Abstract
High-grade B-cell lymphoma with concurrent MYC and BCL2/BCL6 rearrangements (HGBL-DHL) is a challenging disease resistant to front-line immunochemotherapies, which urgently requires novel therapeutic approaches. Herein, combination of chidamide and anlotinib demonstrated potential synergistic anti-lymphoma effects against HGBL-DHL. The cooperative effect of cell proliferation inhibition, apoptosis induction, and cell cycle arrest were demonstrated in cell lines through Cell Counting Kit-8, Annexin V/PI staining, and PI staining respectively. Moreover, in an HGBL-DHL-xenografted mouse model, the combination therapy markedly reduced tumor burden without inducing fatal toxicity. Mechanistically, chidamide suppressed HDAC3, while anlotinib inhibited VEGFR2, both leading to down-regulation of the PI3K/AKT signaling cascade. This dual suppression occurred as a result of their synergistic interaction. Downstream targets of the PI3K/AKT signaling pathway including proliferation-associated protein c-Myc, anti-apoptotic proteins BCL2 and MCL1, and cell cycle-associated protein cyclin A2 were all synergistically down-regulated, which coincided with the phenotypic outcomes. Collectively, our preclinical findings underscored the synergistic activity of chidamide and anlotinib combination against HGBL-DHL, warranting further clinical evaluation of this regimen for treating this challenging entity.
Similar content being viewed by others
Introduction
According to the World Health Organization (WHO) classification, high-grade B-cell lymphoma (HGBL) is a distinct category that includes aggressive mature B-cell neoplasms such as double-hit lymphoma (DHL) and triple hit lymphoma (THL)1,2. Among these subtypes, HGBL-DHL is particularly challenging and exhibits morphological, immunophenotypic, and genetic features that lie between those of Burkitt’s lymphoma (BL) and diffuse large B-cell lymphoma (DLBCL). Despite therapeutic advancements, HGBL-DHL continues to be associated with a poor prognosis, often resulting in treatment failure even with front-line immunochemotherapy3,4,5. Specifically, when treated with the R-CHOP regimen (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone), patients with HGBL-DHL harboring concurrent MYC and BCL2 rearrangements have a worse prognosis compared to those with DLBCL6,7,8,9. A meta-analysis10 revealed that, compared to R-CHOP, intensified chemotherapy regimens such as cyclophosphamide, vincristine, doxorubicin, high-dose methotrexate alternating with ifosfamide, etoposide, and high-dose cytarabine (CODOX-M/IVAC), or cyclophosphamide, vincristine, doxorubicin, dexamethasone, cytarabine combined with methotrexate (HyperCVAD) did not improve the survival of HGBL-DHL patients. Moreover, even with stem cell transplantation, which is regarded as an underlying curative choice for this entity, the prognosis for HGBL-DHL patients has not substantially improved, with a median overall survival of less than 2 years11. Thus, there is a compelling need to develop new therapeutic methods tailored to address the unique challenges posed by HGBL-DHL.
Histone deacetylases (HDACs) act as epigenetic regulators by removing acetyl groups from histones and other proteins, thereby influencing the transcription of various genes, including those involved in apoptosis and cell cycle regulation. HDAC1, HDAC2, and HDAC3 are substantially overexpressed in patients with DLBCL compared to healthy controls. Additionally, DLBCL samples frequently exhibit alterations such as gene amplification and/or mRNA overexpression of HDAC3, indicating that abnormal HDAC expression is a common feature in DLBC12. Notably, in B-cell lymphomas harboring CREB (cAMP Response Element-Binding Protein) binding protein mutations, HDAC3 forms a deacetylase complex with BCL6 to maintain the deacetylated state of gene enhancer regions, thereby promoting tumorigenesis13. As an innovative histone deacetylase inhibitor (HDACi), chidamide can selectively target HDAC1,2,3 and 10. Previous studies have demonstrated its potential efficacy and tolerability in relapsed/refractory DLBCL (R/R DLBCL)14 and THL15 when used in combination with chemotherapy and other targeted agents. However, many R/R DLBCL patients show poor clinical response to chidamide, with resistance being common16. These observations suggest that while chidamide may exhibit activity against HGBL-DHL, its therapeutic benefit might be limited due to unfavorable genetic features such as MYC and BCL2/BCL6 rearrangements, which exacerbate the treatment challenges associated with this disease. Therefore, we aim to identify other targeted drugs that can enhance the efficacy of chidamide and prevent disease progression in HGBL-DHL patients.
In DLBCL, the expression of vascular endothelial growth factor receptor 2 (VEGFR2) in lymphoma cells is associated with reduced overall survival (OS). Moreover, phosphorylated VEGFR2 expression correlates with shorter progression-free survival and tends to be related to shorter OS in DLBCL patients treated R-CHOP17. Anlotinib, a potent multi-tyrosine kinase inhibitor, effectively targets c-kit, platelet-derived growth factor receptor (PDGFR), and vascular endothelial growth factor receptor (VEGFR). Accordingly, anlotinib combined with rituximab, gemcitabine, and oxaliplatin has demonstrated promising efficacy and well-tolerated safety features in R/R DLBCL18. Given the adverse prognostic impact of VEGFR2 in DLBCL and the efficacy of anlotinib in R/R DLBCL, anlotinib may emerge as a potent therapeutic option to enhance the efficacy of chidamide in HGBL-DHL patients.
Phosphatidylinositol 3-kinases (PI3Ks) are a family of crucial lipid kinases that are widely expressed in mammalian cells. The Phosphatidylinositol 3-kinase/Protein Kinase B (PI3K/AKT) pathway is intricately involved in cell growth, survival, apoptosis, and cell cycle progression, highlighting its multifaceted role in the pathogenesis and progression of DLBCL. Dysregulation of this pathway is frequently observed in DLBCL, contributing to uncontrolled cell proliferation, resistance to apoptosis, and enhanced angiogenesis19,20. Aberrant activation of the PI3K/AKT pathway leads to increased expression of BCL2, thereby promoting cell survival in DLBCL21. In DHL, there is a close correlation between PI3K/AKT pathway activation and elevated cell proliferation rates22. Conversely, inhibition of this pathway substantially reduces cell viability and induces apoptosis23. These observations underscore the critical role of the PI3K/AKT pathway in DHL and its potential as a therapeutic target.
