Abstract
Due to high resistance to medicines, multidrug-resistant (MDR) bacterial pathogens, particularly MRSA (methicillin-resistant Staphylococcus aureus) and VRE (vancomycin-resistant enterococci), are a significant public health concern for treating nosocomial infections. Researchers are developing novel compounds responding to the global rise in MDR infections. This study aimed to extract, purify, and characterize bioactive metabolites from Streptomyces levis strain HFM-2, a human gut isolate, exhibiting strong antimicrobial activity against several MDR pathogenic bacteria and fungal phytopathogens. Ethyl acetate extract of S. levis strain HFM-2 was purified using silica-gel column chromatography and reverse-phase high-performance liquid chromatography. Structure elucidation of the purified antimicrobial compound was done by performing detailed analyses including MS, IR, and NMR. The bacteriostatic activity of the compound revealed interesting values against broad-spectrum MDR pathogens. The bacterial cell destruction was recorded through SEM and fluorescence microscopy analyses. HFM-2P is displayed to be non-mutagenic and non-cytotoxic to the normal cell line. However, dose-dependent cytotoxicity was observed against the HeLa cancer cell line and exhibited antimutagenic activity against Salmonella Typhimurium strains (TA98 and TA100). This study is the first to report antiproliferative, DNA protective potential, antimutagenic properties, and antimicrobial activity of a 2,6-disubstituted chromone derivative isolated from S. levis strain HFM-2 against drug-resistant MRSA, VRE, and fungal phytopathogens. Therefore, this essential compound could be a candidate for future research in the pharmaceutical and agricultural sectors.
Similar content being viewed by others
Introduction
The emergence of multidrug-resistant pathogenic bacteria is a major concern in the healthcare system. They are responsible for the development of serious infectious diseases in humans, causing substantial health risks, especially in the aged and immunocompromised individuals, resulting in significant mortality and morbidity1,2,3,4. The life-threatening and most widespread antibiotic-resistant bacteria are methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococci, multidrug-resistant Pseudomonas aeruginosa, Klebsiella pneumoniae, and Extended-Spectrum Beta-Lactamase producing bacteria (ESBL)1,5,6. According to Peterson and Kaur2, these bacteria have been rapidly growing resistant to all antibiotic medicines currently available in the market, including macrolides, aminoglycosides, fluoroquinolones, and vancomycin. Antibiotic usage in humans and food animals has gradually raised antibiotic resistance in a variety of microorganisms7,8,9. The rise of drug-resistant disease strains exceeds the development of novel medications and treatments. To discover new antibiotics, many scientists and pharmaceutical corporations have been actively involved in isolating and screening actinobacteria from diverse unscreened environments10. The naturally occurring compounds are found to be excellent sources of novel antibacterial agents that have been effectively employed for protecting human health against infectious illnesses11. Finding novel actinobacterial strains with antimicrobial properties through unexplored habitat selection is an interesting approach12,13. Actinobacteria are the major family of microorganisms, producing 39% of all the known natural antibiotics. They have structural complexity and a varied range of biological activities. Streptomyces is the most significant genus and attractive resource among actinobacteria, having the biggest number of species and the potential to synthesize around 80% of bioactive metabolites (compounds) like antibiotics14,15,16.
In this study, an antimicrobial compound was purified and characterized from S. levis strain HFM-2. The purified compound (HFM-2P) exhibited strong antimicrobial activity against various bacterial pathogens, with notable efficacy against VRE and MRSA. Scanning electron microscopy (SEM) and fluorescence microscopy analyses revealed that HFM-2P induced cell deformities and leakage of intracellular contents in the tested pathogens. The biosafety of HFM-2P was assessed using the Ames assay and DNA nicking assay. In addition, HFM-2P demonstrated antiproliferative activity against the HeLa cancer cell line.
Material and methods
Test pathogens
Different test pathogenic bacteria, Klebsiella pneumoniae sub sp. pneumoniae (MTCC 109), Escherichia coli (MTCC 1885), Staphylococcus aureus (MTCC 96), Staphylococcus epidermidis (MTCC 435), Enterobacter aerogenes (MTCC 111), Bacillus subtilis (MTCC 619), and fungal phytopathogens viz. Fusarium oxysporum (MTCC 284), Alternaria brassicicola (MTCC 2102), and Cladosporium herbarum (MTCC 351) were obtained from the Microbial Type Culture Collection (MTCC) and GenBank, CSIR-Institute of Microbial Technology (IMTECH), Chandigarh, India. Colletotrichum gleosporoides is a lab isolate. MRSA (resistant to imipenem, methicillin, and clindamycin), VRE (resistant to methicillin, clindamycin, vancomycin, and imipenem), and Escherichia coli S1-LF (resistant to cefoperazone, cefotaxime, rifampicin, ciprofloxacin, and clindamycin) were collected from resident hospitals. The bacterial and fungal cultures were maintained at 4ºC on nutrient agar (NA) and potato dextrose agar (PDA) slants, respectively.
Fermentation and recovery of antimicrobial metabolites
S. levis strain HFM-2 produced antibiotic metabolites as described by Verma et al.17. Following inoculation of the production medium with Streptomyces culture, fermentation was carried out in Erlenmeyer flasks on a rotary shaker at 180 rpm. For the development of the seed culture, a 7-day-old Streptomyces culture was inoculated in 100 mL of SCNB (starch casein nitrate broth, pH 6). After 24 h, the inoculum (2%) was aseptically transferred to Erlenmeyer flasks (250 mL) containing the same seed medium (50 mL) and cultured for five days at 28 °C at 180 rpm. After fermentation, the culture broth was centrifuged at 10,000×g for 20 min at 4 °C to separate the cell-free supernatant. Active metabolites from the cell-free supernatant were recovered using ethyl acetate.
Purification of the antimicrobial compound from the S. levis strain HFM-2
The Ethyl acetate (EtOAc) extract was subjected to column chromatography using silica gel (60–120 mesh) to purify the antimicrobial compound. The sample was eluted stepwise using linear gradients of chloroform: ethyl acetate at a flow rate of 2 mL/min after pre-equilibration of the column with chloroform. Approximately 200 mL was eluted with each solvent system, and a total of 70 fractions were collected, each containing 25 mL. All fractions were concentrated and re-dissolved using the same solvent ratio. Antimicrobial activity was assessed for each fraction using the disc diffusion method, followed by evaluation through thin-layer chromatography (TLC) with ethyl acetate and chloroform (8:2, v/v) as the solvent system. The air-dried TLC plates were removed and visualized under UV light and stained with iodine. The pooled active fractions, showing antibacterial activity against MRSA, were further subjected to preparative Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC) (Shimadzu Microsorb MV, 100 mm × 10 mm ID, 10 µm) at a flow rate of 2 mL/min, using acetonitrile: water as the mobile phase and UV detection at 290 nm to achieve the final purification of the active component. A fraction collector, connected with the HPLC system, was used for the collection of chromatogram peaks. The obtained peaks were concentrated and then screened for antimicrobial activity.
