sterilized seeds were cultivated on a Murashige-Skoog (MS) medium (Sigma-Aldrich, US) solidified with 0.25% gellan-gum at 22 °C under a 16 h light condition in a growth chamber. For infiltration using a needle-less syringe, Arabidopsis Col-0 plants were cultivated in soil (Promix, Rivièredu-Loup, Canada) supplemented with vermiculite at a ratio of 2:1 at 22 °C under a 8 h light condition.
Synthesis of mRNA
Template DNA for in vitro transcription was amplified by PCR using Arabidopsis 1 st strand cDNA with p1 and p2 for FT (Sequences of primers are listed in Table S1). For FT-Citrine, template DNA was amplified by PCR with p3 and p4, and p5 and p6 for FT and Citrine, respectively, and subsequently the PCR products were mixed and then fused by overlapping PCR with p3 and p6. mRNA was synthesized by in vitro transcription using the template DNA (Fig. S5) and in vitro Transcription Kit (TaKaRa, Japan) or HiScribe T7 ARCA mRNA Kit (with Tailing) (New England BioLabs, US). The synthesized mRNA was purified by a column. Purity of mRNA was analyzed by Bioanalyzer 2100 (Agilent, US).
Complexation of mRNA with peptides and physicochemical evaluation of mRNA-peptide complex
BP100-KH9 [KKLFKKILKYLKHKHKHKHKHKHKHKHKH: 3809.8 Da] was mixed with mRNA at various N/P ratios, and then left them for 20 min at room temperature. For gel shift assay, the complexes were analyzed by electrophoresis in a 1% agarose gel with 0.5 x TBE. The hydrodynamic diameter of the complex was measured by Dynamic Light Scattering using Zeta Sizer Nano ZS instrument (Malvern Instruments Ltd., UK) after filtering the complex solution.
mRNA protection assay
FT mRNA (200 ng) was mixed with BP100-KH9 at various N/P ratios, and then incubated at room temperature for 20 min. Following addition of 0.16 ng RNaseA (QIAGEN, Germany) to the mRNA-peptide complex solution, the mixture was incubated at room temperature for 20 min. After adding a loading buffer containing SDS to the solution to stop RNase activity and dissociate the peptide from the mRNA, the RNA was analyzed by agarose gel electrophoresis.
Observation of FT-Citrine in leaves
For delivery of FT-Citrine mRNA into leaf cells, 1 µg of FT-Citrine mRNA was complexed with 0.125 µg of BP100-KH9 in 100 µl solution, and then infiltrated into 2 months-old Arabidopsis leaf by infiltration using needleless syringe as described previously33. For biolistic delivery of FT gene, FT-Citrine used for the template for in vitro transcription was placed downstream of a constitutive P35S promoter in pUC19. The resulting plasmid was introduced into 2 months-old Arabidopsis leaves by biolistic particle delivery system PDS-1000 (Bio-Rad, US)50. The fluorescence of the FT-Citrine in the infiltrated and bombarded leaves was observed by a confocal laser scanning microscopy LSM700 (Zeiss, Germany) with chlorophyll autofluorescence.
Peptide-mediated delivery of mRNA into Arabidopsis seedlings
For delivery of FT mRNA into Arabidopsis seedlings, 10 µg of FT mRNA was mixed with 1.25 µg of BP100-KH9 in 1 ml solution, and the complex was then incubated at room temperature for 20 min. Arabidopsis seedlings cultivated for eight days on a solidified medium were dipped in 0.02% (v/v) Silwet L-77 (PhytoTechnology Laboratories, Shawnee Mission, USA) and then submerged in the mRNA-peptide complex solution (~ 40 seedlings in the 1 ml complex solution). The seedlings were subjected for vacuum at −0.08 MPa for 1 min and compression at + 0.08 MPa for 1 min for infiltration of the complex. After 1 h incubation in the solution, the seedlings were washed with water, and then cultivated on a solidified medium. For the analysis of abundance of the internalized FT mRNA in Arabidopsis seedlings, the seedlings were washed three times 1, 2 and 3 days after introduction, and then total RNA was extracted by RNeasy Plant Mini kit (QIAGEN, Germany).
Expression analysis of FT and floral genes
For expression analysis of the floral genes, FT mRNA was introduced into 7, 9, or 11 DAG Arabidopsis seedlings, and 2 days after the introduction, shoot apex of the seedlings were collected. Total RNA was extracted from the shoot apex by RNeasy Plant Mini kit (QIAGEN, Germany), and first strand cDNA was synthesized with the total RNA and ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Japan). Quantitative real-time PCR analyses of floral genes were performed by using Applied Biosystems StepOnePlus Real-Time PCR Systems (Thermo Fisher, US), the cDNA and primers p7 and p8 for AP1, p9 and p10 for LFY, p11 and p12 for SEP3, and p13 and p14 for SOC1, and p15 and p16 for an internal control gene PEX4. For analysis of FT, primers p17 and p18 were used, and the amount of FT mRNA was normalized with that of PEX4. Quantification was performed with standard curves for each target gene.
