Abstract
Chickpea productivity remains low due to limited genetic variability and susceptibility to biotic and abiotic stresses. To address these challenges, the introgression of novel genes from wild relatives and multi-environment evaluations are essential for identifying high-yielding, stable interspecific derivatives (ISDs). Despite the availability of several statistical tools, few chickpea studies have used a comprehensive multi-model approach for stability analysis. This study fills the void by applying four models- AMMI, GGE, WAASB, and MTSI for the first time in chickpea to evaluate multi-trait stability across diverse environments. 19 ISDs developed from four interspecific crosses (BGD 72 × ILWC 229, PBG 5 × ILWC 229, BGD 72 × ILWC 246, and PBG 5 × ILWC 246), along with two checks (PBG 7 and PBG 8), were evaluated across six environments. Trials were conducted over three consecutive rabi seasons (2020–2023) at Ludhiana (ENV1–ENV3), followed by multi-location testing during rabi 2023–24 at Ludhiana (ENV4), Karnal (ENV5), and Kanpur (ENV6). Key yield traits, including number of pods per plant (NPP), 100-seed weight (HSW), and seed yield per plot (SYPP), were analyzed using all four models. Significant effects of genotype, environment, and genotype × environment interactions (GEIs) were detected through pooled ANOVA, and positive correlations were observed among all key traits. Genotypes GL21-1 (G1) and GL21-54 (G8) were superior performers with high trait values and stable performance across all environments, with yield variability up to 20% under stress-prone conditions. Among the models, GGE and WAASB were found effective for individual traits, while MTSI was superior for multi-trait selection. These promising ISDs can be released as high-yielding varieties or used as donor parents in climate-resilient chickpea breeding programs, addressing the rising global demand for chickpea production.
Similar content being viewed by others
Introduction
Chickpea (Cicer arietinum L.), also known as garbanzo bean or Bengal gram, is the world’s second most important legume crop after common bean (Phaseolus vulgaris L.). It is a diploid (2n = 2x = 16) self-pollinated crop with a genome size of 738.09 Mb1. Its origin date back to around 7,500-6,800 BC, with historical evidence pointing to south-eastern Turkey and parts of Syria as its primary centers of domestication2,3. Chickpea is highly valued for its nutritional content, often referred to as “poor man’s meat” due to its rich protein content and the presence of essential minerals and vitamins4. Chickpea covers around 14.81 million hectares area worldwide with 18.09 million tonnes production and 1221.8 kg/ha productivity. India holds the top position globally in chickpea production and consumption, with an output of approximately 13.56 million tonnes (75%) from an area of 10.74 million hectares5. To achieve self-sufficiency in pulse production by 2050, India aims to increase chickpea production to 16.0–17.5 million tonnes from an area of 10.5 million hectares3. The relatively low productivity of chickpea compared to rabi cereal crops is primarily due to the limited genetic variability and susceptibility to various biotic and abiotic stresses. To address this limitation, it is essential to explore closely related wild species to enhance both yield and resistance. Introgressing novel genes from wild relatives into cultivated chickpea can broaden the genetic base, making the crop more resilient to diseases, pests, drought and other environmental challenges. This can ultimately contribute to higher productivity and sustainability in chickpea cultivation.
Chickpea has several wild relatives primarily found in Turkey, Iran, Afghanistan and Central Asia. The number of recognized Cicer species has increased to 46, including 36 perennials and 10 annuals6. C. reticulatum and C. echinospermum are mainly found in south-eastern Turkey, with C. echinospermum typically confined to basaltic soils at lower altitudes, while C. reticulatum favours sandstone or granite substrates7. Due to early flowering and substantial barriers to crossability after fertilization, only a few interspecific hybrids have been reported within the genus Cicer8,9. The cultivated chickpea (C. arietinum) and C. reticulatum are cross compatible, sharing similar seed protein compositions and morphological traits3. Interspecific hybridization is the method of crossing cultivated types with their wild relatives to incorporate new genetic traits. The transfer of genes from C. reticulatum and C. echinospermum into cultivated chickpeas has resulted in significant yield improvements10,11, resilience to drought and heat12 and improved disease resistance3,13.
To achieve stability and adaptability in a crop, it must demonstrate the ability to thrive in specific environments. Due to the significant influence of variability in environmental conditions, crops are often subject to genotype × environment interactions (GEIs). These interactions arise due to variations in rainfall, temperature, soil composition, moisture levels, soil types, fungal and viral pathogens, nematodes and bacteria14. The complex interplay between environmental factors and the genetic composition of the genotypes, makes GEI a crucial aspect to consider in crop improvement15. The major goals in crop improvement include; minimizing risks, ensuring stable yields, reducing costs and maximizing economic returns16.
To tackle the challenges posed by GEIs, breeders employ a range of statistical models and tools, including AMMI (additive main effects and multiplicative interaction), GGE (genotype main effect + genotype × environment interaction), WAASB (weighted average of absolute scores for the BLUPs (best linear unbiased predictions)) and MTSI (multi-trait stability index). The AMMI model quantifies the additive main effects associated with genotypes and environments by combining analysis of variance (ANOVA) and principal component analysis (PCA), for the identification of stable genotypes across environments as well as those best suited to specific conditions17,18. However, AMMI focuses on single trait and may not perform well under unbalanced data. GGE biplots visualize the main effect of genotype, the magnitude and nature of the GEI19, though they can be sensitive to data scaling and may oversimplify complex interactions. WAASB, on the other hand, combines genotype stability and trait performance by utilizing the WAASBY index, based on BLUPs. It effectively integrates both adaptability and performance, handles unbalanced data, and provides more accurate genotype rankings in multi-environment trials20. Nevertheless, WAASB may be less efficient for traits strongly influenced by non-additive gene actions. MTSI is also a BLUP-based model that offers the selection of genotypes by considering multiple traits, to identify those with higher mean performance and greater stability21. MTSI is especially critical under changing climate conditions. This helps breeders identify genotypes that are both stable and high-yielding across diverse environments. Hence, making it a valuable tool for developing climate-resilient varieties to achieve India’s goal of 16–17.5 million tonnes of chickpea production by 2050. By combining these approaches, breeders can achieve a more comprehensive understanding of the genotype’s performance for the selection of the most promising ones for utilization in further improvement programs.
This study aims to assess the stability of three yield traits in interspecific chickpea derivatives by employing AMMI, GGE, WAASB and MTSI analysis. These methodologies seek to identify high-yielding, stable genotypes that can be used to enhance chickpea breeding programs. Further, ultimately it would contribute to the development of improved cultivars well-suited to diverse environmental conditions.