Mechanistically, chidamide can downregulate the PI3K/AKT signaling pathway in DLBCL cells, leading to the inhibition of cell proliferation and the induction of apoptosis12. In ovarian carcinoma cells, overexpression of HDAC3 substantially upregulates the expression levels of PI3K, phospho-PI3K, AKT, and phospho-AKT, whereas HDAC3 knockout markedly decreases expression levels of these proteins24. Both in DLBCL25 and BL26, HDAC3 knockdown has been shown to reduce the phosphorylation level of AKT. Regarding anlotinib, it exhibits a potential anti-leukemic effect in both Philadelphia chromosome-negative and -positive B-cell acute lymphoblastic leukemia by blocking the PI3K/AKT cascade. Additionally, in intrahepatic cholangiocarcinoma, anlotinib suppresses tumor progression by disrupting the VEGFR2/PI3K/AKT signaling pathway27.
Given the dual inhibitory effects of chidamide and anlotinib on the PI3K/AKT signaling pathway, we aimed to explore the combined effects and underlying molecular mechanisms of these two agents in HGBL-DHL. Herein, we demonstrated that anlotinib substantially enhanced the cytotoxic effects of chidamide both in vitro and in vivo. This synergistic effect was observed in human cell lines and a cell-derived xenograft (CDX) mouse model of HGBL-DHL, and was achieved through dual blockade of the PI3K/AKT cascade.
Methods
Reagents and cell lines
Chidamide (chidamide; HBI-8000) was purchased from Shenzhen Chipscreen Biosciences (Shenzhen, China). Anlotinib was provided by Chia Tai Tianqing Pharmaceutical Group Co. Ltd (Nanjing, China). For in vitro experiments, stock solutions were obtained by dissolving each drug respectively in sterile DMSO (Sigma, MO, USA), both at a concentration of 50 mM and stored at − 20 °C. For oral gavage, chidamide solution was produced by suspending powder in 0.5% (w/v) CMC-Na, and anlotinib solution was acquired by suspending stock solution in 0.5% (w/v) CMC-Na by ultrasonic processing.
TMD8, Toledo, MCA, and LR cells, all of which were HGBL-DHL cell lines, were obtained from MD Anderson Cancer Center Laboratory. The incubator condition was set at 37 °C with 5% CO2. TMD8, MCA, and LR cell lines was cultured in RPMI-1640 (Hyclone, Thermo Scientific, Logan, UT, USA) adding 10% fetal bovine serum (Gibco, Thermo Scientific, Grand Island, NY, USA). Toledo cell line was cultured in RPMI-1640 (Gibco, Thermo Scientific, Grand Island, NY, USA) adding 10% fetal bovine serum (Superculture, Dakewe Biotech., China). Both culture solutions were supplemented with 100 units/ml penicillin and 100 mg/ml Streptomycin.
Cell viability assessment
Cell Counting Kit-8 (CCK-8, APExBIO, Texas, USA) was adopted for cell viability analysis. Briefly, TMD8, Toledo, MCA, and LR cells were cultured in 100 µl medium at 2 × 105 cells/ml in 96-well plates with DMSO, chidamide, anlotinib, or combination of chidamide and anlotinib for 24–48 h. For TMD8 and Toledo cells, we set the concentrations of chidamide as 0 µM, 1 µM, 2 µM, 4 µM, 8 µM and 16 µM. The concentration of anlotinib was fixed at 10 µM. For MCA and LR cells, the concentration of chidamide were both set as 0.25, 0.5, 1, and 2 µM, the concentration of anlotinib were both set as 0.5, 1, 2, and 4 µM. Then we added 10 µl CCK-8 reagent per well and incubated for an additional 2 h. Bio-Rad microplate reader (Bio-Rad, CA, USA) was utilized to detect absorbance at 450 nm. The inhibition rate was calculated according to the following formula (1):
The fraction affected (Fa) and the drug combination index (CI) for each treatment group were computed using CompuSyn software, and the Fa-CI curve was subsequently generated.
Apoptosis analysis via flow cytometry
For apoptosis assessment, TMD8 and Toledo cells were exposed to DMSO, chidamide, anlotinib, or chidamide combined with anlotinib for 24–48 h. Then we collected and washed the cells with ice-cold phosphate-buffered saline (PBS; Gibco BRL, Rockville, MD, USA). The harvested cells were then stained with APC Annexin V (556454, BD Bioscience, CA, USA) and FITC Annexin V Apoptosis Detection Kit (4201822 ,BD Bioscience, CA, USA) under dark and room temperature for 15 min by following the manufacturer’s instructions. Lastly, the processed samples were detected within 1 h after staining via flow cytometry (Quanteon, ACEA Biosciences, USA). In the dot plot, Annexin V-positive cells in the right quadrant are indicative of apoptosis.
Cell cycle analysis via flow cytometry
After being starved in an FBS-free medium for 24 h, 2 × 105 TMD8 cells per well were treated with DMSO, 2 µM chidamide, 1.6 µM anlotinib or 2 µM chidamide combined with 1.6 µM anlotinib in complete medium for 24 h. 2 × 105 Toledo cells were also starved and treated in the same condition as TMD8 cells, except that chidamide concentration was 750 nM, and anlotinib concentration was 1.6 µM. After treatment, cells were collected and washed with ice-cold phosphate-buffered saline (PBS; Gibco BRL, Rockville, MD, USA). 70% ethanol was utilized to fix the cells at 4 °C overnight. Then we resuspended fixed cells in PBS supplemented with RNase and stained cells with propidium iodide (PI) for 30 min under dark. The cell cycle was analyzed via flow cytometry (Quanteon, ACEA Biosciences, USA).
Network pharmacological analysis
Firstly, we downloaded 3D structure of chidamide and anlotinib from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). PharmMapper (https://lilabecust.cn/.
pharmmapper/) was used to identify a large number of possible targets for chidamide and anlotinib. Chidamide and anlotinib target gene sets were obtained after gene symbol annotation by utilizing Uniprot (https://www.uniprot.org/). Then, we tried to search OMIM, Phenopedia, and TTD databases using “Double hit lymphoma” and “High-grade B-cell lymphoma” as keywords to explore potential disease-related targets. However, no related results were obtained. Thus, we only applied the drug target gene sets to build the PPI network through the STRING database. The parameter was set at 0.400 which represented moderate confidence. Next, we imported the PPI network into Cytoscape 3.9.1 to evaluate the vital network. ClusterProfler GO and clusterProfler KEGG of R 4.1.2 were applied to perform GO and KEGG enrichment analysis28 after obtaining the core targets. Both software packages can be obtained from the website (https://www.bioconductor.org/). GO enrichment analysis primarily investigates the biological processes, cellular components, and molecular functions of the targets, while KEGG enrichment assesses the potential biological pathways and functions associated with the targets. For filtering, an adjusted P-value < 0.05 and a Q-value < 0.05 were adopted.