Structure elucidation of the purified HFM-2P compound
The structure of the purified antimicrobial compound was elucidated using various spectroscopic techniques. Using chloroform as the reference solvent, the UV–visible spectrum was recorded using a Shimadzu UV–visible spectrophotometer in the wavelength range of 200–800 nm. The NMR (nuclear magnetic resonance) spectroscopy was performed in chloroform-d [99.8% D, containing 0.1% (v/v) TMS (tetramethylsilane)], while the FT-IR (Fourier transformation infrared spectroscopy) was performed with a Perkin-Elmer Spectrum RX-IFTIR spectrophotometer in the range 400–4000 cm and a 5 mm double channel solution state probe on a 500 MHz AVANCE III Bruker spectrometer at 25 °C.
Antimicrobial activity and bioautography of purified HFM-2P compound
Using the standard Kirby-Bauer disc diffusion method, the HFM-2P compound was tested for antimicrobial activity against MRSA, VRE, S. aureus, S. epidermidis, E. aerogenes, E. coli, B. subtilis, K. pneumoniae sub sp. pneumoniae; C. herbarum, F. oxysporum, A. brassicicola, C. gleosporoides. The Mueller Hinton Agar (MHA) and PDA plates were seeded with test bacteria and fungi, respectively, and discs holding 30 µg/disc of the HFM-2P compound were put on the medium’s surface, followed by diffusion for 30 min at 4 °C. After incubating the plates (at 37 °C for bacteria and 28 °C for fungi), the zones of inhibition were measured in millimeters (mm).
The bioautography of antimicrobial compound was performed by thin layer chromatography (TLC) using ethyl acetate: chloroform (8:2, v/v) solvent system. The generated chromatogram was visualized under iodine vapors and UV light. To identify the antimicrobial compound, TLC strips were aseptically placed on MHA and PDA plates previously inoculated with test cultures. The plates were left at 4 °C for an hour to allow the active metabolite to diffuse into the agar. Additionally, the plates were incubated at 37 °C for bacteria and 28 °C for fungi. The inhibition zone, which indicates the compound that is active in the extract, was detected.
Minimum inhibitory concentration (MIC) of purified HFM-2P compound
A 96-well microtiter plate dilution experiment was performed to determine the MIC of the HFM-2P compound against bacterial pathogens like MRSA, VRE, B. subtilis, E. coli, S. epidermidis, S. aureus, E. aerogenes, and K. pneumoniae sub sp. pneumoniae [0.5 optical density (OD) at 595 nm]. Concentrations of the HFM-2P compound (1.97, 3.95, 7.56, 15.12, 31.25, 62.5, 125 µg/mL) were prepared in distilled water. A 100 µL of test culture was mixed with 100 µL of compound in various concentrations, control blanks contained 100 µL of test compound in various concentrations with 100 µL of nutrient broth (NB), a positive control well contained 100 µL of bacterial culture and 100 µL of NB, and a negative control contained 200 µL of NB only. The plates were incubated at 37 °C for 24 h before the OD was determined by an ELISA microplate reader at 595nm (Bio-Rad, Model 680XR).
The MIC of a fungal spore suspension obtained by scraping spores from 5-day-old PDA slants and adding them to potato dextrose broth was determined after 48 h of incubation at 28 °C. 100 µL of fungal culture mixed with various concentrations of purified compound, for control blanks, 100 µL of purified compound and 100 µL of PDB broth, for positive control wells, 100 µL of fungal culture and 100 µL of PDB, and for negative control wells, 200 µL of PDB only. The plates were then incubated at 28 °C for 48 h. The OD was measured with an ELISA microplate reader at 595 nm.
Scanning electron microscopic (SEM) study to observe the effect of purified HFM-2P compound on bacterial cell morphology
Morphological changes in the tested bacteria (MRSA, VRE, E. coli, and B. subtilis) caused by HFM-2P and standard antibiotics vancomycin, teicoplanin, and gentamicin were examined through SEM. The turbidity of the bacterial suspensions was adjusted to 0.5 McFarland standard, and the cells were centrifuged at 10,000×g for 15 min. The pellets were resuspended in 50 µL of phosphate-buffered saline (PBS) (pH 7.4) after discarding the supernatant. The bacterial cell suspensions were incubated at 37 °C for 24 h following treatment with the compound and standard antibiotics. After incubation, the cell suspensions were centrifuged and resuspended in 30 µL of PBS for both treated and control bacteria. SEM slides (10 mm) with thin smears were made and air-dried. The smears were initially fixed at room temperature for 1 h with a 4% (v/v) glutaraldehyde solution. After that, the fixed smears were washed 3–5 times with MilliQ distilled water, each for 5–10 min. Following subsequent fixation with a 4% (v/v) glutaraldehyde solution for 2 h (3–5 times), they were rinsed again with MilliQ distilled water. The fixed smears were then dehydrated for 10 min each in 25%, 50%, 75%, and 95% ethanol, with the final dehydration stage (100% ethanol) repeated twice for 1 min each. The slides were air-dried and placed on silver-coated SEM stubs before being viewed under a scanning electron microscope (Carl Zeiss EVOLS 10 model). Positive controls comprised drugs such as vancomycin for MRSA, teicoplanin for VRE, and gentamicin for E. coli and B. subtilis, whereas negative controls were untreated cells.
The effect of purified HFM-2P compound on cell membrane permeability using fluorescence microscopy
The membrane damage of the tested pathogens was assessed using the double-staining approach as described by Costanza et al.18, with minor adjustments. The prepared inoculum was centrifuged at 1118×g for 10 min at 4 °C. The bacterial pellets were rinsed with sterile distilled water and then treated with purified HFM-2P compound. Every cell suspension was incubated at 37 °C with an average of 50 rpm agitation. After 24 h, the cells were centrifuged at 1118×g for 10 min at room temperature. The bacterial cell pellets were cleaned with sterile double-distilled water, and the supernatant was removed. The bacterial pellets were suspended in sterile distilled water, then propidium iodide (5 μg/mL in water) was added, and the pellets were incubated for 15 min in the dark. After rinsing to remove the excess PI solution, the pellets were suspended in sterile double-distilled water and incubated with 4′,6-diamidino-2-phenylindole (DAPI) (10 μg/mL in water) in the dark for 15 min. In the same way, the pellets were cleaned with sterile double-distilled water to get rid of the extra DAPI solution. The pellets were incubated for 15 min at room temperature after fixation with 2.5% (v/v) glutaraldehyde solution (EM grade) in 0.1 M phosphate buffer (pH 7.2). Following fixation, the bacterial pellets were washed and resuspended in sterile distilled water. The cell suspensions were then placed on slides and analyzed using a fluorescence microscope.