Materials and methods
Plant materials
In this study, advanced chickpea interspecific derivatives (ISDs) from crosses between the cultivated chickpea varieties BGD 72 and PBG 5 (C. arietinum) and two wild annual Cicer species: C. reticulatum (ILWC 229) (EC720438) and C. echinospermum (ILWC 246) (EC720481)3,22 were involved. The 19 ISDs were derived from four different crosses viz., BGD 72 × ILWC 229 (9 ISDs), PBG 5 × ILWC 229 (3 ISDs), BGD 72 × ILWC 246 (6 ISDs) and PBG 5 × ILWC 246 (1 ISD). These were evaluated along with the checks PBG 7 and PBG 8 in multiple environments for yield and its contributing traits. A detailed list of chickpea genotypes and their parental derivations, assessed across various environments, is provided in Supplementary Table 1.
Field experimental sites and design
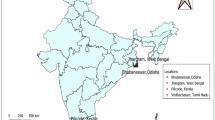
The experiment was conducted across multiple locations during different rabi seasons, with each site exhibiting distinct variations in temperature, rainfall, humidity and soil types. At Punjab Agricultural University (PAU), Ludhiana (30°54’N, 75°48’E), the experiments were carried out for three consecutive rabi seasons from 2020-21 to 2022-23, with each season referred to as ENV1 (2020-21), ENV2 (2021-22) and ENV3 (2022-23). Additionally, multi-location trials were conducted during the rabi 2023-24 season at PAU, Ludhiana, designated as ENV4, Central Soil Salinity Research Institute (CSSRI), Karnal (29°43’N, 76°58’E), designated as ENV5, and at the Indian Institute of Pulses Research (IIPR), Kanpur (20°27’N, 80°14’E), designated as ENV6. Ludhiana and Karnal are located in zone 6 (Transgangetic plains), while Kanpur falls under zone 5 (Upper Gangetic plains), representing two distinct agro-climatic zones of India. Geographical details, including the coordinates of the three test locations, are shown in Fig. 1. A total of 19 ISDs, along with two checks (PBG 7 and PBG 8), were sown in paired rows, each 2 m long, with 30 cm spacing between them. The experiments were laid out in a randomized complete block design (RCBD) across all the locations. The recommended agronomic practices such as field preparation, land clearing, weeding, irrigation and fertilization were meticulously followed to ensure optimal crop growth. The details of climatic conditions observed during the cropping season are presented in Table 1.
Data collection
The key yield traits assessed in the study were: number of pods per plant (NPP), 100-seed weight (HSW) and seed yield per plot (SYPP). To ensure accurate and representative data, NPP was measured by randomly selecting five plants from each plot while HSW was recorded by weighing 100 seeds from the harvested seed lot. SYPP was recorded on plot basis to ensure that the data accurately reflects the overall yield potential of each genotype.
Statistical analysis
The statistical analyses for this study were performed using various packages within the R environment23. Analysis of variance (ANOVA) was performed after checking the normality of the data using the Shapiro-Wilk test at 5% level of significance. The quantitative traits were then subjected to ANOVA for assessing variations among genotypes, environments and GEIs. This analysis was conducted using the ‘agricolae’ and ‘lme4’ packages24. The phenotypic distribution of yield traits was visualized using the ‘ggplot2’ library in R, to have a clear and effective representation of the data. Correlations among traits were determined using BLUPs obtained from six environments using the META-R tool25, these correlations were visualized using the ‘psych’ library26 in R.
Stability analysis across the six environments were performed using four indices: AMMI, GGE, WAASB and MTSI. The AMMI model estimated the additive main effects of genotypes and environments by combining ANOVA and PCA, as per the statistical model described by Zobel et al.17. Two fundamental biplots were generated as a part of the AMMI analysis. The AMMI1 biplot is based on the genotypic main effects and the scores of the first Interaction Principal Component (IPC1), while the AMMI2 biplot incorporates both the first and second Interaction Principal Component (IPCA1 and IPCA2) scores, focusing on the multiplicative components18,27. Further, genotypes were classified as stable performers based on the calculation of the AMMI Stability Value (ASV) as defined by Purchase et al.28. The Simultaneous Selection Index (SSI) was calculated by adding the genotype rank based on the mean of the yield trait (Y) and the ASV. GGE biplots were utilized to visualize the genotype main effect with the magnitude and nature of the GEI. These biplots were constructed following the site regression model18,29. WAASB was utilized as a stability parameter integrating statistics from both the AMMI and BLUP models. To test the significance of random effects, a likelihood ratio test was conducted by comparing models with and without random effects. Genotype stability was evaluated by Singular Value Decomposition (SVD) applied to the BLUPs matrix of GEI effects, facilitating the calculation of Weighted Average of Absolute Scores (WAAS) to quantify the stability of each genotype. Additionally, using WAASB, a new simultaneous selection index WAASBY was calculated for the ith genotype, assigning equal weight (50%) to both performance and stability20,30. MTSI was calculated based on the WAASBY of NPP, HSW and SYPP for the identification of desirable genotypes with high performance and stability. Later, a 15% selection intensity was applied for the selection of top-performing genotypes21. All these analyses (AMMI, GGE, WAASB and MTSI) were performed using the ‘metan’ package in R31.
Results
ANOVA, mean performances and correlations of chickpea genotypes for yield parameters
The combined ANOVA across multi-test environments revealed highly significant variation for all three traits. The mean sum of squares for each trait indicated significant differences among environments, genotypes and GEIs (Table 2). The percentage of variation attributed to total variability (GEN + ENV + GEN × ENV) by the environment was highest for SYPP (12.50%), followed by NPP (7.21%) and HSW (2.24%). Similarly, the variation attributed to genotype was greater for HSW (91.30%), followed by NPP (67.82%) and SYPP (61.57%). The interaction between genotype and environment was higher for SYPP (19.82%) followed by NPP (19.20%) and HSW (5.18%) (Table 2). The performance of 21 chickpea genotypes (19 ISDs + 2 checks) across six environments for three yield traits- NPP, HSW and SYPP have been presented in Supplementary Table 2. The average NPP across the environments ranged from 41.3 to 71.5, with an overall mean of 53.6. HSW varied from 13.2 to 28.3 g, with a mean of 19.8 g. Similarly, SYPP ranged from 478.4 to 600.4 g, with an average of 531.8 g across the environments. Genotypes G1, G8, G19 and G18 for NPP; G1, G8, G5, G6 and G18 for HSW; and G1, G8, G19, G4, G18, G6, G10, G12, G13 and G9 for SYPP showed higher values than the overall mean and exceeded the checks PBG7 and PBG8 across all environments. Interestingly, G1, G8 and G18 were found to have mean values higher than the overall average across the six environments for all the yield traits.
Phenotypic distributions (a. Number of pods per plant (NPP); b. 100-seed weight (HSW) (g); c. Seed yield per plot (SYPP) (g)) and d. correlation matrix (In the pair plot, bivariate scatter plots are presented below the diagonal, histograms along it, and correlation coefficients above the diagonal (*** indicating P ≤ 0.001)) of yield traits.
The phenotypic distributions of all traits for six environments, along with Pearson’s correlation coefficients from a combined analysis of six environments are shown in Figure 2. The combined environments (CENV) reflected a balanced overall performance across all traits (Fig. 2a, 2b and 2c). A strong positive correlation was observed between NPP and SYPP, with a coefficient of 0.79 (significant at p < 0.001). A moderate positive correlation was found between HSW and SYPP (0.41), while the correlation between NPP and HSW was weak (0.23) (Fig. 2d).