RNA-sequencing
TMD8 cells were treated with 375 nM chidamide, 1 µM anlotinib, or 375 nM chidamide combined with 1 µM anlotinib for 24 h, and then first lysed with TRIzol reagent (Invitrogen, NY, USA) and further extracted following the manufacturer’s guidelines. We constructed next-generation sequencing libraries by performing sample quality control, mRNA isolation, mRNA fragmentation, cDNA synthesis, end repair, add A and adaptor ligation, PCR, library quality control, and circularization in turn. Finally, we performed sequencing through combinatorial Probe-Anchor Synthesis (cPAS) in accordance with the manufacturer’s recommendations. The Dr.TOM II online interface was employed to analyze the data. Sequentially, Data filtering, RNA identification, gene quantification differential expression analysis were carried out for subsequent gene annotation. To observe phenotypic change, we adopted differential genes that had been annotated to carry out GO (http://www.geneontology.org/) and KEGG (https://www.kegg.jp/) enrichment analysis29 by Phyper based on the Hypergeometric test. Terms and pathways with Q value ≤ 0.05 were deemed as substantially enriched. Detailed RNA sequencing protocols for each step were demonstrated in supplementary document 1. In this RNA-seq experiment, we conducted three biological replicates.
Western blot analysis
1 × 106 TMD8 and Toledo cells were inoculated onto a 6 cm cell culture dish. TMD8 cells were treated with DMSO, 1.5 µM chidamide, 1.6 µM anlotinib, and 1.5 µM chidamide combined with 1.6 µM anlotinib for 24 h. Toledo cells were treated with DMSO, 375 nM chidamide, 1.6 µM anlotinib, and 375 nM chidamide combined with 1.6 µM anlotinib for 24 h. Then TMD8 and Toledo cells were harvested by centrifugation at 300 g for 5 min 4 °C. Each sample was lysed with 200 µL RIPA Lysis buffer (Thermo, USA) for 10 min on ice. Whole-cell extracts were obtained by collecting the supernatant after centrifugation at 9500 rpm for 10 min at 4°C, which was subsequently subjected to electrophoresis on 8% or 10% gels and blotted onto PVDF membranes (Millipore, Billerica, MA, USA). 5% nonfat milk in TBST solution was applied as a blocking agent for the transblotted membranes which were treated with specific primary antibodies, followed by incubation with HRP-conjugated anti-rabbit secondary antibodies (HuaAn Biotechnology, Zhejiang, China) or HRP Goat anti-mouse secondary antibodies (ABclonal Technology, Hubei, China). The primary antibodies against VEGFR2 (9698 S) and CyclinA2 (4656T) were purchased from Cell Signaling Technology (Danvers, Massachusetts, USA). The primary antibodies against HDAC3 (A19537), PI3K85α(A4992), phospho-PI3KP85α/P55γ/P85β-Y467/Y199/.
Y464 (AP0854), c-Myc (A9032) nd phospho-VEGFR2-Y1175 (AP0382) were adopted from ABclonal Technology (Hubei, China). The primary antibodies against PI3K110δ (ET1705-46), AKT1 (ET1609-47), phospho-AKT1 (Ser473) (ET1607-73), MCL-1 (ET1606-14), and BCL-2 (ET1702-53) were purchased from HuaAn Biotechnology (Zhejiang, China). Antibodies against GAPDH (HA721136, 1:10000, HuaAn Biotechnology, China) was applied as a loading control. Protein visualization was achieved by utilizing ECL Western Blotting Detection Kit (Gene-Flow, Staffordshire, UK) and Bio-Rad ChemiDoc XRS + detection system (Bio-Rad, CA, USA). Under the same experimental conditions, we performed three replicate western blot experiments.
Plasmids and transfection
The pCMV-HDAC3 plasmid and pCMV-VEGFR2 plasmid were obtained from Miaoling Biological Engineering Co., Ltd. (Shanghai, China). In each well, approximately 5 × 10⁵ TMD8 and Toledo cells are plated in 4 mL of growth medium and cultured for 24 h prior to transfection. Subsequently, 4 µg and 12 µg of DNA are dissolved in 400 µL of Opti-MEM media (Gibco, Thermo Scientific, Grand Island, NY, USA) for TMD8 and Toledo cells, respectively. Subject the Lipofectamine™ 3000 (Thermo Fisher Scientific, Waltham, MA) to a brief vortex, then add 2 µL to the diluted DNA and mix immediately by pipetting or vortexing. Allow the mixture to incubate at room temperature for 15 min. 400 µL of the Lipofectamine™ 3000/DNA mixture drop-wise was added to each well immediately after 1.5 µM chidamide and 10 µM anlotinib were added. After the addition of the transfection reagent mixture, immediately gently agitate the plate to evenly distribute the complexes. Maintain the culture at 37 °Cin a CO2 incubator for 24 h. Then cell viability assessment and apoptosis analysis via flow cytometry were performed according to procedures recorded above.
In-vivo efficacy of chidamide combined with anlotinib in HGBL-DHL cell-derived xenograft mouse models
The CB17-SCID mice (8–10 weeks old) applied in the current study were acquired from Xiamen University Animal Care and raised in aseptic conditions. The Animal Care and Use Committee and Ethics Committee of Xiamen University approved all animal investigations, and we carried out all animal experiments complying with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. For HGBL-DHL cell-derived xenograft models, mice were injected subcutaneously with 2 × 107 TMD8 cells after exposure to a 1.5-Gy total body irradiation dose. 9 Days after injection, we randomly divided mice into four distinct groups (n = 10) and consecutively administrated mice for 7 days: (i) control group (0.5% Carboxymethylcellulose sodium); (ii) chidamide group (oral gavage, d10-11 10 mg/kg/d, d12 12 mg/kg/d, d13-16 15 mg/kg/d ); (iii) anlotinib group (oral gavage, d10-11 10 mg/kg/d, d12-16 8 mg/kg/d); (iv) combination of chidamide (dose same as group ii) and anlotinib (dose same as group iii) group. Daily measurements of tumor sizes and body weights were conducted, and tumor volumes were determined using the specified Eq. (2) displayed below. Specifically, “L” represented the greatest longitudinal diameter and “W” was defined as the greatest transverse diameter, both of which were measured by caliper. On the 17th day, five mice per group were euthanized with CO2, and the tumors were collected, weighed, and then assigned into two parts: one was fixed with 4% paraformaldehyde for immunohistochemistry (IHC) and hematoxylin and eosin (HE) staining, and the remaining part was frozen at − 80 °C for western blot. Primary antibodies targeting HDAC3 (A19537, 1:100, ABclonal, Wuhan, China) and VEGFR2 ( 9698 S, 1:800, CellSignalingTechnology, Boston, USA) were dripped on tumor slides, which were then incubated overnight at 4 °C. Then, DAB (ZLI-9018, ZSGB Biotechnologies, Beijing, China) was applied at room temperature and incubated until the microscope slide displayed a color change. Moreover, to investigate the potential toxicity of the drug on the liver and kidney, we conducted HE staining using liver and kidney tissues sections from normal tumor- free mice with the same dosing regimen. Both IHC and HE analyses were performed under a fluorescence microscope (CX23, OLYMPUS, Tokyo, Japan). The remaining five CDX mice per group continued to be administrated with previous therapeutic regimens and monitored for survival analysis. Tumor volume exceeding 1500 mm3 was deemed as end point.