Safety evaluation of purified HFM-2P compound using Ames assay
Mutagenicity studies
The Ames (mutagenicity test) assay, first described by Maron and Ames19, was slightly modified to evaluate the effect of purified compound on mutagenicity using Salmonella histidine point mutants. To 2 mL of top agar, 100 µL of activated S. Typhimurium strains (TA98/TA100), 100 µL of purified compound at various concentrations (50, 100, and 250 µg/plate), and 200 µL of 0.5 mM histidine-biotin mixture (1:1) were added. After that, top agar was poured onto the minimal agar plate and incubated at 37 °C for 48 h. In positive controls, 4-Nitro-o-phenylenediamine (NPD; 20 μg/0.1 mL per plate) for strain TA98, and sodium azide (2.5 µg/100 µL/plate) for strain TA100 were used, whereas in negative controls, 0.5% Dimethyl sulfoxide (DMSO) (100 µL/plate) was used. The spontaneous reversion of test strains TA98 and TA100 was determined by counting revertant his+ colonies after incubation. Evaluation of the mutagenic potential of the purified compound was carried out by comparison of the count of colonies with control plates lacking the test compound, or a mutagen was added. Additionally, pre- and co-incubation techniques were used to assess the antimutagenicity of the purified compound.
Co-incubation method
In the co-incubation test, 100 μL of NPD (20 μg/0.1 mL per plate) and sodium azide (2.5 μg/0.1 mL per plate), used as direct-acting mutagens for TA98 and TA100, respectively, along with 100 μL of purified compound at various concentrations were added in 2 mL of top agar (at 45 °C).
Pre-incubation method
To 2 mL of top agar, 100 μL of sodium azide/NPD and purified compound (50, 100, and 250 μg/100 µL per plate) were added along with 100 μL of TA100/TA98 strains, respectively. The mixture was incubated at 37 °C for 30 min. The top agar was then poured and evenly spread on minimum agar plates, which were incubated at 37 °C for 48 h. Three control experiments were performed: one positive (100 µL mutagen + 100 µL bacterial culture), one spontaneous (100 µL bacterial culture), and one negative (100 µL bacterial culture + 100 µL test chemical). Following a percentage reduction of reverse mutations that represents the antimutagenic effect of purified compounds:
a = The number of histidine revertants generated by the mutagen (NPD/Sodium azide) alone; b = The number of histidine revertants produced by the mutagen in the presence of the test chemical; c = The number of histidine revertants induced in the absence of mutagen.
DNA protective potential of purified HFM-2P compound
This experiment was carried out according to Lee et al.20. This process was used to test the ability of the HFM-2P compound from S. levis strain HFM-2 to maintain the supercoiled pBR322 plasmid against Fenton’s reagent’s hydroxyl radicals. A reaction mixture consisting of 2 µL of plasmid DNA (50 µg/100 µL), 10 µL of Fenton’s reagent (80 mM Ferric Chloride (FeCl₃), 30 mM hydrogen peroxide (H₂O₂), and 50 mM ascorbic acid), varying concentrations of the HFM-2P compound (2–10 µg/mL), and Milli-Q water was prepared to a final volume of 20 µL. In the negative control, an equal volume of MQ water was substituted in place of Fenton’s reagent, whereas rutin was used as a positive control. The reaction mixture was incubated for 30 min at 37 °C. DNA was separated using 1% agarose gel electrophoresis. Using Image J software, a quantitative examination of DNA damage was conducted. The percentage ratio of native supercoiled (Form I), single-stranded nicked (Form II), and linear (Form III) DNA was calculated.
Cytotoxic activity of purified HFM-2P compound
Cell line maintenance and growth conditions
The National Centre for Cell Science (NCCS), Pune, India, provided the HeLa cervical cancer and L929 (murine fibroblast) normal cell lines. They were grown in DMEM (Dulbecco’s modified Eagle’s medium) culture medium supplemented with 10% FBS (Fetal bovine serum) and antibiotics (100 units/mL penicillin and 100 µg/mL streptomycin). The cell lines were maintained at 37 °C in an environment with 5% CO2 and 90% relative humidity. A bright field inverted microscope was used to check the cultures for confluency and the presence or absence of bacterial and fungal contaminations21.
MTT assay
The cytotoxicity of HFM-2P compound on HeLa cancer cell line was assessed using the MTT assay 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide) described by Mosmann22. Cells (5000 per well) were seeded in a 96-well microtiter plate and left to adhere overnight. HFM-2P was added to each well at different concentrations (1.56, 3.12, 6.25, 12.5, 25, 50, 100 µg/mL) and incubated for 24 h. After that, each well received 100 µL of 0.5 mg/mL MTT dye (Sigma-Aldrich), and the plates were incubated for 4 h at 37 °C in a humidified atmosphere with 5% CO2. The MTT reaction yielded blue formazan crystals that were dissolved in 100 µL of DMSO. The color was measured with a microplate reader.
where Ao denotes the absorbance of untreated cells (only medium) and Ae denotes the absorbance of treated cells.
Statistical analysis
All statistical analyses were carried out in duplicate. The results were given as mean ± standard error (SE). Statistical significance was determined at p < 0.05. SPSS software analyzed the data using one-way ANOVA and Tukey’s test (p < 0.05).
Results
Fermentation and recovery of antimicrobial compound
Ethyl acetate was the best-suited solvent to extract antimicrobial metabolites from the cell-free supernatant because it provides the highest recovery of metabolites in terms of inhibition zones. EtOAc extract showed pronounced activity against different test bacterial pathogens, especially drug-resistant MRSA, VRE, S. aureus, S. epidermidis, E. aerogenes, E. coli, B. subtilis, K. pneumoniae sub sp. pneumoniae, and fungal phytopathogens viz., C. herbarum, F. oxysporum, A. brassicicola, and C. gleosporoides (15–28 mm), which is represented in Fig. S1 and Table S1.
Purification of antimicrobial compound from S. levis strain HFM-2
The reddish-brown ethyl acetate (EtOAc) extract was subjected to silica-gel column chromatography to purify an antimicrobial compound. Seventeen fractions (1–17), eluted with chloroform: ethyl acetate (80:20, v/v), showed antimicrobial activity against different tested pathogens. These active fractions were pooled based on the TLC pattern and finally subjected to semi-preparative RP-HPLC. During semi-preparative RP-HPLC analysis, some peaks were detected, mainly with retention times of 9.124, 10.554, 11.157, 14.169-, and 15.540 min using acetonitrile: water (90:10) as a solvent system (Fig. 1a). The active peak (14.169 min) was further chromatographed with the same solvent system ratio. A single peak with a retention time of 14.238 min and also showing antimicrobial activity was found, confirming the compound’s purity (Fig. 1b). The purified compound was termed HFM-2P.
Antimicrobial activity and bioautography of the purified HFM-2P compound
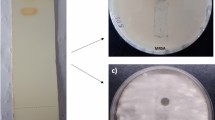
The HFM-2P compound exhibited potent antibacterial activity against both Gram-positive and Gram-negative bacteria, including MRSA, VRE, E. coli, S. epidermidis, S. aureus, K. pneumoniae sub-sp. pneumoniae, E. aerogenes, B. subtilis, and fungal phytopathogens, viz. C. herbarum, F. oxysporum, A. brassicicola and C. gleosporoides. Notably, the compound was more potent against MRSA and VRE compared to standard antibiotics (zones of inhibition of vancomycin 18 mm, teicoplanin 15 mm against MRSA and VRE, respectively) with zones of inhibition of 30.0 ± 0.25 mm and 29.0 ± 0.35 mm, respectively (Fig. 2 and Table S1). Bioautography of the HFM-2P compound also confirmed the presence of one antimicrobial compound with an Rf value of 0.65 against MRSA, VRE, and F. oxysporum (Fig. 3).