AMMI analysis
As the pooled ANOVA revealed a significant influence of the environments and their interaction with genotype, AMMI analysis was conducted for all the yield traits to partition the variability into genotype, environment and GEI. The AMMI ANOVA for yield traits in chickpea genotypes tested across six environments has been presented in Table 3. The analysis revealed that the yield traits were significantly (p < 0.001) affected by environments, genotypes and GEI. The interaction component was divided into five IPCs. For NPP, IPC1 contributed 56.5% of the variation, followed by IPC2 (23.5%), IPC3 (10.6%), IPC4 (5.9%), and IPC5 (3.5%). For HSW, IPC1 explained 76.2% of the variation, followed by IPC2 (18.3%), IPC3 (3.7%), IPC4 (1.4%) and IPC5 (0.5%). For SYPP, IPC1 accounted for 59.5% of the variation, followed by IPC2 (20.2%), IPC3 (14.3%), IPC4 (3.5%) and IPC5 (2.5%). These results from the AMMI ANOVA clearly indicate the substantial influence of the environment and GEI on all three traits.
AMMI biplots, used for analysing multi-environment trials (METs), represents the GEIs to aid in the selection of stable and high-performing cultivars. AMMI1 biplot plotted genotype and environment main effects on the x-axis and IPC1 on the y-axis (Fig. 3a, b and c). The environmental influence on genotype performance revealed significant variations for all the traits. ENV6 indicated strong interaction forces for all the traits, as it was located far from the origin with longer vectors. This was followed by ENV4 and ENV5 for NPP and SYPP, and by ENV5 and ENV4 for HSW. In contrast, shorter vectors close to the origin in ENV1, followed by ENV2 and ENV3 for NPP; ENV1, followed by ENV3 and ENV2 for HSW; and ENV2, followed by ENV1 and ENV3, for SYPP, showed weak interaction forces. Genotypes: G17 and G14 for NPP; G4, G17, G9 and G7 for HSW and G9, PBG8 and G17 for SYPP were positioned near to the origin. These genotypes have broad adaptability as they exhibited low sensitivity to environmental variations. The overall average performance of these genotypes was close to the general mean across different environments.
The AMMI2 biplot augmented IPC2 on the y-axis, providing a more detailed view of specific GEIs. The first two IPCs of the interaction in the AMMI2 biplot model, accounted for 80.0%, 94.5% and 79.7% of GEI for NPP (Supplementary Figure. 1a), HSW (Supplementary Figure. 1b), and SYPP (Supplementary Figure. 1c), respectively. In the polygon, dotted lines connecting the vertex of the genotypes, represents maximum or minimum values and specific adaptation to certain environments. Genotypes viz., G13, G12, G1, G5, G10 and G14 for NPP, G16, G2, G19, PBG8, G6, G5 and G4 for HSW and G5, G7, G10, PBG8, G16, G18, G6 and G11 for SYPP, in the plot, exhibited either higher or lower values and specific adaptation to particular environments.
The SSI for each genotype was calculated by summing the ranks derived from the mean of yield traits and the AMMI stability values (Table 4). Genotypes with lower selection index ranks were considered more stable. In this study, G19, G8, PBG7, G1 and G17 for NPP; G8, G17, G1, G4 and G9 for HSW; and G8, G1, G9, G17 and G19 for SYPP were the top-performing stable genotypes based on the lowest SSI values in ascending order across different environments (Table 4).
GGE biplot analysis
GGE biplot analysis was performed to examine the diversity of genotypes and environments, focusing on GEI. The main effects of the genotypes were plotted on the horizontal axis (PC1), while the GEI effects on the vertical axis (PC2)29. GGE analysis for NPP (Fig. 4a), HSW (Fig. 4b) and SYPP (Fig. 4c) showed that the first two principal components (PC1 and PC2) explained 90.66%, 94.76% and 90.63% of the total variation, respectively.
Which-Won-Where biplot
The GGE biplot’s ‘Which-one-where’ approach was used to select the best-performing genotypes for target-specific environments and the visual grouping of environments based on their interactions with superior genotypes. For NPP (Fig. 4a), genotypes G1 and G8 had the uppermost rank in ENV1, while G18 performed best in ENV4. With respect to HSW (Fig. 4b), the biplot identified two mega-environments (ME). ME1 comprised of ENV1, ENV2, ENV3 and ENV4, while ME2 had ENV5 and ENV6. Genotype G5 performed best in the ME1 whereas genotypes G1 and G8 excelled in the ME2. For SYPP (Fig. 4c), genotype G18 performed best in ENV4, while G1 and G4 in ENV5. Notably, genotypes G13, G11, G5 and G10 for NPP; PBG7 and G2 for HSW; and G5, G15, G11, G12 and G6 for SYPP were occupied by none of the environments in the biplot, indicating their poor performance owing to low ranking across the environments.
Discriminativeness vs. representativeness biplot
In the discriminativeness vs. representativeness biplot, the length of the vectors represents the discriminative ability of the environments, while the angles between the vectors indicate their representativeness. Environments; ENV4, ENV3 and ENV6 for NPP (Fig. 5a), ENV1, ENV3, ENV4 and ENV2 for HSW (Fig. 5b), as well as ENV6, ENV1 and ENV3 for SYPP (Fig. 5c), displayed longer vectors, indicating a strong discriminative ability. Conversely, environments ENV5 and ENV2 for NPP; ENV5 and ENV6 for HSW; and ENV5 and ENV2 for SYPP exhibited the least discriminative power. The angles between all six environments were acute (< 90 degrees) for all traits, indicating a strong positive correlation and similarities with better representativeness among the test environments. ENV1 for NPP and HSW; and ENV2 for SYPP represented the ideal test environment, with vector closer to the average environment coordinate (AEC).
Mean vs. stability biplot
The AEC view of the GGE biplot for NPP, HSW and SYPP visualized the mean performance and stability of the genotypes. The genotypes placed toward the arrow on the AEC abscissa (single directional arrow) had above-average performance across the environments. Accordingly, for NPP (Fig. 6a) genotypes such as G1, G8, G19, G18, PBG7 and PBG8; for HSW (Fig. 6b) G1, G8, G5, G6 and G18; and for SYPP (Fig. 6c) G1, G8, G19, G4, G18 and G6 displayed above average mean performance. Contrastingly, genotypes G5, G3, G11 and G7 for NPP; PBG7, PBG8, G16 and G12 for HSW; and G5, G15, G3 and G15 recorded below average mean performance as they were placed farther to AEC abscissa. The ordinate (a line perpendicular to the AEC) represented interaction effects and stability. The genotypes having shorter projections were more stable, while those with longer projections as less stable. Thus, genotypes G1, G8, G19, PBG7 and PBG8 for NPP; G1, G8, G4 and G18 for HSW; and G1, G8, G19, G4, G9 and PBG8 for SYPP, with shorter projections exhibited more stable performance, while genotypes G13, G18 and G5 for NPP; G2, G6, G16 and G5 for HSW, along with G5, G18, G6 and G12 for SYPP, with longer projections displayed greater variability, indicating a high level of instability across all the test environments.