Statistical analyses
Experiments were executed in triplicate across three independent replicates, with values shown as mean ± SD. Prism software v6.0 (GraphPad Software, La Jolla, CA, USA) was adopted for statistical analyses. For comparisons between two experimental conditions, student’s t-test was applied. For analyses involving multiple groups, two-way ANOVA was utilized. Statistical significance was set at P < 0.05. “*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001” are used to denote the significance levels for all statistical results presented in the manuscript.
All methods were performed in accordance with the relevant guidelines and regulations.
Results
Chidamide combined with anlotinib synergistically inhibit HGBL-DHL by proliferation inhibition, apoptosis and cell cycle arrest inducement in vitro
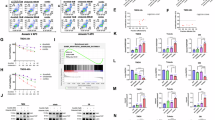
Herein, we detected the combined effects of chidamide and anlotinib on the viability, apoptosis, and cell cycle progression of HGBL-DHL cell lines including TMD8, Toledo, MCA and LR. Initially, chidamide treatment alone resulted in a dose-dependent attenuation of cell viability. Interestingly, the addition of anlotinib substantially enhanced the inhibitory impact of chidamide on cell proliferation in all cell lines, both after exposed for 24 h (Fig. 1a, Supplementary Fig. 1a) and 48 h (Supplementary Fig. 1b, 1c). The combination index (CI) values assessed via the inhibition rate of drug treatment were mostly less than 1.0 for 24 h (Fig. 1a), except for two specific cases for TMD8 cells. Meanwhile, chidamide treatment at a series of concentrations induced remarkable apoptosis in a concentration-dependent manner. Anlotinib could substantially enhance the apoptosis induction of chidamide, both after exposed for 24 h (Fig. 1b, c) and 48 h (Supplementary Fig. 1d, e). For instance, the addition of anlotinib (1.6 µM) can obviously increase the apoptosis rate of chidamide (375 nM) from 25.03 to 42.9% in TMD8 cell after 24 h. The combination achieved an effect approximately 1.7-fold higher. In Toledo cell, apoptosis rate increased by 337.9% in the combination (chidamide at 12 µM, anlotinib at 2.5 µM) group compared to anlotinib monotherapy.
Previous reports have demonstrated that genes that play important parts in cell cycle regulation can be impaired by aberrant HDAC expression30,31. Thus, chidamide may exert cytotoxicity on HGBL-DHL cells partly by inducing cell cycle arrest. Therefore, we further examined chidamide-mediated cytotoxicity and whether anlotinib would potentiate the effect in cell cycle arrest. As demonstrated in Fig. 1d, e, the percentage of cells in the S phase were substantially reduced in both TMD8 and Toledo cells. The percentage of cells in the G1 phase was substantially increased in TMD8 cells by combination treatment (Fig. 1d). Figure 1e demonstrated that in comparison with the control group, chidamide or anlotinib alone, combination treatment for 24 h substantially elevated the proportion of cells in the G2 phase in Toledo cells.
Thus, chidamide combined with anlotinib exerted a potent anti-lymphoma effect by synergistically inhibiting proliferation, inducing apoptosis, and cell cycle arrest in TMD8, Toledo, MCA or LR cells.
Chidamide and anlotinib combination exerted a synergistically anti-lymphoma effect on HGBL-DHL in vitro. (a) The proliferation and combination index (CI) values was assessed after treating human TMD8 and Toledo cells with chidamide and anlotinib for 24 h. (b, c) Typical flow cytometric image for Annexin V/PI staining (b left column, c left column) and percentages of apoptosis (b right column, c right column) in TMD8 and Toledo cells were measured. (d, e) Percent of TMD8 (d) and Toledo cells (e) arrest in G1, S, and G2 after treatment with chidamide and anlotinib for 24 h. Values were indicated as mean ± SD of at least three independent experiments investigated in triplicate. Two-way ANOVA was utilized. Statistical significance was set at P < 0.05. “*P < 0.05, **P < 0.01, ***P < 0.001, and ****P <.0001” are used to denote the significance levels.
Chidamide combined with anlotinib mostly affects the PI3K/AKT signaling pathway in silico and in transcriptional level in vitro
To predict the potential targets and joint pathways of chidamide and anlotinib we performed network pharmacology study in silico, attempting to select enriched pathways for further confirmation through molecular biology experiments. By utilizing PharmMapper, 475 potential drug targets of chidamide and anlotinib were obtained. Then, we visualized the compound-target interaction network by using Cytoscape3.9.1 (Supplementary Fig. 2a). Generally, one gene was targeted by at least one drug, while one drug could target more than one gene. Interactions between targets were also presented. Lastly, we acquired 20 substantially KEGG pathways (Fig. 2a, Supplementary Fig. 2b) and 14 substantially enriched GO terms (Supplementary Fig. 2c, d) by clusterProfler KEGG and ClusterProfler GO respectively. Specifically, the KEGG pathway enrichment results marked the PI3K/AKT signaling pathway as the most enriched pathway as demonstrated in the bubble plot of the most substantial 20 KEGG pathways (Fig. 2a).
Accordingly, next, we verified these in silico enrichment results by a high-throughput RNA sequencing analysis in TMD8 cells. Firstly, as shown in Fig. 2B and Supplementary Table 1, chidamide combined with anlotinib synergistically down-regulated transcripts encoding PI3K/AKT signaling pathway effectors, including AKT1, proliferation-associated gene MYC, anti-apoptotic gene BCL2 (Fig. 2b). A substantial decline of MCL1 mRNA expression was also observed in the combination group by contrast to anlotnib alone (Supplementary Table 1).