Antibacterial activity of EtOAc extract and HFM-2P compound against (a) MRSA, (b) VRE, (c) E. coli, (d) S. aureus, (e) S. epidermidis, (f) K. pneumoniae sub sp. pneumoniae; (g) E. aerogenes, (h) B. subtilis, (i) C. herbarum, (j) F. oxysporum, (k) A. brassicicola, (l) C. gleosporoides E: EtOAc extract, P: Purified compound, T: Teicoplanin (30 µg/disc), M: Methicillin (10 µg/disc), V: Vancomycin (30 µg/disc).
MIC values of purified HFM-2P compound
The MIC values were determined by the 96-well plate method. The HFM-2P compound was found to be potent with low MICs against different test bacteria, ranging from 1.95 to 125 μg/mL. It was found to be more potent against VRE and MRSA with MICs of 3.9 and 1.95 μg/mL, respectively, than against S. aureus, K. pneumoniae sub sp. pneumoniae, E. coli, E. aerogenes, S. epidermidis, and B. subtilis with MIC values of 15.6, 31.25, 31.25, 125, 31.25, and 62.5 μg/mL, respectively. In the case of fungal phytopathogens, the MIC values for C. herbarum, F. oxysporum, A. brassicicola, and C. gleosporoides were 125, 125, 62.5, and 125 μg/mL, respectively.
Structure characterization of purified HFM-2P compound from the S. levis strain HFM-2
The antimicrobial compound (30 mg) with a retention time of 14.238 min was characterized as a derivative of 2,6-disubstituted chromone, using spectroscopic techniques viz. LC–MS (Fig. S2), UV–visible, FT-IR (Fig. 4), and 1H-NMR (Table S2 and Fig. 5). It was soluble in methanol, ethyl acetate, DMSO, and acetonitrile, but sparingly soluble in chloroform and water. The maximum ultraviolet (UV) absorption bands of the purified compound were noted at 246 and 338 nm. FTIR spectrum of the compound showed bands at 3324 and 1654 cm−1 due to the presence of hydroxyl and conjugated carbonyl groups, respectively. The absorption bands at 2944 and 2832 cm−1 corresponded to aromatic C–H and alkyl C–H stretching, respectively, while those at 1654 and 1504 cm−1 were attributed to C=C ring stretching. The molecular formula was calculated as C35H59NO12 for the ion peak at m/z 685.37 (M) (Molecular weight: C:12.011u; O:15.999u, N:14.0; H:1.00u) (Fig. 6).
The 1H NMR spectrum displayed signals δ 7.22dd, 6.97dd, and 6.85dd, indicating three aromatic protons, whereas δ 6.65 s confirmed the γ-pyrone ring, typical of a 2,6-disubstituted chromone derivative. Additionally, the singlet signal for δ 6.65 s (H-3) showed the presence of a chromone ring due to the substituent at C-2. The other protons of the isolated compound appeared at δ 5.38–5.48 m, 4.59–4.92 m, 3.29–3.39 m, and 1.24–1.48 m. Also, in the side chains, the presence of multiplets at δ 1.24–1.48 and δ 0.86t in 1H NMR spectra showed the presence of moieties of CH2 and CH3 groups, respectively.
Scanning electron microscopic (SEM) observation
SEM was used to detect treated bacterial cells and analyze any physical or morphological changes in the cells. SEM studies indicated that all four tested bacteria were physically damaged and had significant structural damage and morphological alterations after exposure to the purified HFM-2P compound . The SEM images (Fig. 7b, c) show MRSA cells treated with HFM-2P compound and standard antibiotic, i.e., vancomycin . These images illustrate the cell aggregation, clumped entities, abnormal cell structures, and destructive effects of the HFM-2P compound and vancomycin on MRSA. Nontreated cells were intact and showed a smooth surface (Fig. 7a). VRE untreated (control) cells appeared smooth-surfaced and spherical (Fig. 7d). However, VRE cells treated with the HFM-2P compound had a distracted cell morphology and clumped together (Fig. 7e). The cells had a rough, creased surface, clumped and deformed, and might be seen as bunches of interconnected cells. Similarly the cells treated with standard antibiotic were flattened and clumped together (Fig. 7f).
The SEM (scanning electron micrographs) display the effect of HFM-2P compound on MRSA, VRE, E. coli, and B. subtilis; Control (untreated cells): (a) MRSA, (d) VRE, (g) E. coli; j) B. subtilis. Cells treated with HFM-2P compound: (b) MRSA, (e) VRE, (h) E. coli, and (k) B. subtilis. Positive control: (c) MRSA treated with vancomycin, (f) VRE treated with teicoplanin, i and l) E. coli and B. subtilis treated with gentamicin.
In the case of E. coli, untreated (control) cells showed a typical rod-like shape with smooth and complete cell surfaces (Fig. 7g). However, E. coli cells treated with the HFM-2P compound had a slanted cell morphology and significant deformation (Fig. 7h). Cell aggregation, clumping, and mass formation of cell debris were detected, similar to that caused by a standard antibiotic (Fig. 7i). As shown by the distorted cellular shape, the HFM-2P compound mostly had a disruptive effect on the bacterial cells, similar to that of traditional antibiotics.
On the other hand, B. subtilis normal (untreated) cells displayed a distinctive morphology and were rod-shaped, smooth on the surface, and had an undamaged cell wall (Fig. 7j). HFM-2P compound-treated cells displayed significant disruption and defects in cellular shapes, such as uneven surfaces, lengthy and clustered together (Fig. 7k). Similar results were obtained with gentamicin, which produced abnormally elongated, irregular cells that differed from normal cell shape (Fig. 7l).
Cell damage and membrane permeability by fluorescence microscopy
In this assay, MRSA, VRE, E. coli, and B. subtilis cells were stained with PI and DAPI. PI can only stain bacterial cells by intercalating nucleic acids in the nucleus after the membranes have been ruptured. In contrast, DAPI can pass through intact bacterial CMs despite vitality. As demonstrated in Fig. S3, cells exhibit intact cell membranes with blue fluorescence following incubation with DAPI, but no fluorescence has been observed for PI after 24 h of incubation. The HFM-2P compound-treated MRSA, VRE, E. coli, and B. subtilis bacterial cells were significantly stained with blue (DAPI) and red (PI) fluorescence, showing that the PI had penetrated the cells. This demonstrated that treatment with the HFM-2P compound damaged the membranes of MRSA, VRE, E. coli, and B. subtilis cells.