WAASB and MTSI analysis
Stability analysis using WAASB model
The study predicted genotype performance as per BLUP-based linear mixed-effect models, across different environments by treating the environment as a fixed effect and the genotype as a random effect. The results of likelihood ratio test showed that all traits were significantly (p < 0.01) affected by both genotypes and GEI (Supplementary Table 3). An exploratory factor analysis identified two principal components (FA1 and FA2), explaining 93.82% of the total variance. As communality represents the variance proportion in each trait explained by the factors identified via factor analysis. The communality value for NPP was 0.92, 0.99 for HSW and 0.90 for SYPP, with an overall average of 0.93 (Supplementary Table 4). A WAASB biplot was plotted with the mean trait value (Y) of genotypes on the x-axis and WAAS values on the y-axis. The centrally positioned vertical line represents the overall average of yield traits. Genotypes positioned to the right of the central vertical line exhibited higher yield levels compared to the overall mean, while those on the left displayed lower yields relative to the mean for the respective trait. The biplots were divided into four quadrants, with genotypes in each quadrant categorized based on their adaptability to different environments. For NPP (Fig. 7a), quadrant I contained 6 genotypes and 1 environment (ENV5), while quadrant II included 2 genotypes and 3 environments (ENV3, ENV4 and ENV6). Quadrant III had 8 genotypes and 1 environment (ENV1), while quadrant IV had 5 genotypes and 1 environment (ENV2). For HSW (Fig. 7b), quadrant I displayed 4 genotypes and 2 environments (ENV5 and ENV6), quadrant II had 2 genotypes and 3 environments (ENV2, ENV3 and ENV4), while quadrant III had only 12 genotypes, and quadrant IV had 3 genotypes and 1 environment (ENV1). For SYPP (Fig. 7c), quadrant I had 4 genotypes and 2 environments (ENV5 and ENV6), quadrant II contained 4 genotypes and 2 environments (ENV3 and ENV4), quadrant III had only 7 genotypes, and quadrant IV had 6 genotypes along with 1 environment (ENV2). Contrastingly, ENV1 was centrally placed on origin in the biplot. Overall, genotypes located in quadrants II and IV exhibited above average value for all traits. The WAASB biplot identified genotypes with near-zero WAAS values as the most stable, while those with values exceeding the mean of yield traits were considered the most desirable (Supplementary Table 5).
Ranking of genotypes based on their WAASBY scores
Simultaneous selection for traits with higher values and stability was conducted using the WAASBY scores, assigning equal weight (50%) to both trait value and stability (Supplementary Table 5). Genotypes with blue circles represent the top-ranked genotypes, while those with red circles indicate the lower ranks (Fig. 8). On the joint interpretation of yield traits (Y) and stability (WAASB), 5 genotypes (G1, G8, G19, PBG7 and PBG8) for NPP (Fig. 8a); 2 (G8 and G1) for HSW (Fig. 8b); and 5 (G1, G8, G19, G4 and G9) for SYPP (Fig. 8c) out-performed (above overall mean) with greater stability across the environments.
MTSI
MTSI was calculated using the genotype-ideotype Euclidean distance, with scores obtained from exploratory factor analysis (FA). The scores of the 21 chickpea genotypes and their ideotype estimates for FA1 and FA2 are provided in Supplementary Table 6. The traits were grouped into two factors: FA1, with NPP and SYPP; and FA2, represented HSW. The selection differential (SD) for genotypes selected based on the WAASBY scores was positive for all yield traits, indicating that this method effectively identified genotypes with both high mean performance and stability. HSW reported an SD of 28.32%, while SYPP and NPP had SD at 11.43% and 19.05%, respectively (Supplementary Table 7). MTSI values were calculated using WAASBY scores for NPP, HSW and SYPP traits. The lower MTSI values specify greater stability and superior mean performance of genotypes, wherein genotypes were selected at a 15% selection intensity (Fig. 9a). Among the 21 genotypes, G1, G8 and G4 were the best performers with greater stability across all environments, as they were located outside the red circle. The MTSI cut-off value for 15% selection intensity was set at 2.42 (Supplemental Table 6). Genotypes exceeding this value were positioned near the origin (marked with black circles), signifying poor performance and stability. The “strengths and weaknesses view” radar plot (Fig. 9b) provided a detailed breakdown of the FA1 and FA2 contributions to the MTSI for the selected genotypes. Genotype G1 exhibited a strong contribution from FA1, and a smaller influence from FA2. In contrast, G8 displayed balanced contributions from both FA1 and FA2, indicating consistent performance across multiple traits. Genotype G4, however, had minimal contributions from both factors, reflecting lower overall performance and stability compared to G1 and G8.
Comparison among four stability models
The ideal stability model was selected by calculating the SD for SYPP among genotypes chosen with a selection intensity of 15% using the AMMI, GGE, WAASB and MTSI models. Genotypes selected from GGE and WAASB models had a higher SD (61.5 g) compared to the MTSI (61.0 g) and AMMI (44.8 g) models (Supplementary Table 8). The difference in SD of GGE and WAASB with MTSI was minimal. Hence, if the criterion is an individual trait, GGE and WAASB can be considered the best models. However, for multiple traits, MTSI is a more suitable model for the selection of genotypes that exhibit both stability and high performance.
Additionally, a venn diagram (Fig. 10) was drawn to identify the common genotypes across the four stability indices: AMMI, GGE, WAASB and MTSI. The genotypes G1 and G8 were the common performers across all four stability indices, while G4 was a common performer in the GGE, WAASB and MTSI analysis (Supplementary Table 9).
Discussion
Chickpea is an important pulse crop, valued for its high protein and micronutrient content, which makes it especially important as a dietary staple in many regions. However, the productivity of chickpea, during the rabi season, is relatively low compared to cereal crops. This is largely due to limited genetic variability and the crop’s susceptibility to various biotic and abiotic stresses. To overcome these limitations, breeding programs have increasingly focused on utilizing wild Cicer species to broaden the genetic base and improve climate resilience3. In the current study, derivative lines from crosses of cultivated chickpea varieties BGD 72 and PBG 5 (C. arietinum) with wild Cicer species- C. reticulatum (ILWC 229) and C. echinospermum (ILWC 246) have been undertaken to access their genetic base and resilience to climate shifts. Environmental factors such as temperature, rainfall, soil conditions, light, and wind, play significant roles in determining crop yield, particularly for chickpea cultivation. These factors can influence flowering, pod development, nutrient availability, and the prevalence of pests and diseases, vital for maximizing yield potential32. Increasing crop productivity through plant breeding programs is essential to ensure the food security amid rising climate variabilities. Multi-environment trials, addressing GEI, allow breeders to identify genotypes with consistent performance across different conditions. Using appropriate statistical tools to analyze GEI is key to selection of stable and adaptable genotypes that can thrive in the face of biotic and abiotic challenges in target environments. In this context, AMMI, GGE, WAASB and MTSI models were employed to identify genotypes with high adaptability and stability. Hence, under varying environmental conditions yield-related traits like NPP, HSW and SYPP were accessed. It is a well-known fact that the comprehension and management of these factors is key to sustainable crop productivity and food security in the face of climate change and other environmental challenges.