As for pathway enrichment analysis, differential genes between control and combination groups were mainly enriched in the cell cycle-related pathway (column plot: Fig. 2c, bubble plot: Supplementary Fig. 3a) by KEGG pathway enrichment analysis. So were the differential genes between anlotinib and combination groups (column plot: Fig. 2d, bubble plot: Supplementary Fig. 3b), or between chidamide and combination groups (column plot: Supplementary Fig. 3c, bubble plot: Supplementary Fig. 3d). Differential genes between anlotinib and combination groups were also enriched in the apoptosis-related pathway (column plot: Fig. 2d, bubble plot: Supplementary Fig. 3b). GO Biological process enrichment analysis showed that both differential genes between anlotinib versus combination groups (Supplementary Fig. 3e, f) and between chidamide versus combination groups (Supplementary Fig. 3g, h) were enriched in cell cycle-related pathways. In addition, KEGG pathway classification (Fig. 2e) and GO pathway classification analysis (Fig. 2f) showed that differential genes between the control and chidamide group were mainly enriched in pathways related to transcription and transcription regulator activity, which coincided with the epigenetic regulation effect of chidamide.
Chidamide showed joint effects with anlotinib on PI3K/AKT associated signaling pathways in silico and at the transcriptional level. (a) Bubble plot of the 20 most substantial KEGG terms. Number of the enriched genes was represented as counts, and the ratio of enriched genes to all target genes was presented as gene ratio. (b) Fold change of mRNA levels of PI3K/AKT downstream genes between different treatment groups. A, C, and Com represent Anlotinib, Chidamide, and Combination respectively. (c, d) KEGG pathway enrichment analysis displayed differential signaling pathways between different drug treatment groups and substantially enriched genes involved in the cell cycle (c, d) and apoptosis (d) in the combination group. (e, f) KEGG pathway classification (e) and GO pathway classification analysis (f) unveiled a chidamide-mediated effect on differential genes enriched in pathways related to transcription regulation. mRNA expression levels were indicated as mean ± SD of three biological replicates. Two-way ANOVA was utilized. Statistical significance was set at P < 0.05. “*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001” are used to denote the significance levels.
Downregulation of HDAC3 and VEGFR2 mediated by chidamide and anlotinib synergistically induces PI3K/AKT signaling pathway inhibition, proliferation inhibition, and apoptosis in vitro
Protein levels of PI3K/AKT signaling pathway effectors were assessed to further verify the inhibitive effect of chidamide combined with anlotinib in TMD8 and Toledo cells. Specifically, we chose to examine the abundance of proliferation-associated protein c-Myc, anti-apoptotic proteins BCL2 and MCL1, and cell cycle-associated protein cyclin A2, all of which were also downstream genes of the PI3K/AKT signaling pathway. Cyclin A2 is known to peak and function in the S phase, which was the vital decreasing phase in the combination group both in TMD8 and Toledo cells. Western blotting was carried out after TMD8 and Toledo cells were treated with DMSO, and chidamide and/or anlotinib for 24 h. The results showed that chidamide and anlotinib combination synergistically reduced the protein level of PI3K110δ, PI3K p85α,phosphor-PI3KP85α/P55γ/P85β-Y467/Y199/.
Y464, AKT1, phosphor-AKT1 (Ser473) and the downstream gene regulated by PI3K/AKT axis, including MCL-1, BCL-2, c-Myc and Cyclin A2 (Fig. 3a). As reported in previous research, HDAC3 seemed to regulate PI3K/AKT signaling pathway in ovarian carcinoma cells24, anlotinib suppressed proliferation, migration, and invasion of intrahepatic cholangiocarcinoma via blocking the VEGFR2/PI3K/AKT cascade32. Thus, we further detected the HDAC3, VEGFR2, and phospho-VEGFR2-Y1175 protein expressions and found that these three proteins were synergistically reduced in the combination treatment. Herein, we speculated that chidamide inhibited HDAC3 protein level, and anlotinib suppressed VEGFR2 and phosphor-VEGFR2-Y1175 protein abundance, which might contribute to dual inhibition of PI3K/AKT signaling pathway.
To further confirm the contribution of HDAC3 and VEGFR2 inhibition to proliferation inhibition and apoptosis induced by chidamide and anlotinib combination, TMD8 and Toledo cells were exposed to 1.5 µM chidamide combined with 10 µM anlotinib and then transfected with the PCMV-HDAC3, PCMV-VEGFR2 or empty vector. Proliferation inhibition induced by chidamide and anlotinib was rescued by the expression of HDAC3 and VEGFR2 in TMD8 and Toledo cells (Fig. 3b). Meanwhile, flow cytometric analysis showed that apoptosis was also rescued (Fig. 3c). Taken together, these results suggest that chidamide and anlotinib exerted an anti-lymphoma effect at least partly by down-regulation of HDAC3 and VEGFR2.
Chidamide and anlotinib mediated anti-lymphoma effect by HDAC3 and VEGFR2 inhibition and subsequent PI3K/AKT blockage. (a) Differential protein levels of relative genes in TMD8 and Toledo cells after chidamide and anlotinib treatment were determined by western blot. (b, c) Rescued expression of HDAC3 and VEGFR2 impaired increased proliferation inhibition (b) and induced apoptotic phenotypes (c) by chidamide and anlotinib in TMD8 and Toledo cells. Values were indicated as mean ± SD of at least three independent experiments investigated in triplicate. Two-way ANOVA was utilized. Statistical significance was set at P < 0.05. “*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001” are used to denote the significance levels.