Measurement of flowering time
For measurement of flowering time, FT mRNA was introduced into 8 DAG seedlings as described above, and after 1 day cultivation on a solidified medium for recovery, the seedlings were transferred to a MS solidified medium. For precise comparison of flowering time, seedlings to be compared were cultivated on a same solidified medium, and such plates were replicated for statistical analysis. The plants were cultivated at 22 °C under a 16 h light condition in a growth chamber (Biotron, NK System, Japan). The light intensity was 100 µmol m−2 s−1. The number of leaves was counted without cotyledons after bolting. The data was statistically evaluated by the Tukey’s multiple comparison test, excluding outliers identified by Smirnov-Grubbs test. During the measurements, dwarf and dark green individuals were occasionally observed, likely as a result of the vacuum–pressure infiltration treatment. Because these plants were abnormal, exhibited delayed flowering, and were identified as outliers that could confound accurate measurement of flowering time, they were excluded from the analyses.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Amasino, R. Seasonal and developmental timing of flowering. Plant. J. 61, 1001–1013 (2010).
Kardailsky, I. et al. Activation tagging of the floral inducer FT. Science 286, 1962–1965 (1999).
Wigge, P. A. FT, a mobile developmental signal in plants. Curr. Biol. 21, R374–378 (2011).
Putterill, J., Robson, F., Lee, K., Simon, R. & Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857 (1995).
Turck, F., Fornara, F. & Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant. Biol. 59, 573–594 (2008).
Takada, S. & Goto, K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant. Cell. 15, 2856–2865 (2003).
Mathieu, J., Warthmann, N., Kuttner, F. & Schmid, M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17, 1055–1060 (2007).
Corbesier, L. et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033 (2007).
Jaeger, K. E. & Wigge, P. A. FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17, 1050–1054 (2007).
Teper-Bamnolker, P. & Samach, A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant. Cell. 17, 2661–2675 (2005).
Abe, M. et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 (2005).
Wigge, P. A. et al. Integration of Spatial and Temporal information during floral induction in Arabidopsis. Science 309, 1056–1059 (2005).
Michaels, S. D., Himelblau, E., Kim, S. Y., Schomburg, F. M. & Amasino, R. M. Integration of flowering signals in winter-annual Arabidopsis. Plant. Physiol. 137, 149–156 (2005).
Yoo, S. K. et al. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant. Physiol. 139, 770–778 (2005).
Wellmer, F. & Riechmann, J. L. Gene networks controlling the initiation of flower development. Trends Genet. 26, 519–527 (2010).
Kaufmann, K. et al. Orchestration of floral initiation by APETALA1. Science 328, 85–89 (2010).
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M. & Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962 (1999).
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S. & Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036 (2007).
Bohlenius, H. et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043 (2006).
Raskin, I. Salicylate, a new plant hormone. Plant. Physiol. 99, 799–803 (1992).
D’Aloia, M. et al. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant. J. 65, 972–979 (2011).
Ionescu, I. A., Moller, B. L. & Sanchez-Perez, R. Chemical control of flowering time. J. Exp. Bot. 68, 369–382 (2017).
Marton, L., Wullems, G., Molendijk, L. & Schilperoort, R. In vitro transformation of cultured cells from Nicotiana tabacum by Agrobacterium tumefaciens. Nature 277, 129–131 (1979).
Klein, R. M., Wolf, E. D., Wu, R. & Sanford, J. C. High-velocity microprojectiles for delivering nucleic acids into living cells. Biotechnology 24, 384–386 (1992). (1987).
Squire, H. J., Tomatz, S., Voke, E., González-Grandío, E. & Landry, M. The emerging role of nanotechnology in plant genetic engineering. Nat. Reviews Bioeng. 1, 314–328 (2023).
Chugh, A., Eudes, F. & Shim, Y. S. Cell-Penetrating peptides: nanocarrier for macromolecule delivery in living cells. Iubmb Life. 62, 183–193 (2010).
Milletti, F. Cell-penetrating peptides: classes, origin, and current landscape. Drug Discovery Today. 17, 850–860 (2012).