The current study assessed a total of 21 chickpea genotypes (19 ISDs + 2 checks) across six different environments (ENV1 to ENV6), encompassing three locations: PAU-Ludhiana, CSSRI-Karnal and IIPR-Kanpur. The availability of trait variation is a fundamental requirement for any genetic enhancement program. Pooled ANOVA results across six environments revealed significant differences among the genotypes, indicating variable performance under different conditions. Additionally, the ENV1, ENV2, ENV3 environments displayed significant differences among themselves over the years, demonstrating the impact of changing environmental conditions on chickpea productivity. The presence of a significant GEI for all evaluated traits highlighted the influence of environmental factors on genotype performance across the tested environments. This interaction underscores the utility of identifying out-performing chickpea genotypes across diverse environments rather than being suited to specific conditions. These observations are consistent with findings from multiple other studies33,34,35,36,37, further reinforcing the notion that GEIs are key factors in influencing yield and related traits. This even highlighted the complexities involved in the evaluation of genotype performance across diverse conditions in chickpea. On comparative analysis of the mean performance of genotypes across all test environments, genotypes G1 (GL21-1) and G8 (GL21-54) were found to be superior as they exhibited the highest mean values for key traits studied (NPP, HSW and SYPP) (Supplementary Table 2). Consequently, GL21-1 and GL21-54 are the best-performing genotypes, with minimal GEI and wider adaptability across different environments. The ISDs GL21-1 and GL21-54 were derived from the crosses PBG5 × ILWC229 and BGD72 × ILWC229, respectively. Interestingly, ILWC229, an accession of C. reticulatum, served as the common donor in both crosses. These observations align with the findings of Vadithya et al.3, highlighting ILWC229 as a valuable source of yield-contributing traits for enhancing chickpea productivity. The wild species C. reticulatum (ILWC229) has proven to be a useful donor for improving yield stability in chickpea by contributing genes associated with stress tolerance, phenological flexibility, and better adaptability to varying environments. Similar outcomes were reported by Singh et al.13, wherein introgression from C. reticulatum led to improvements in seed size. Singh et al.11 demonstrated enhanced yield and resistance to both biotic and abiotic stresses in derivatives involving wild chickpea species. Together, these studies, along with the present findings, underline the value of ILWC229 as a rich source of useful alleles for broadening the genetic base and development of stable, high-performing chickpea varieties.
Knowledge of trait associations facilitates the formulation of effective breeding strategies for genetic enhancement. Traits that are positively correlated exhibit a strong response to selection, enabling simultaneous improvement. Although seed yield is influenced by multiple components, analyzing trait relationships can help enhance yield through the targeted selection of key traits. In the present study, the NPP and SYPP showed a strong positive correlation across environments (Fig. 2d), consistent with earlier reports3,38. This indicates that an increase in NPP is closely associated with higher seed yield, possibly due to shared genetic factors or tight linkage between genes controlling these traits30. In contrast, the weak correlation between NPP and HSW suggests that these traits are largely independent3. This implies that increasing pod number does not necessarily affect seed weight, and vice versa. The independence likely reflects distinct genetic and physiological controls—NPP is associated with reproductive traits, while HSW is governed by assimilate partitioning during seed development. This distinction allows for the simultaneous improvement of both traits through targeted selection, supporting the development of high-yielding chickpea genotypes with both high pod numbers and bold seeds.
Evaluation of a genotype’s performance for various traits and across the environments is crucial for ascertaining its suitability for cultivation21,39. However, this process is often complicated by the confounding effects of GEI and the additive influences that genotype and environment have on each other39,40. As an ideal crop variety needs to exhibit both stability and high yield across different environments, GEI poses challenges to plant breeders. However, selection based on stability alone is insufficient, as the most stable genotypes may not always provide the highest yield30,41. Hence, for the selection of genotypes, breeders require statistical tools to evaluate their stability and yield performance. To tackle this, models like AMMI and GGE have been developed to quantify GEI42. Significant results for GEI, along with the combined ANOVA and IPCs from the AMMI and GGE biplot, indicated the presence of crossover interactions, affecting the response and ranking of the genotypes across the different environments29,43. Hence the genotypes viz., G17 (GL21-80) and G14 (GL21-76) for NPP; G4 (GL21-37), G17 (GL21-80), G9 (GL21-56) and G7 (GL21-49) for HSW; and G9 (GL21-56), PBG8 and G17 (GL21-80) for SYPP were positioned close to the biplot origin, with IPCA1 scores close to zero (Fig. 3). This indicated the environment-insensitivity of these genotypes as they had stable mean values across the environments. In contrast, genotypes (environmentally sensitive) positioned farther from the biplot origin exhibited strong interactions in specific environments, as indicated by their higher positive or negative IPCA1 scores (Fig. 3). These observations aligned with the findings of Hinsta et al.33, Gebremedhin et al.34, Ojiewo et al.44, Singamsetti et al.43, Mekonnen et al.45, Madhusudhana et al.30 and Appunu et al.46, wherein the significant influence of environmental factors on yield variations in chickpea genotypes were reported. Several AMMI-based statistics have been proposed for the simultaneous selection of yield traits and stability. SSI, a combination of ASV and yield traits (Y), was calculated to evaluate and identify stable and superior genotypes (Table 4). Lower SSI values indicated higher stability and greater productivity of the genotypes30. Among 21 genotypes; G1 (GL21-1), G8 (GL21-54) and G17 (GL21-80) demonstrated high mean performance and stability across three yield traits studied.