Chidamide combined with anlotinib synergistically inhibits tumor growth in a HGBL-DHL xenograft mouse model
Thrilled by the favorable in vitro outcomes shown above, we performed further in vivo studies to confirm the synergetic anti-lymphoma efficacy of chidamide combined with anlotinib in a mouse xenograft model bearing TMD8 cells. Firstly, after sublethally radiated for 1.5 Gy, mice were subcutaneously injected with TMD8 cells (2 × 107) on the right side. After 9 days, mice bearing tumors were randomly separated into four groups: vehicle, chidamide, anlotinib, and combination treatment groups. Vehicle or drugs were orally given continuously daily for seven days according to the protocol demonstrated in Fig. 4a. During the course of treatment, we measured the tumor volume of mice daily to evaluate therapeutic effects. After drug treatment for 7 days, we euthanized 20 mice in all and collected tumors from 5 mice per group. Volume, photograph, and weight of tumors were demonstrated in Fig. 4b, c, and d, respectively. Herein, we observed that either chidamide or anlotinib monotherapy could substantially suppress the in vivo proliferation of TMD8 cells. Moreover, the inhibition tendency was further enhanced in the combination group by contrast to chidamide or anlotinib alone (Fig. 4b). Then, tumor weights were dramatically alleviated in the combination therapy compared with the vehicle and chidamide groups, except that the tumor weight decline between the combination group and anlotinib monotherapy was not statistically substantial (Fig. 4d), which could be compensated by the substantial tumor volume variances between these two groups (Fig. 4b). Both chidamide and anlotinib alone substantially prolonged survival and the combination group further enhanced the survival benefit (Fig. 4e). Besides, no substantial body weight differences were observed among the four groups (Fig. 4f). Accordingly, the histopathology analysis of mouse tumor tissues after 7 days of treatment was performed. HE staining of tumor tissues (Fig. 4g) taken from the vehicle group displayed abundant tumor cells and vigorous tumor growth, tumor tissues taken from the chidamide and anlotinib monotherapy group showed diminished tumor cell density plus more necrosis. As for chidamide combined with the anlotinib group, we found minimal tumor cell density and most necrosis, which verified the synergetic effect again. Liver and kidney tissues collected from normal tumor-free mice were subjected to sectioning and HE staining. The results demonstrated that chidamide and anlotinib did not exhibit obvious hepatotoxicity or nephrotoxicity (Supplementary Fig. 4). Altogether, these results argued strongly that the therapy combining chidamide and anlotinib was remarkably synergistically active and tolerable for the treatment of HGBL-DHL in vivo.
Chidamide and anlotinib combination synergistically leads to tumor growth inhibition in xenograft mouse models. (a) Cell transfer protocol in a HGBL-DHL tumor xenograft model. (b, c) Tumor volumes (b) and tumor images (c) of mice were monitored daily. (d) Tumor weights from TMD8-bearing xenograft mice after respective treatments (n = 5). Tumor volumes and weights were indicated as mean ± S.D to investigate the synergistic effects of chidamide and anlotinib. (e) Kaplan-Meier assessment of overall survival in mice harboring tumor xenografts. (f) Body weights of mice were indicated as mean ± S.D to investigate the toxicity of chidamide and anlotinib. (g) HE staining of tumor tissue slices. Tumor samples collected from the vehicle, chidamide, anlotinib, and combination group were fixed, sliced and HE stained. Images were acquired under an OLYMPUS fluorescence microscope (original magnification,×200). Two-way ANOVA was utilized. Statistical significance was set at P < 0.05. “*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001” are used to denote the significance levels.
Inhibition of HDAC3 and VEGFR2 induced by chidamide and anlotinib dually led to PI3K/AKT signaling pathway inhibition in vivo
To ascertain the in-vivo inhibitive effect of chidamide and anlotinib on HDAC3 and VEGFR2 protein expression respectively, immunohistochemical (IHC) staining (Fig. 5a, b) and western blot experiments (Fig. 5c) were performed to assess the expression of HDAC3 and VEFGR2 in tumor tissues of CDX mice. Compared with the vehicle and monotherapy group, the combination treatment group demonstrated substantially fewer HDAC3 positive tumor cells (upper row of Fig. 5a, right column of Fig. 5b) and VEGFR2 positive vascular endothelial cells (lower row of Fig. 5a marked by the red box, left column of Fig. 5b). Western blot experiments based on tumor tissue of CDX mice were carried out to testify the protein abundance of HDAC3, VEGFR2, phosphor-VEGFR2-Y1175, PI3K p85α, phosphor-PI3KP85α/P55γ/P85β-Y467/Y199/.
Y464, AKT1, phospho-AKT1(Ser473) and the downstream genes regulated by PI3K/AKT axis, such as MCL-1, BCL-2, c-Myc and Cyclin A2. As shown in Fig. 5c, chidamide combined with anlotinib markedly suppressed the expression of these proteins compared to the other three groups.
Chidamide and anlotinib synergistically suppressed the PI3K/AKT cascade in vivo by inhibiting HDAC3 and VEGFR2 respectively. (a) Chidamide combined with anlotinib synergistically inhibited HDAC3 and VEGFR2 expression in vivo. Tumor slices were immunohistochemically incubated with HDAC3 (upper row of a) and VEGFR2 ( lower row of a) antibodies. (b) Percent of HDAC3 positive tumor cells (left column) was calculated with ImageJ Software. VEGFR2 positive vascules (right column) were counted. Both were then used for statistical analysis. Values were indicated as mean ± SD of three independent experiments. Two-way ANOVA was utilized. Statistical significance was set at P < 0.05. “*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001” are used to denote the significance levels. (c) Protein levels of HDAC3, VEGFR2 and PI3K/AKT cascade of tumor tissues were determined by western blot.
Discussion
HGBL-DHL represents a particularly challenging subtype, often refractory and resistant to front-line immunochemotherapies. This underscores the urgent need to explore novel therapeutic strategies. In this study, we demonstrated that the combination of chidamide and anlotinib exhibited synergistic activity in HGBL-DHL, which was correlated with the dual downregulation of the PI3K/AKT signaling pathway via HDAC3 and VEGFR2 inhibition. Specifically, the protein levels of downstream targets of the PI3K/AKT signaling pathway, including c-Myc, MCL-1, BCL-2, and cyclin A2, were downregulated both in vitro and in vivo, ultimately leading to proliferation inhibition, apoptosis, and cell cycle arrest (Fig. 6).
Resistance to chidamide in R/R DLBCL has been associated with elevated expression of CREBBP and subsequent increased stability of Aurora Kinase A (AURKA)16. The phosphorylation, nuclear translocation, and trans-activating activity of CREB are dependent on VEGFR2 signaling in endothelial cells. VEGFR2 inhibitors can fully prevent CREB activation33. Moreover, anlotinib treatment has been shown to substantially reduce AURKA protein levels in ovarian cancer cells34. The regulatory effects of anlotinib on the targets that induce resistance to chidamide may represent parts of the pharmacological mechanisms underlying its enhancement of chidamide therapeutic efficacy against HGBL-DHL.