Numata, K. et al. Library screening of cell-penetrating peptide for BY-2 cells, leaves of arabidopsis, tobacco, tomato, poplar, and rice callus. Sci. Rep. 8, 10966 (2018).
Lakshmanan, M., Kodama, Y., Yoshizumi, T., Sudesh, K. & Numata, K. Rapid and efficient gene delivery into plant cells using designed peptide carriers. Biomacromolecules 14, 10–16 (2013).
Midorikawa, K., Kodama, Y. & Numata, K. Vacuum/Compression Infiltration-mediated permeation pathway of a Peptide-pDNA complex as a Non-Viral carrier for gene delivery in planta. Sci. Rep. 9, 271 (2019).
Thagun, C., Motoda, Y., Kigawa, T., Kodama, Y. & Numata, K. Simultaneous introduction of multiple biomacromolecules into plant cells using a cell-penetrating peptide nanocarrier. Nanoscale 12, 18844–18856 (2020).
Miyamoto, T. & Numata, K. Advancing Biomolecule Delivery in Plants: Harnessing Synthetic Nanocarriers to Overcome Multiscale Barriers for Cutting-edge Plant Bioengineering. Bulletin Chem. Soc. Japan 96(9), (2023).
Watanabe, K., Odahara, M., Miyamoto, T. & Numata, K. Fusion Peptide-Based biomacromolecule delivery system for plant cells. Acs Biomater. Sci. Eng. 7, 2246–2254 (2021).
Virlouvet, L. & Fromm, M. Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New. Phytol. 205, 596–607 (2015).
Shen, L., Kang, Y. G., Liu, L. & Yu, H. The J-domain protein J3 mediates the integration of flowering signals in Arabidopsis. Plant. Cell. 23, 499–514 (2011).
Kazan, K. & Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 67, 47–60 (2016).
Bonfanti, A. et al. Stiffness transitions in new walls post-cell division differ between Marchantia polymorpha gemmae and Arabidopsis thaliana leaves. Proc. Natl. Acad. Sci. U S A. 120, e2302985120 (2023).
Sanagi, M. et al. Low nitrogen conditions accelerate flowering by modulating the phosphorylation state of FLOWERING BHLH 4 in Arabidopsis. Proc Natl. Acad. Sci. U S A 118, e2022942118 (2021).
Notaguchi, M. et al. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant. Cell. Physiol. 49, 1645–1658 (2008).
Callis, J., Fromm, M. & Walbot, V. Expression of mRNA electroporated into plant and animal cells. Nucleic Acids Res. 15, 5823–5831 (1987).
Jackson, S. D. & Hong, Y. Systemic movement of FT mRNA and a possible role in floral induction. Front. Plant. Sci. 3, 127 (2012).
Yu, Z. et al. Mobile flowering locus T RNA - Biological relevance and biotechnological potential. Front. Plant. Sci. 12, 792192 (2021).
Wu, H. et al. WUSCHEL triggers innate antiviral immunity in plant stem cells. Science 370, 227–231 (2020).
Sanda, S., John, M. & Amasino, R. Analysis of flowering time in ecotypes of Arabidopsis thaliana. J. Hered. 88, 69–72 (1997).
Blumel, M., Dally, N. & Jung, C. Flowering time regulation in crops-what did we learn from arabidopsis?? Curr. Opin. Biotechnol. 32, 121–129 (2015).
Yan, L. et al. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U S A. 103, 19581–19586 (2006).
Lifschitz, E. et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. U S A. 103, 6398–6403 (2006).
Meng, X., Muszynski, M. G. & Danilevskaya, O. N. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant. Cell. 23, 942–960 (2011).
Mulligan, M. J. et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586, 589–593 (2020).
Ueki, S., Lacroix, B., Krichevsky, A., Lazarowitz, S. G. & Citovsky, V. Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat. Protoc. 4, 71–77 (2009).
Acknowledgements
We thank Kanako Saga, Kumiko Tachikawa and Aili Ailizati for their technical support.
Funding
This work was supported by Japan Science and Technology Agency (JST) Exploratory Research for Advanced Technology (ERATO; KN, JPMJER1602), JST COI-NEXT (KN, JPMJPF2114), and the Japan Society for the Promotion for Scientific Research (JSPS) (MO, 22K05625).
Author information
Authors and Affiliations
Contributions
MO, SK and KN conceived the research, MO and MM performed the experiments, MO, SI, SK and KN analyzed the data, and MO and KN wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Odahara, M., Mori, M., Ishio, S. et al. Induction of flowering in Arabidopsis through functional peptide-mediated FT mRNA delivery. Sci Rep 15, 22088 (2025). https://doi.org/10.1038/s41598-025-05773-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-05773-9