The evaluation of a large number of genotypes without graphical representations of the data across different environmental conditions can be challenging while determining the patterns of genotypic responses. Similar to AMMI, GGE biplot is the graphical method for the simultaneous representation of genotypes and environments47. Therefore, the GGE biplot was utilized to illustrate the main effects of genotypes with GEIs in multi-environment data46,48. GGE biplot technique effectively identified the promising chickpea genotypes with high PC1 and low PC2 values. The polygon view of the ‘which-won-where’ pattern biplot enables the division of environments into distinct MEs and recommendation of suitable cultivars within each ME43,49. In ‘which-won-where’ pattern of present study, the genotypes specific to individual environments as well as to MEs were identified (Fig. 4). These results highlight the genotypes that are well-suited to specific environments and demonstrated their adaptability across multiple environments, aligning with the findings of Yan and Rajcan50 and Singamsetti et al.43. Furthermore, a two-dimensional visualization of yield and stability based on PC1 and PC2 scores in the biplot pattern was similarly reported by Yan et al.48 and Singamsetti et al.43. The GGE biplot analysis highlighted the test environment’s discriminative ability and representativeness based on vector length and angle. For traits; NPP, HSW and SYPP, the environments viz., ENV4, ENV3 and ENV6; ENV1, ENV3, ENV4 and ENV2 while ENV6, ENV1 and ENV3, respectively, displayed longer vectors (Fig. 5). This indicated a strong discriminative power of the test environments, implying their suitability for stability trials. ENV6 showed strong discriminative power, likely due to its clay loam soil and relatively higher temperatures. These conditions created a more stressful environment to distinguish genotypes more clearly based on their performance. According to Yan51 and Singamsetti et al.43, the angle between environmental vectors; explains the associations between them. In this study, all six environments displayed acute angles for each trait, indicating a strong positive correlation and high representativeness. This positive correlation implies the consistent performance of genotypes across test environments46,52. Further, the mean vs. stability biplot provided a clear depiction of genotype’s mean performance and their stability across environments. Genotypes with shorter projections toward the AEC arrow, exhibited superior mean performance with higher stability (Fig. 6). This positioning demonstrated above-average performance of these genotypes with less influence to environmental fluctuations. Hence, making them ideal candidates for environments where consistency is crucial. The similar trends were also stated in the findings by Shim et al.53 and Khan et al.54.
A recently proposed statistic, WAASB, meticulously integrates the stability measures from both AMMI and BLUP models. The weighted average of absolute scores (WAAS) has been utilized to evaluate genotype stability across different environments using linear mixed models20,43. The Y × WAAS biplot was divided into four quadrants (Fig. 7). Genotypes or environments positioned in quadrant I were identified as unstable, displaying high discrimination ability with trait values below the grand mean. Quadrant II included unstable genotypes having trait values above the grand mean, warranting particular attention due to their strong discrimination ability and high trait values. Quadrant III genotypes, although less productive than the grand mean, were stable due to their lower WAAS values. The environments in this quadrant were classified as low in productivity and discrimination ability. In contrast, genotypes in quadrant IV exhibited both high productivity and stability, with broad adaptability as indicated by their low WAAS values (Fig. 7)43. Simultaneous selection based on both mean trait values and stability was conducted using the WAASBY scores, assigning equal weight (50%) to both30. A higher WAASBY value reflected better genotype performance in terms of both trait value (Y) and stability (WAASB). On the joint interpretation of yield traits and stability, greater stability across environments was exhibited by different genotypes, with higher WAASBY scores and mean for yield traits (Fig. 8). However, AMMI, GGE, and WAASB can only analyze one trait at a time. This limits their usefulness in real breeding programs, where breeders need to improve several traits together, like yield, seed weight, and pod number. MTSI solves this problem by combining the performance and stability of multiple traits into one simple value using BLUPs, making it more efficient for the selection of genotypes with overall superiority.
The success of a cultivar proposal is greater when it emphasizes both the mean performance and stability of desirable yield traits. The MTSI analysis allowed effective interpretation by using genotype-ideotype distance as a selection criterion. In this study, a 15% selection intensity was applied to identify the top-performing genotypes based on their MTSI scores across all traits (Fig. 9a). This threshold was chosen following the recommendations of Olivoto et al.21 and Reddy et al.18, as 15% offers a good balance between selection pressure and the retention of genetic diversity. It enables the selection of superior genotypes without excessively narrowing the genetic base. Similar thresholds ranging from 10–20% have been reported in other multi-trait selection studies, depending on population size and breeding goals55,56,57. The selected genotypes G1 (GL21-1), G8 (GL21-54), and G4 (GL21-37) also showed stable performance in the ‘mean vs. stability’ biplot (Fig. 6). Additionally, genotype G17 (GL21-80), with an MTSI score of 2.44 and located near the selection threshold, showed potential for further evaluation. The concept and application of the MTSI method have been well documented by Olivoto et al.21 and Singamsetti et al.43. MTSI provides a significant advantage by enabling simultaneous selection for mean performance and stability across traits, and its effectiveness has been demonstrated in several studies55,58.
This study is the first in chickpea to integrate MTSI with WAASB, offering a novel and comprehensive approach to evaluate genotype(s) across traits and environments. This integrated framework would strengthen the reliability of selection decisions, offering breeders clearer, more practical insights for improving complex traits in the face of climate stress.
Conclusion
To identify novel sources with high yield potential, 21 chickpea genotypes (19 ISDs + 2 checks) were screened across multiple locations in two agro-climatic zones (5 and 6) of India. The magnitude of the GEI effects varied between AMMI and GGE models. AMMI biplots facilitated the analysis of specific GE interactions, whereas GGE biplots effectively captured both genotype and GEI effects across environments. WAASBY analysis identified genotypes with high mean performance and stability, while MTSI enabled effective multi-trait selection. Overall, GGE and WAASB were most effective for individual trait selection, whereas MTSI was superior when multiple traits were targeted simultaneously. Notably, genotypes G1 (GL21-1) and G8 (GL21-54) consistently showed stable and superior performance across all analytical models. These genotypes can be advanced as promising candidate varieties for enhancing chickpea productivity and may serve as valuable parental lines in breeding programs for the development of improved chickpea cultivars to meet the demands of farmers under changing environmental conditions.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Abbreviations
- ISDs:
-
Interspecific derivatives
- GEIs:
-
Genotype × environment interactions
- AMMI:
-
Additive main effects and multiplicative interaction
- GGE:
-
Genotype main effect + genotype × environment interaction
- WAASB:
-
Weighted average of absolute scores for the BLUPs (best linear unbiased predictions)
- MTSI:
-
Multi-trait stability index
- ANOVA:
-
Analysis of variance
- PCA:
-
Principal component analysis
- PAU:
-
Punjab Agricultural University
- CSSRI:
-
Central Soil Salinity Research Institute
- IIPR:
-
Indian Institute of Pulses Research
- RCBD:
-
Randomized complete block design
- NPP:
-
Number of pods per plant HSW:100-seed weight SYPP: Seed yield per plot
- IPC:
-
Interaction Principal Component
- ASV:
-
AMMI Stability Value
- SSI:
-
Simultaneous Selection Index
- Y:
-
Yield trait
- SVD:
-
Singular Value Decomposition
- WAAS:
-
Weighted Average of Absolute Scores
- CENV:
-
Combined environments
- METs:
-
Multi-environment trials
- ME:
-
Mega-environment
- AEC:
-
Average environment coordinate
- SD:
-
Selection differential
References
Varshney, R. K. et al. Draft genome sequence of Chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246 (2013).
Van der Maesen, L. J. G. Origin, history and taxonomy of Chickpea. In The Chickpea (eds Saxena, M. C. & Singh, K. B.) 11–34 (CAB International, 1987).
Vadithya, A. S. et al. Evaluation and identification of advanced inter-specific derivatives from crosses of Cicer arietinum with C. reticulatum and C. echinospermum for agro-morphological, quality traits and disease resistance. Front. Plant Sci. 15, 1461280. https://doi.org/10.3389/fpls.2024.1461280 (2024).