Protein leakage of bacterial cells treated with purified HFM-2P compound
The antibacterial mode of action of the HFM-2P compound against MRSA, VRE, E. coli, and B. subtilis cells was further confirmed by protein leakage assay. After 24 h of incubation, treated cells leaked 88.6, 78.8, 58.0, and 55.0 µg/mL of protein content in the supernatant, respectively. These results displayed that the HFM-2P compound had a potential inhibitory effect against the tested pathogens, especially drug-resistant MRSA and VRE.
Safety evaluation of the purified HFM-2P compound by Ames assay
The HFM-2P compound from S. levis strain HFM-2 was found to be non-mutagenic at concentrations, i.e. 50,100, and 250 µg/plate against TA100 and TA98 (S. Typhimurium strains). In the positive control (presence of mutagen), revertant colonies of TA100 and TA98 were found to be 983.33 ± 0.75 and 1084.33 ± 0.96, respectively. In the presence of the HFM-2P compound (without mutagen), the revertant colonies observed were: 22.66 ± 0.48 against TA98 and 110.66 ± 0.67 for TA100, which were comparable to spontaneous revertant colonies (79.33 ± 0.41 for TA98, and 120.66 ± 0.78 for TA100), indicating the non-mutagenic character of the HFM-2P compound.
However, HFM-2P compound from the S. levis strain HFM-2 displayed antimutagenic activity against S. Typhimurium strains. The two experiments (pre-incubation and co-incubation) were carried out independently to check the antimutagenic activity of HFM-2P compound. In terms of percentage inhibition, HFM-2P exhibited a strong antimutagenic effect against both mutagens. The compound exhibited significant antimutagenicity with inhibition of 73.63% and 87.39% against NPD (TA98) and sodium azide (TA100), respectively, in co-incubation, and inhibition of 77.20% and 86.36%, respectively, was observed during pre-incubation (at 250 µg/100 µL concentration). (Table S3 and Fig. 8 and 9).
DNA protective potential of purified HFM-2P compound
The DNA protective ability of the HFM-2P compound was assessed using a DNA nicking assay. The results showed that the presence of Fenton’s reagent caused the pBR322 plasmid DNA to degrade from supercoiled to single-stranded, nicked, and linear forms. However, as seen in lanes 4–8 (Fig. S4), addition of the HFM-2P compound to the reaction mixture reduced the amount of DNA damage. It was found that Form I was destroyed in the presence of Fenton’s reagent, whereas Form II (81.78%) and Form III (18.21%) increased concurrently. However, densitometric examination revealed that the total quantity of Form I was 46.32%, 51.14%, 57.14%, 68.35% and 71.47%, respectively, in the presence of Fenton’s reagent and HFM-2P compound (2–10 μg/mL) (Fig. 10). The results were compared with the native supercoiled DNA amount present in the positive control (Fenton’s reagent + rutin) with 54.72% Form I and 45.27% Form II, showing that the purified compound is more protective than rutin Table.S4).
Cytotoxic activity of purified HFM-2P compound using MTT assay
The HFM-2P compound induced significant (p ≤ 0.05), dose-dependent cytotoxicity in HeLa cancer cells, while exhibiting minimal effects on L929 normal cell lines (Fig. S5). At the highest tested concentration (100 µg/mL), HFM-2P inhibited 82.75 ± 1.65% of HeLa cell proliferation, with an IC₅₀ value of 17.24 µg/mL. In contrast, the compound exhibited insignificant cytotoxicity against L929 cells, with only 10.15 ± 0.65% inhibition observed at the maximum tested concentration.
Discussion
The emergence of antibiotic resistance has been recognized as a worldwide health-threatening problem23. The search for novel, effective antibiotics against antibiotic-resistant pathogens is the most vital issue for the treatment of infectious diseases24. Natural products are the main source of antimicrobial agents, most of which are produced by Streptomyces14,16,25. Therefore, there has been increasing interest among researchers in screening new bioactive metabolites from Streptomyces species to overcome antibiotic-resistant pathogens. The compound efficiency in the treatment of different infections in humans has been demonstrated by calculating the inhibition zones and MIC values. In the current study, a potent antimicrobial compound was detected in EtOAc extract from S. levis strain HFM-2. During the prediction of the structure of the purified antimicrobial compound, 1H-NMR data and strong absorbance at 246 and 338 nm suggested the presence of a ‘chromone’ nucleus in the active compound. The compound has been confirmed to be a 2,6-disubstituted chromone derivative based on NMR, LC–MS, and FTIR data, comparable to the published literature25,26,27,28.
Previous studies have identified eleven compounds from the soil Streptomyces levis with the same molecular formula. These included 2-piperidinone, oleandomycin, and compounds of erythromycin or tylonolide. However, none of the described compounds included a ‘chromone’ moiety29. Recently, Singh et al.28 reported a 2,6-disubstituted chromone derivative from the soil Streptomyces levis strain and its activity against Pseudomonas aeruginosa MTCC 741, Staphylococcus aureus MTCC 96, and Klebsiella pneumoniae MTCC 109, with the zones of inhibition of 20, 24, and 23 mm, respectively.
In the present study, the antimicrobial compound HFM-2P isolated from S. levis strain HFM-2 was found to be more potent, with zones of inhibition of 27.0 ± 0.25, 26.0 ± 0.5, 25.1 ± 0.5, 12.0 ± 0.5, and 15.0 ± 0.5 against K. pneumoniae sub sp. pneumoniae, S. aureus, S. epidermidis, E. aerogenes, and B. subtilis, respectively which are higher than those reported by Singh et al.28.
In addition, the present study also demonstrated the antimicrobial activity of a 2,6-disubstituted chromone derivative (HFM-2P), isolated from S. levis strain HFM-2, against drug-resistant MRSA and VRE with zones of inhibition of 30 ± 0.25 mm and 29.0 ± 0.35 mm, respectively. The compound is more potent than standard antibiotics vancomycin and teicoplanin, which showed inhibition zones of 18 mm and 15 mm against MRSA and VRE, respectively.
Besides antibacterial activity, the compound HFM-2P also showed activity against fungal phytopathogens, with zones of inhibition of 18.0 ± 0.25, 20.0 ± 0.25, 22.5 ± 0.1, and 24.3 ± 0.57 mm against C. herbarum, C. gleosporoides, F. oxysporum and A. brassicicola, respectively.
The compound HFM-2P demonstrated higher potency against VRE and MRSA, with MIC values at very low concentrations, i.e., 3.9 μg/mL and 1.95 μg/mL, respectively than against S. aureus, K. pneumoniae sub sp. pneumoniae, E. coli, E. aerogenes, S. epidermidis, and B. subtilis with 15.6 μg/mL, 31.25, 31.25 μg/mL, 125 μg/mL, 31.25 μg/mL, and 62.5 μg/mL MICs, respectively. In addition, the MIC values against fungal phytopathogens, viz., C. herbarum, F. oxysporum, A. brassicicola, and C. gleosporoides, were 125 μg/mL, 125 μg/mL, 62.5 μg/mL, 125 μg/mL, respectively. However, Singh et al.28 reported MIC values at 12.5, 6.25, and 6.25 μg/mL against Pseudomonas aeruginosa MTCC 741, Staphylococcus aureus MTCC 96, and Klebsiella pneumoniae MTCC 109, respectively.