According to the drug instruction, recommended dosing regimen for chidamide in patients with MYC and BCL2 positive DLBCL is 20 mg on days 1, 4, 8, and 11 for a 3-week cycle. The dosing regimen of anlotinib for solid tumors is 12 mg once daily for 2 weeks, followed by a 1-week break with a 3-week cycle. Based on the pharmacokinetic characteristics of chidamide and anlotinib, we preliminarily recommend exploring the administration of these agents according to the two regimens proposed by drug instruction. Specifically, after one single dose, chidamide reaches its peak concentration at 4 h after the first dose. As its concentration gradually declines, anlotinib begins to reach its peak concentration at 6–11 h. As chidamide is constantly being cleared with a average terminal half-life (T1/2) of 17 h, anlotinib exerts a continuous exposure with a T1/2 of 95–116 h. On day 11, chidamide is temporarily discontinued, while anlotinib continues to be administered until day 14, when the plasma concentration reaches its peak after continuous dosing. Thus, anlotinib compensates for the exposure decline and dosing gap of chidamide. Both drugs have a large apparent volume of distribution and are widely distributed in the body, which ensures sustained release into blood and efficacy prolongation. Moreover, both drugs exhibit high plasma protein binding rates, which can reach approximately 90%. Their combined use facilitates competition for plasma protein binding, which can increase the free concentration of the other drug, thereby enhancing therapeutic efficacy.
Based on the dosing regimens of chidamide and anlotinib above, the synergistic pharmacological effects and complementary pharmacokinetic advantages of this combination regimen ensure the maximization of clinical efficacy when the two drugs are used in combination, and lay a theoretical and pre-clinical experimental foundation for conducting clinical trials of this regimen for the treatment of HGBL-DHL.
Nevertheless, several unresolved issues warrant further investigation in future studies. In the present research, we have not elucidated the underlying mechanisms by which HDAC3 and VEGFR2 regulate the PI3K/AKT signaling cascade in HGBL-DHL. Thus far, we have only demonstrated the preclinical efficacy and safety of the combination of chidamide and anlotinib. Their effects on primary HGBL-DHL cells and in clinical settings remain to be determined. Therefore, additional cellular and molecular studies, as well as clinical trials are essential to address these unresolved questions.
In conclusion, this study provides compelling evidence that the PI3K/AKT signaling pathway is a promising therapeutic target for HGBL-DHL. The dual inhibition of PI3K/AKT by the combination of chidamide and anlotinib effectively targets HGBL-DHL without inducing fatal toxicity, thus representing a potential anti-lymphoma strategy. The favorable preclinical efficacy and safety profile of this combination regimen underscore the necessity to further evaluate its clinical potential in the management of HGBL-DHL.
Data availability
Data will be made available upon request to Bing Xu. The datasets generated and/or analysed during the current study are available in the GEO repository: Series GSE292250, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE292250.
References
Swerdlow, S. H. et al. The 2016 revision of the world health organization classification of lymphoid neoplasms. Blood 127, 2375–2390. https://doi.org/10.1182/blood-2016-01-643569 (2016).
Sesques, P. & Johnson, N. A. Approach to the diagnosis and treatment of high-grade B-cell lymphomas with MYC and BCL2 and/or BCL6 rearrangements. Blood 129, 280–288. https://doi.org/10.1182/blood-2016-02-636316 (2017).
Sarkozy, C., Traverse-Glehen, A. & Coiffier, B. Double-hit and double-protein-expression lymphomas: Aggressive and refractory lymphomas. Lancet Oncol. 16, e555–e567. https://doi.org/10.1016/S1470-2045(15)00005-4 (2015).
Scott, D. W. et al. High-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with diffuse large B-cell lymphoma morphology. Blood 131, 2060–2064. https://doi.org/10.1182/blood-2017-12-820605 (2018).
Riedell, P. A. & Smith, S. M. Double hit and double expressors in lymphoma: Definition and treatment. Cancer 124, 4622–4632. https://doi.org/10.1002/cncr.31646 (2018).
Horn, H. et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood 121, 2253–2263. https://doi.org/10.1182/blood-2012-06-435842 (2013).
Johnson, N. A. et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J. Clin. Oncol. 30, 3452–3459. https://doi.org/10.1200/Jco.2011.41.0985 (2012).
Green, T. M. et al. Immunohistochemical Double-Hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J. Clin. Oncol. 30, 3460–3467. https://doi.org/10.1200/Jco.2011.41.4342 (2012).
Hu, S. M. et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: A report from the international DLBCL rituximab-CHOP consortium program. Blood 121, 4021–4031. https://doi.org/10.1182/blood-2012-10-460063 (2013).
Howlett, C. et al. Front-line, dose-escalated immunochemotherapy is associated with a significant progression-free survival advantage in patients with double-hit lymphomas: A systematic review and meta-analysis. Br. J. Haematol. 170, 504–514. https://doi.org/10.1111/bjh.13463 (2015).
Petrich, A. M. et al. Impact of induction regimen and stem cell transplantation on outcomes in double-hit lymphoma: A multicenter retrospective analysis. Blood 124, 2354–2361. https://doi.org/10.1182/blood-2014-05-578963 (2014).
Wu, C., Song, Q., Gao, S. & Wu, S. Targeting HDACs for diffuse large B-cell lymphoma therapy. Sci. Rep. 14, 289. https://doi.org/10.1038/s41598-023-50956-x (2024).
He, M. Y. et al. GNAS knockout potentiates HDAC3 Inhibition through viral mimicry-related interferon responses in lymphoma. Leukemia 38, 2210–2224. https://doi.org/10.1038/s41375-024-02325-4 (2024).
Li, Q., Huang, J., Ou, Y., Li, Y. & Wu, Y. Progressive diffuse large B-cell lymphoma with TP53 gene mutation treated with chidamide-based chemotherapy. Immunotherapy 11, 265–272. https://doi.org/10.2217/imt-2018-0083 (2019).
Kang, J. et al. Modified conditioning regimen with chidamide and high-dose rituximab for triple-hit lymphoma. J. Cell. Mol. Med. 25, 10770–10773. https://doi.org/10.1111/jcmm.16999 (2021).
Sun, Y. et al. CREBBP cooperates with the cell cycle machinery to attenuate chidamide sensitivity in relapsed/refractory diffuse large B-cell lymphoma. Cancer Lett. 521, 268–280. https://doi.org/10.1016/j.canlet.2021.09.002 (2021).
Gratzinger, D. et al. Lymphoma cell VEGFR2 expression detected by immunohistochemistry predicts poor overall survival in diffuse large B cell lymphoma treated with immunochemotherapy (R-CHOP). Br. J. Haematol. 148, 235–244. https://doi.org/10.1111/j.1365-2141.2009.07942.x (2010).