Jukanti, A. K., Gaur, P. M., Gowda, C. L. L. & Chibbar, R. N. Nutritional quality and health benefits of Chickpea (Cicer arietinum L.): a review. Br. J. Nutr. 108, 11–26 (2012).
FAO. Statistical Database of the United Nations Food and Agriculture Organization (FAO) Statistical Division, Rome. Available online at: (2024). https://www.fao.org/faostat/en/#data/QCL (Accessed 26 August 2024).
Toker, C. et al. Cicer turcicum: A new Cicer species and its potential to improve Chickpea. Front. Plant Sci. 12, 662891 (2021).
Von Wettberg, E. J. et al. Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nat. Commun. 9, 649 (2018).
Bassiri, A., Ahmad, F. & Slinkard, A. E. Pollen grain germination and pollen tube growth following in vivo and in vitro self and interspecific pollinations in annual Cicer species. Euphytica 36, 667–675 (1987).
Ahmad, F., Slinkard, A. E. & Scoles, G. J. Investigations into the barrier(s) to interspecific hybridization between Cicer arietinum L. and eight other annual Cicer species. Plant. Breed. 100, 93–98 (1988).
Singh, K. B. & Ocampo, B. Exploitation of wild Cicer species for yield improvement in Chickpea. Theor. Appl. Genet. 95, 418–423 (1997).
Singh, A., Singh, I., Bindra, S., Singh, M. & Singh, P. Assessment of genetic diversity in interspecific derivatives of Chickpea (Cicer arietinum L). Agricultural Res. J. 61, 70–74. https://doi.org/10.5958/2395-146X.2024.00010.7 (2024).
Canci, H. & Toker, C. Evaluation of annual wild Cicer species for drought and heat resistance under field conditions. Genet. Resour. Crop Evol. 56, 1–6 (2009).
Singh, M. et al. Identification of promising Chickpea interspecific derivatives for agro-morphological and major biotic traits. Front. Plant Sci. 13, 1035878. https://doi.org/10.3389/fpls.2022.941372 (2022).
Oladosu, Y. et al. Principle and application of plant mutagenesis in crop improvement: a review. Biotechnol. Biotechnol. Equip. 30, 1–16. https://doi.org/10.1080/13102818.2015.1087333 (2016).
Karimizadeh, R. et al. GGE biplot analysis of yield stability in multi-environment trials of lentil genotypes under rainfed condition. Notulae Scientia Biologicae. 5, 256–262. https://doi.org/10.15835/nsb529067 (2013).
Oladosu, Y. et al. Genotype × environment interaction and stability analyses of yield and yield components of established and mutant rice genotypes tested in multiple locations in Malaysia. Acta Agriculturae Scand. Sect. B—Soil Plant. Sci. 67, 590–606. https://doi.org/10.1080/09064710.2017.1321138 (2017).
Zobel, R. W., Wright, M. J. & Gauch, H. G. Statistical analysis of a yield trial. Agron. J. 80, 388–393. https://doi.org/10.2134/AGRONJ1988.00021962008000030002X (1988).
Reddy, S. S. et al. Spatio-temporal evaluation of drought adaptation in wheat revealed NDVI and MTSI as powerful tools for selecting tolerant genotypes. Field Crops Res. 311, 109367. https://doi.org/10.1016/j.fcr.2024.109367 (2024).
Yan, W. & Kang, M. S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists (in CRC, 2002).
Olivoto, T. et al. Mean performance and stability in multi-environment trials I: combining features of AMMI and BLUP techniques. Agron. J. 111, 2949–2960. https://doi.org/10.2134/agronj2019.03.0220 (2019a).
Olivoto, T., Lúcio, A. D., da Silva, J. A., Sari, B. G. & Diel, M. I. Mean performance and stability in multi-environment trials II: selection based on multiple traits. Agron. J. 111, 2961–2969. https://doi.org/10.2134/agronj2019.03.0221 (2019b).
Singh, M., Rani, S., Malhotra, N., Katna, G. & Sarker, A. Transgressive segregations for agronomic improvement using interspecific crosses between C. arietinum L.× C. reticulatum ladiz. And C. arietinum L.× C. echinospermum Davis species. PloS One. 13, e0203082. https://doi.org/10.1371/journal.pone.0203082 (2018).
R Core Team. R: a Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020).
Bates, D., Mächler, M., Bolker, B. M. & Walker, S. C. Fitting linear mixed-efects models using lme4. J. Stat. Softw. 67, 1–48. https://doi.org/10.18637/jss.v067.i01 (2015).
Alvarado, G. et al. META-R (Multi Environment Trail Analysis with R for Windows) Version 6.04. CIMMYT Research Data & Software Repository Network, V23, 2015. https://hdl.handle.net/11529/10201
William, R. Psych: Procedures for Psychological, Psychometric, and Personality Research (Northwestern University, 2023). https://CRAN.R-project.org/package=psych
Gauch, H. G. & Zobel, R. W. Predictive and postdictive success of statistical analyses of yield trials. Theor. Appl. Genet. 76, 1–10. https://doi.org/10.1007/BF00288824 (1988).
Purchase, J. L., Hatting, H. & Van Deventer, C. S. Genotype× environment interaction of winter wheat (Triticum aestivum L.) in South africa: II. Stability analysis of yield performance. South. Afr. J. Plant. Soil. 17, 101–107. https://doi.org/10.1080/02571862.2000.10634878 (2000).
Yan, W. & Tinker, N. A. Biplot analysis of multi-environment trial data: principles and applications. Can. J. Plant Sci. 86, 623–645. https://doi.org/10.4141/P05-169 (2006).
Madhusudhana, R. et al. Genetic variability, G× E interaction and stability for iron and zinc content in sorghum grains in advanced breeding lines. J. Cereal Sci. 110, 103653. https://doi.org/10.1016/j.jcs.2023.103653 (2023).
Olivoto, T. & Lúcio, A. D. C. Metan: an R package for multi-environment trial analysis. Methods Ecol. Evol. 11, 783–789. https://doi.org/10.1111/2041-210X.13384 (2020).
Sharma, S. et al. Meta-analysis of QTLs for drought, heat, and salinity tolerance identifies high-confidence MQTLs and candidate genes in Chickpea (Cicer arietinum L). J. Biosci. 50, 19. https://doi.org/10.1007/s12038-025-00499-2 (2025).
Hinsta, G., Hailemariam, A. & Belay, T. Genotype-by-environment interaction and grain yield stability of early maturing bread wheat (Triticum aestivum L) genotypes in the drought prone areas of Tigray region, Northern Ethiopia. Ethiop. J. Appl. Sci. Technol. 2, 51–57 (2011).
Gebremedhin, W., Firew, M. & Tesfye, B. Stability analysis of food barley genotypes in Northern Ethiopia. Afr. Crop Sci. J. 22, 145–153 (2014).
Erdemcı, İ. Investigation of genotype× environment interaction in Chickpea genotypes using AMMI and GGE biplot analysis. Turkish J. Field Crops. 23, 20–26 (2018).