To the best of our knowledge, this is the first report of the antifungal activity of the 2,6-disubstituted chromone derivative (HFM-2P) from S. levis strain HFM-2 (isolated from the human gut) against the mentioned fungi, as well as its potent antibacterial activity against drug-resistant MRSA and VRE. Additionally, this compound is being reported for the first time for its DNA-protective, antimutagenic, and antiproliferative properties. In the development of antibacterial drugs, elucidation of the antibacterial mechanism of action is a key point. This information enables the prediction of challenges with clinical safety and bacterial resistance in the context of antibacterial medications29. Microbial factors affecting morphological changes include the cell wall structure of the Gram-positive and Gram-negative bacterial pathogens. When bacteria are exposed to an antibiotic agent, they undergo morphological and ultrastructural alterations30.
In this study, several morphological alterations were observed through SEM analysis in drug-resistant bacteria, including MRSA, VRE, B. subtilis, and E. coli, following treatment with the HFM-2P antimicrobial compound. The treated bacterial cells exhibited deformities in the cell wall, cellular abnormalities, irregular shapes, and significant surface collapse. These alterations affect the integrity and function of the membrane, resulting in cell lysis and death. Hartmann et al.31 reported bactericidal action by gramicidin S and PGLa produced by Aneurinibacillus migulanus DSM 5759 and observed extensive bacterial cell wall damage with multiple blisters, deep craters, and holes in E. coli DSM 1103 and S. aureus DSM 1104, respectively. Similarly, Aruldass et al.32 reported morphological alterations (rupture of the cell membrane, irregular shapes, and lysis of cell walls) in S. aureus and MRSA, and the inhibitory effect of violacein, a violet pigment produced by Chromobacterium violaceum. Recently, Devi et al.33 also reported some morphological changes like abnormalities in cellular morphology, disintegrated cell wall, permeability, and leaked cytoplasmic material in the bacterial cells treated with plicacetin (antimicrobial compound) isolated from endophytic Streptomyces sp. SP5.
Furthermore, disruption of the cell membrane (CM) integrity, which leads to bacterial growth inhibition and cell death, is a crucial factor34. Using DAPI and PI dyes, we demonstrated permeability and damage to the cell membrane under a fluorescence microscope, which was further confirmed. In this study, MRSA, VRE, E. coli, and B. subtilis cells treated with purified HFM-2P compound formed clusters, indicating membrane damage and loss of membrane potential, which caused the cells to aggregate.
Niu et al.35 demonstrated membrane disruption of B. subtilis and E. coli membranes using fluorescent microscopy analyses. Cells of both bacteria, when treated with lipo-γ-AApeptides, were strongly stained with DAPI and PI. Li et al.36 demonstrated that DAPI and PI dyes were able to stain S. aureus cells treated with γ-AA peptide, indicating that the γ-AA peptide had damaged the bacterial membrane. Cui et al.37 observed that Salvia sclarea essential oil reduced the blue (DAPI) fluorescence intensity of S. aureus after treatment, suggesting a decrease in the DNA content of S. aureus, most likely as a result of cell membrane injury. Aruldass32 demonstrated strong binding of PI and DAPI stains to treated bacterial cell membranes, indicating that the treatment compromised membrane integrity and permitted PI entry into the nuclei of S. aureus ATCC 29,213 and MRSA ATCC 43,300 cells. Mendes et al.38 showed cytoplasmic membrane disruption in different bacterial cells treated with ZnO NPs, using DAPI and PI staining and observing by fluorescence microscopy.
Additionally, the integrity of the cell membrane was assessed by measuring the release of protein into the supernatant from MRSA, VRE, E. coli, and B. subtilis cells treated with the HFM-2P compound purified from S. levis strain HFM-2. The leakage of protein by MRSA, VRE, E. coli, and B. subtilis (88.6, 78.8, 58.0, 55.0 µg/mL in supernatant, respectively) after 24 h of treatment indicated that the compound gradually altered cell surfaces and was able to enter the peptidoglycan layers and the CM. Francis et al.39 reported that the mesosomes promote swelling of S. aureus ATCC 29,213 and MRSA ATCC 43,300 cells due to turgor pressure in creating bigger holes when treated with lysostaphin, an enzyme. This swelling results in cell lysis and protein leaking from the cytoplasm. Aruldass32 reported, 64.0 ± 1.8 and 93.8 ± 1.3 μg/mL of protein leaked by S. aureus ATCC 29,213 and MRSA ATCC 43,300 cells, treated with 3.9 μg/mL of violet fraction, at 12 and 24 h, respectively that was able to enter the peptidoglycan layers and the CM as the CM creates a selective permeability barrier.
Mutations are mostly caused by external sources, like chemical and physical substances known as mutagens. In addition, mutations may also occur spontaneously as the consequence of errors in DNA replication, repair, and recombination40. Mutagens can be mitigated by antimutagenic substances41. The Ames mutagenicity and DNA nicking assays are the simplest and reliable methods for verifying a compound’s biosafety. Ames19 established the Ames mutagenicity assay, commonly known as the reverse mutation assay, which is a widely used approach in experimental research to identify compound mutagenesis. In this experiment, S. Typhimurium strains TA98 and TA100 with a histidine gene mutation (his- mutant) are used to assess if tested chemicals cause base-pair substitution and frameshift mutations in DNA (GC-base pairs). The antimutagenic activity of the chemical was found to be dose-dependent, which is consistent with recent findings that indicated a concentration-dependent antimutagenic response of various natural extracts against Salmonella strains42,43,44. Kaur et al.45 reported the antimutagenic potential of the antifungal compound SH2 produced by Streptomyces hydrogenans strain DH16 that showed 70% suppression at 250 g/100 µL against S. Typhimurium strains TA98 and TA100. Similarly, Sharma et al.46 displayed 47.87–88.11% and 41.10–88.53% antimutagenic activity of salvianolic acid B purified from Streptomyces M4 for TA98 and TA100 strains, respectively, at the highest dose (250 μg/100 μL). Bouguellid et al.44 reported mutagenic and antimutagenic activities of Fraxinus angustifolia Vahl extracts against TA98 and TA100 S. Typhimurium strains. Similarly, Kaur et al.47 reported antimutagenicity of 2-aminofuorene (2-AF) from the endophytic fungus Aspergillus fumigatus isolated from Moringa oleifera against the reactive carcinogenic ester generating mutagen. Devi et al.33 described 47.87–88.11% and 41.10–88.53% antimutagenic activity of the antimicrobial compound plicacetin, purified from endophytic Streptomyces sp. SP5, isolated from citrus lemon, for TA98 and TA100 strains, respectively at the highest dose (250 μg/100 μL). In this study, the purified HFM-2P compound of S. levis strain HFM-2 was non-mutagenic against S. Typhimurium strains TA98 and TA100. But, in the presence of mutagens, the HFM-2P compound showed antimutagenic activity of 77.20% and 86.36% against sodium azide, 73.63% and 87.39% against NPD for TA100 and TA98, respectively, at the highest dose (250 µg/100 µL). So, the antimutagenic substances (HFM-2P) produced by the S. levis strain HFM-2 may be useful in combating the effects of environmental mutagens.