Wu, M. et al. Spatial multi-omics treatment paradigms in a phase II clinical study of early relapsed/refractory diffuse large B-cell lymphoma. Blood 144, 2996–2996. https://doi.org/10.1182/blood-2024-203388 (2024).
Wu, C. et al. Chidamide and orelabrutinib synergistically induce cell cycle arrest and apoptosis in diffuse large B-cell lymphoma by regulating the PI3K/AKT/mTOR pathway. J. Cancer Res. Clin. Oncol. 150, 98. https://doi.org/10.1007/s00432-024-05615-7 (2024).
Wang, J. et al. AKT hyperactivation and the potential of AKT-targeted therapy in diffuse large B-cell lymphoma. Am. J. Pathol. 187, 1700–1716. https://doi.org/10.1016/j.ajpath.2017.04.009 (2017).
Xiaojie Liang, J. G., Luo, B., Lu, W., Chen, Q. & Deng, Y. Refining prognostic assessment of diffuse large B-cell lymphoma: Insights from multi-omics and single-cell analysis unveil SRM as a key target for regulating immunotherapy. J. Big Data12, 1–25 (2025).
Gao, G. C. et al. Single-cell RNA sequencing in double-hit lymphoma: IMPDH2 induces the progression of lymphoma by activating the PI3K/AKT/mTOR signaling pathway. Int. Immunopharmacol. 125. https://doi.org/10.1016/j.intimp.2023.111125(2023).
Pan, G. C. et al. Orelabrutinib and venetoclax synergistically induce cell death in double-hit https://doi.org/10.1007/s00432-022-04473-5lymphoma by interfering with the crosstalk between the PI3K/AKT and p38/MAPK signaling. J. Cancer Res. Clin. 149, 5513–5529. (2023).
Lou, T., Y Zhuang, H., D Liu, C. & Y Zhang, Z. HDAC3 positively regulates HE4 expression to promote ovarian carcinoma progression. Arch. Biochem. Biophys. 675. https://doi.org/10.1016/j.abb.2019.07.009 (2019).
Gupta, M. et al. Inhibition of histone deacetylase overcomes rapamycin-mediated resistance in diffuse large B-cell lymphoma by inhibiting Akt signaling through mTORC2. Blood 114, 2926–2935. https://doi.org/10.1182/blood-2009-05-220889 (2009).
Dong, L. H. et al. Histone deacetylase inhibitor potentiated the ability of MTOR inhibitor to induce autophagic cell death in Burkitt leukemia/lymphoma. J. Hematol. Oncol. 6, 53. https://doi.org/10.1186/1756-8722-6-53 (2013).
Song, F. et al. Anlotinib suppresses tumor progression via blocking the VEGFR2/PI3K/AKT cascade in intrahepatic cholangiocarcinoma. Cell Death Dis 11. https://doi.org/10.1038/s41419-020-02749-7 (2020).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Wei, H. et al. KDELR2 promotes breast cancer proliferation via HDAC3-mediated cell cycle progression. Cancer Commun. (Lond). 41, 904–920. https://doi.org/10.1002/cac2.12180 (2021).
Jang, Y. G., Hwang, K. A. & Choi, K. C. Rosmarinic acid, a component of rosemary tea, induced the cell cycle arrest and apoptosis through modulation of HDAC2 expression in prostate cancer cell lines. Nutrients 10. https://doi.org/10.3390/nu10111784 (2018).
Song, F. et al. Anlotinib suppresses tumor progression via blocking the VEGFR2/PI3K/AKT cascade in intrahepatic cholangiocarcinoma. Cell. Death Dis. 11, 573. https://doi.org/10.1038/s41419-020-02749-7 (2020).
Corsini, M. et al. Cyclic adenosine monophosphate-response element-binding protein mediates the proangiogenic or Proinflammatory activity of Gremlin. Arterioscler. Thromb. Vasc Biol. 34, 136–145. https://doi.org/10.1161/ATVBAHA.113.302517 (2014).
Wang, H. & Wang, Y. Anlotinib induces apoptosis and second growth/mitosis phase block in cisplatin-resistant ovarian cancer cells via the aurora kinase A/p53 pathway. Hum. Exp. Toxicol. 42, 9603271231185774. https://doi.org/10.1177/09603271231185774 (2023).
Funding
This research was funded by the National Natural Science Foundation of China (U22A2090 and 82170180 granted for Bing Xu; 82470187 granted for Jie Zha), the Natural Science Foundation of Fujian Province (2023J06054 granted for Jie Zha), Xiamen Municipal Bureau of Science and Technology (3502Z20234001 granted for Jie Zha; 3502Z20244015 granted for Bing Xu), Young Investigator Research Program of Xiang’an Hospital of Xiamen University (XM02070002 granted for Jiazhen Lin), Open Innovation Fund for undergraduate students of Xiamen University (KFJJ-202410 granted for Bozhang Chen) and XMU Training Program of Innovation and Enterpreneurship for Undergraduates (granted for Mingxin Zhuang), Xiamen Medical and Health Guidance Project (3502Z20224ZD1014 granted for Xinguo Zhuang), the Natural Scientific Foundation of Xiamen (3502Z20227340 granted for Xinguo Zhuang), and the Fujian Natural Science Foundation of China (2022J011372 granted for Xinguo Zhuang).
Author information
Authors and Affiliations
Contributions
Conceptualization, X.B., Z.J. and Y.L.; Data curation, L.J.Z. and Z.X.G.; Formal analysis, J.S.M. and Q.D.M.; Funding acquisition, X.B. and Z.J.; Investigation, Z.J.; Methodology, L.J.Z. and Z.X.G. ; Resources, X.B., Z.J. and Y.L.; Software, L.J.Z. and J.S.M.; Supervision, X.B. and Z.J.; Validation, L.J.Z., Z.X.G. and J.S.M. Writing—original draft, L.J.Z.; Writing—review and editing, Z.H. and L.J.Z. All authors have read and agreed to the published version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The animal study was performed in line with ARRIVE guidelines. Approval of animal experiment was granted by the Animal Care and Use Committee and Ethics Committee of Xiamen University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, J., Zhuang, X., Jia, S. et al. Chidamide and anlotinib synergistically inhibit high grade B-cell lymphomas via PI3K/AKT signaling pathway. Sci Rep 15, 29526 (2025). https://doi.org/10.1038/s41598-025-10334-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10334-1