Pouresmael, M. et al. Stability of Chickpea (Cicer arietinum L.) landraces in National plant gene bank of Iran for drylands. J. Agricultural Sci. Technol. 20, 387–400 (2018).
Karimizadeh, R., Pezeshkpour, P., Mirzaee, A., Barzali, M., Sharifi, P., Khoshkhoy Nilash, E. A., … Safari Motlagh, M. R. (2023). Identification of stable chickpeas under dryland conditions by mixed models. Legume Science, 5, e206.
Johnson, P. L., Sharma, R. N. & Nanda, H. C. Genetic diversity and association analysis for yield traits in Chickpea (Cicer arietinum L.) under rice-based cropping system. Bioscan 10, 879–884 (2015).
Ansari, A. A., Akhatar, J., Sharma, S., Banga, S. S. & Atri, C. Integrating multiple statistical indices to measure the stability of photosynthetic pigment content and composition in Brassica juncea (L.) Czern germplasm under varying environmental conditions. Photosynth. Res. 162, 63–74. https://doi.org/10.1007/s11120-024-01116-3 (2024).
Crossa, J. et al. Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. https://doi.org/10.1016/j.tplants.2017.08.011 (2017).
Gebeyehu, C., Bulti, T., Dagnachew, L. & Kebede, D. Additive main effect and multiplicative interactions (AMMI) and regression analysis in sorghum [Sorghum bicolor (L). Moench] varieties. J. Appl. Biosci. 136, 13877–13886 (2019).
Gauch, H. G. A simple protocol for AMMI analysis of yield trials. Crop Sci. 53, 1860–1869. https://doi.org/10.2135/cropsci2013.04.0241 (2013).
Singamsetti, A., Shahi, J. P., Zaidi, P. H., Seetharam, K., Vinayan, M. T., Kumar,M., … Madankar, K. (2021). Genotype × environment interaction and selection of maize(Zea mays L.) hybrids across moisture regimes. Field Crops Research, 270, 108224. https://doi.org/10.1016/j.fcr.2021.108224.
Ojiewo, C., Monyo, E., Desmae, H., Boukar, O., Mukankusi-Mugisha, C., Thudi, M., …Varshney, R. K. (2019). Genomics, genetics and breeding of tropical legumes for better livelihoods of smallholder farmers. Plant Breeding, 138, 487–499.
Mekonnen, T. W., Mekbib, F., Amsalu, B., Gedil, M. & Labuschagne, M. Genotype by environment interaction and grain yield stability of drought tolerant Cowpea landraces in Ethiopia. Euphytica 218, 57 (2022).
Appunu, C. et al. Evaluation of sugarcane genotypes (Saccharum sp. hybrid) for multi-trait stability analysis across diverse environments. Industrial Crops and Products 219, 118993. https://doi.org/10.1016/j.indcrop.2024.118993 (2024).
Gabriel, K. R. The biplot graphic display of matrices with application to principal component analysis. Biometrika 58, 453–467. https://doi.org/10.1093/biomet/58.3.453 (1971).
Yan, W., Cornelius, P. L., Crossa, J. & Hunt, L. Two types of GGE biplots for analyzing multi-environment trial data. Crop Sci. 41, 656–663. https://doi.org/10.2135/cropsci2001.413656x (2001).
Vaezi, B. et al. Integrating different stability models to investigate genotype x environment interactions and identify stable and high-yielding barley genotypes. Euphytica 215, 63. https://doi.org/10.1007/s10681-019-2386-5 (2019).
Yan, W. & Rajcan, I. Biplot analysis of test sites and trait relations of soybean in Ontario. Crop Sci. 42, 11–20. https://doi.org/10.2135/cropsci2002.1100 (2002).
Yan, W. Singular value partitioning in biplot analysis of multi-environment trial data. Agron. J. 94, 990–996. https://doi.org/10.2134/agronj2002.0990 (2002).
Sserumaga, J. P. et al. Genotype by environment interactions and agronomic performance of doubled haploids testcross maize (Zea mays L.) hybrids. Euphytica 207, 353–365. https://doi.org/10.1007/s10681-015-1549-2 (2016).
Shim, K. B. et al. Interpretation of genotype × environment interaction of Sesame yield using GGE biplot analysis. Korean J. Crop Sci. 60, 349–354 (2015).
Khan, M. M. H. et al. DNA fingerprinting, fixation-index (Fst), and admixture mapping of selected Bambara groundnut (Vigna subterranea [L.] verdc) accessions using ISSR markers system. Sci. Rep. 11, 14527. https://doi.org/10.1038/s41598-021-93867-5 (2021).
Kendal, E. Comparing durum wheat cultivars by genotype x yield x trait and genotype x trait biplot method. Chil. J. Agricultural Res. 79, 512–522. https://doi.org/10.4067/S0718-58392019000400512 (2019).
Bocianowski, J., Niemann, J. & Nowosad, K. Genotype-by-environment interaction for seed quality traits in interspecific cross-derived Brassica lines using additive main effects and multiplicative interaction model. Euphytica 215, 7. https://doi.org/10.1007/s10681-018-2328-7 (2019).
Koundinya, A. V. V. et al. Genetic parameters, stability and selection of cassava genotypes between rainy and water stress conditions using AMMI, WAAS, BLUP and MTSI. Scientia Hortic. 281, 109949. https://doi.org/10.1016/j.scienta.2021.109949 (2021).
Kizilgeci, F., Albayrak, O. & Yildirim, M. Evaluation of thirteen durum wheat (Triticum durum desf.) genotypes suitable for multiple environments using GGE biplot analysis. Fresenius Environ. Bull. 28, 6873–6882 (2019).
Acknowledgements
The authors extend their gratitude to all cooperating partners, including PAU, Ludhiana; CSSRI, Karnal; IIPR, Kanpur and all contributing staff for their essential support in conducting field trials and contributing to this study.
Funding
This work was supported by ICAR projects “Molecular genetic analysis of resistance/tolerance to different stress in Rice, Wheat, Chickpea and Mustard including sheath blight complex genomics Chickpea Component” and “Molecular breeding for improvement of tolerance to biotic and abiotic stresses, yield and quality traits in crops- Chickpea”.
Author information
Authors and Affiliations
Contributions
ASV was responsible for data curation, formal analysis, investigation, methodology, software, validation, visualization, writing the original draft, and review and editing. SB contributed to conceptualization, funding acquisition, investigation, methodology, supervision, validation, and writing the original draft, as well as review and editing. NS handled data curation, investigation, and review and editing. SKS focused on investigation and methodology. SPS also contributed to investigation and methodology. IS participated in investigation and review and editing. CB was involved in conceptualization and investigation. MS provided resources and worked on methodology. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vadithya, A.S., Bindra, S., Sharma, N. et al. Multi-model statistical approaches for assessing the stability of Cicer interspecific derivatives in the trans and upper gangetic regions of India. Sci Rep 15, 22230 (2025). https://doi.org/10.1038/s41598-025-06886-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06886-x