DNA nicking assay demonstrates the effect of the purified HFM-2P compound in an oxidative stress-induced DNA damage model. In the present study, the HFM-2P compound produced by S. levis strain HFM-2exhibited DNA protective potential and reduced the conversion of pBR322 plasmid DNA into single-stranded nicked (Form II), linear forms of DNA (Form III generated by Fenton’s reagent) along with increasing native supercoiled form (Form I) with 46.32%, 51.14%, 57.14% 68.35% and 71.47%, at 2–10 μg/mL. Similar results were observed in our previous study17. The absence of DNA Form III formation in the HFM-2P compound might be responsible for the reduction in the production of free hydroxyl radicals48,49,50,51. Thenmozhi et al.52 showed partial protection of the DNA damaged by H2O2 at high concentration, i.e., 50 μg/mL, by the ethyl acetate extract of Streptomyces sp. VITSTK7. Similarly, Karthik et al.53 reported DNA protection from oxidative stress by crude extracts of Streptomyces cultures isolated from marine sediment. Rani et al.51 showed DNA protection against oxidative stress induced via free hydroxyl radicals by EtOAc extract of S. cellulose strain TES17, demonstrating an increase in native supercoiled form (Form I) and a decrease in Form II (single-stranded or double-stranded nicked) and Form III (linear form). Similarly, Devi et al.33 reported DNA protection against free hydroxyl radicals caused by Fenton’s reagent in the presence of purified plicacetin antimicrobial compound isolated from Streptomyces sp. SP5.
Oxidative stress can cause biological molecule alterations that increase the rate of mutation, which can lead to cancer growth54. In this study, the compound HFM-2P purified from S. levis strain HFM-2 exhibited significant cytotoxicity against HeLa cancer cells with 82.83 ± 1.65% reduction at the maximum concentration of 100 µg/mL, and the IC50 value was recorded at 17.69 µg/mL. According to Saraswathi et al.55, ethyl acetate extract of strain Streptomyces cangkringensis showed 71.01 ± 0.24% inhibition and IC50 value against HeLa cells at high concentrations, i.e., 1000 µg/mL and 410.5 µg/mL, respectively. Similarly, Bhat et al.56 reported anticancer activity of ethyl acetate extract isolated from Streptomyces sp. strain KF15 and showed 64.32 ± 1.76 inhibition at 125 µg/mL, and the IC50 value was found at 99.85 µg/mL against HeLa cancer cells. Recently, Bhat et al.57 reported a crude extract isolated from Streptomyces sp. strain YC69 with 92.05 ± 1.79% inhibition against HeLa cancer cells at 125 µg/mL, and the IC50 value was found at 41.98 µg/mL.
Conclusion
The current study reports, for the first time, the purification of the 2,6-disubstituted chromone derivative (HFM-2P) from S. levis strain HFM-2, an antibiotic-producing bacterium isolated from the human gut, which exhibits activity against drug-resistant bacteria and fungal phytopathogens. The HFM-2P compound was non-mutagenic and non-toxic to normal cell line. However, it displayed cytotoxicity against the HeLa (the human cervical cancer) cell line. Therefore, our results suggest that HFM-2P has promising antimicrobial and anticancer properties and is effective against multidrug-resistant bacteria and HeLa cancer cells.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Change history
29 September 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-19333-8
Abbreviations
- SCNA:
-
Starch casein nitrate agar
- SEM:
-
Scanning electron microscope
- MTCC:
-
Microbial type culture collection
- TLC:
-
Thin layer chromatography
- HPLC:
-
High-performance liquid chromatography
- IMTECH:
-
Institute of Microbial Technology
- NPD:
-
4-Nitro-o-phenylenediamine
- LCMS:
-
Liquid chromatography-mass spectroscopy
- NMR:
-
Nuclear magnetic resonance
References
Ganesan, G., Velayudhan, S. S. & David, J. S. R. Statistical optimization of medium constituents and conditions for improved antimicrobial compound production by marine Streptomyces sp. JRG-04. Arch. Biol. Sci. 69, 723–731 (2017).
Peterson, E. & Kaur, P. Antibiotic resistance mechanisms in Bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 9, 2928 (2018).
Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. The Lancet 399(10325), 629–655 (2022).
Willems, R. P. et al. Incidence of infection with multidrug-resistant Gram-negative bacteria and vancomycin-resistant enterococci in carriers: A systematic review and meta-regression analysis. Lancet Infect. Dis. (2023).
Chernov, V. M., Chernova, O. A., Mouzykantov, A. A., Lopukhov, L. L. & Aminov, R. I. Omics of antimicrobials and antimicrobial resistance. Expert Opin. Drug Discov. 14, 455–468 (2019).
Singh, R. & Dubey, A. K. Isolation and characterization of new endophytic actinobacterium Streptomyces californicus strain ADR1 as a promising source of anti-bacterial, anti-biofilm and antioxidant metabolites. Microorganisms 8, 929 (2020).
Allcock, S. et al. Antimicrobial resistance in human populations: Challenges and opportunities. Glob. Heal. Epidemiol. Genomics 2–4 (2017).
Saber, T. et al. Methicillin-and vancomycin-resistant Staphylococcus aureus from humans and ready-to-eat meat: Characterization of antimicrobial resistance and biofilm formation ability. Front. Microbiol. 12, 735494 (2022).
Xu, K., Li, X. Q., Zhao, D. L. & Zhang, P. Antifungal secondary metabolites produced by the fungal endophytes: Chemical diversity and potential use in the development of biopesticides. Front. Microbiol. 12, 689527 (2021).
Mazumdar, R., Dutta, P. P., Saikia, J., Borah, J. C. & Thakur, D. Streptomyces sp. strain PBR11, a forest-derived soil actinomycetia with antimicrobial potential. Microbiol. Spectr. 11, 03489–03522 (2023).
Driche, E. H. et al. A new Saharan strain of Streptomyces sp. GSB-11 produces maculosin and N-acetyltyramine active against multidrug-resistant pathogenic bacteria. Curr. Microbiol 79, 298 (2022).
Hamedi, J., Mohammadipanah, F. & Ventosa, A. Systematic and biotechnological aspects of halophilic and halotolerant actinomycetes. Extremophiles 17, 1–13 (2013).
Al-Dhabi, N. A., Esmail, G. A., Duraipandiyan, V., Arasu, M. V. & Salem-Bekhit, M. M. Isolation, identification, and screening of antimicrobial thermophilic Streptomyces sp. Al-Dhabi-1 isolated from Tharban hot spring, Saudi Arabia. Extremophiles 20, 79–90 (2016).
Benndorf, R. et al. Natural products from Actinobacteria associated with fungus-growing termites. Antibiotics 7, 83 (2018).
Quinn, G. A., Banat, A. M., Abdelhameed, A. M. & Banat, I. M. Streptomyces from traditional medicine: Sources of new innovations in antibiotic discovery. J. Med. Microbiol 69, 1040–1048 (2020).
Chanthasena, P. et al. Isolation and Identification of Bioactive Compounds from Streptomyces actinomycinicus PJ85 and their in vitro antimicrobial activities against methicillin-resistant Staphylococcus aureus. Antibiotics 11, 1797 (2022).
Verma, J., Attri, S., Arora, S. & Manhas, R. K. Antioxidant and chemoprotective potential of Streptomyces levis strain isolated from human gut. AMB Express 13, 69 (2023).
Costanza, F. et al. Investigation of antimicrobial PRG-poly (amino acid). RSC Adv. 4, 2089–2095 (2014).
Ames, B. N. Dietary carcinogens and anticarcinogens: Oxygen radicals and degenerative diseases. Science 221, 1256–1264 (1983).
Lee, C. H. et al. Novel anticancer agent, benzyldihydroxyoctenone, isolated from Streptomyces sp. causes G1 cell cycle arrest and induces apoptosis of HeLa cells. Cancer Sci. 98, 795–802 (2007).
Hsu, C. L., Huang, S. L. & Yen, G. C. Inhibitory effect of phenolic acids on the proliferation of 3T3-L1 preadipocytes in relation to their antioxidant activity. J Agric. Food Chem. 54, 4191–4197 (2006).
Mosmann, T. Benzimidazole based Pt(II) complexes with better normal cell viability than cisplatin: Synthesis, substitution behavior, cytotoxicity, DNA binding and DFT study. J. Immunol. Methods 65, 55–63 (1983).
Ventola, C. L. The antibiotic resistance crisis: Part 1: causes and threats. Pharm. Ther. 40(4), 277 (2015).
Abdelaziz, R. et al. Bioactive metabolites of Streptomyces misakiensis display broad-spectrum antimicrobial activity against multidrug-resistant bacteria and fungi. Front. Cell. Infect. Microbiol. 13, 1162721 (2023).
Griffiths, P. & Ellis, G. Benzopyrones VI: The ultraviolet absorption spectra of chromone and 2-substituted chromones. Spectrochimica Acta Part A Mol. Spectrosc. 28, 707–713 (1972).
Cao, L. H. & Cui, P. Y. Synthesis of 2-dihydrooxadiazolinyl chromones. J. Chin. Chem. Soc. 50, 903–908 (2003).
Singh, V. et al. Isolation, screening, and identification of novel isolates of actinomycetes from India for antimicrobial applications. Front. Microbiol. 7, 1921 (2016).
Singh, V. et al. Isolation and purification of an antibacterial compound from Streptomyces levis collected from soil sample of north India. PLoS ONE 13, 0200500 (2018).
Silver, L. L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24, 71–109 (2011).
Chen, K., Sun, G. W., Chua, K. L. & Gan, Y. H. Modified virulence of antibiotic-induced Burkholderia pseudomallei filaments. Antimicrob. Agents Chemother. 49, 1002–1009 (2005).
Hartmann, M. et al. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. AAC 54, 3132–3142 (2010).
Aruldass, C. A., Masalamany, S. R. L., Venil, C. K. & Ahmad, W. A. Antibacterial mode of action of violacein from Chromobacterium violaceum UTM5 against Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA). Environ. Sci. Pollut. Res. 25, 5164–5180 (2018).
Devi, S., Sharma, M. & Manhas, R. K. Purification and biological analysis of antimicrobial compound produced by an endophytic Streptomyces sp. Sci. Rep. 13, 15248 (2023).
Diao, W. R., Hu, Q. P., Zhang, H. & Xu, J. G. Chemical composition, antibacterial activity, and mechanism of action of essential oil from seed of fennel (Foeniculum vulgare Mill.). Food Control 35, 109–116 (2014).
Niu, Y. et al. Lipo-gamma-AApeptides as a new class of potent and broad-spectrum antimicrobial agents. J. Med. Chem. 55, 4003–4009 (2012).
Li, Y. et al. Lipidated cyclic γ-AApeptides display both antimicrobial and anti-inflammatory activity. ACS Chem. Biol. 9, 211–217 (2014).
Cui, H., Zhang, X., Zhou, H., Zhao, C. & Lin, L. Antimicrobial activity and mechanisms of Salvia sclarea essential oil. Bot. Stud. 56, 1–8 (2015).
Mendes, C. R. et al. Antibacterial action and target mechanisms of zinc oxide nanoparticles against bacterial pathogens. Sci. Rep. 12, 2658 (2022).
Francius, G., Domenech, O., Paule, M., Mingeot-Leclercq, P. & Dufrene, Y. F. Direct observation of Staphylococcus aureus cell wall digestion by lysostaphin. J. Bacteriol. 190, 7904–7909 (2008).
Weakley, S. M. et al. Role of somatic mutations in vascular disease formation. Expert. Rev. Mol. Diagn. 10, 173–185 (2010).
Sloczynska, K., Powroźnik, B., Pękala, E. & Waszkielewicz, A. M. Antimutagenic compounds and their possible mechanisms of action. J. Appl. Genet. 55, 273–285 (2014).
Kaur, S., Arora, S., Kaur, K. & Kumar, S. The in vitro antimutagenic activity of Triphala an Indian herbal drug. FCT 40, 527–534 (2002).
Aqil, F. et al. Antimicrobial, antioxidant, and antimutagenic activities of selected marine natural products and tobacco cembranoids. Drug Chem. Toxicol 34, 167–179 (2011).
Bouguellid, G. et al. Antimutagenic, antigenotoxic, and antiproliferative activities of Fraxinus angustifolia Vahl. leaves and stem bark extracts and their phytochemical composition. PLoS ONE 15, 0230690 (2020).
Kaur, T., Kaur, A., Sharma, V. & Manhas, R. K. Purification and characterization of a new antifungal compound 10-(2, 2-dimethyl-cyclohexyl)-6, 9-dihydroxy-4, 9-dimethyl-dec-2-enoic acid methyl ester from Streptomyces hydrogenans strain DH16. Front. Microbiol. 7, 1004 (2016).
Sharma, M. & Manhas, R. K. Purifcation and characterization of salvianolic acid B from Streptomyces sp. M4 possessing antifungal activity against fungal phytopathogens. Microbiol. Res. 237, 126478 (2020).
Kaur, N., Arora, D. S., Kalia, N. & Kaur, M. Bioactive potential of endophytic fungus Chaetomium globosum and GC–MS analysis of its responsible components. Sci. Rep. 10, 1–10 (2020).
Russo, A. et al. Bioflavonoids as antiradicals, antioxidants, and DNA cleavage protectors. CBT 16, 91–98 (2000).
Kaur, R., Arora, S. & Singh, B. Antioxidant activity of the phenol rich fractions of leaves of Chukrasia tabularis A. Juss. Bioresour. Technol. 99, 7692–7698 (2008).
Kaur, J. & Arora, S. Actinobacteria from soil as potential free radical scavengers. Malay J. Microbiol. 217–227 (2017).
Rani, R., Arora, S., Kaur, J. & Manhas, R. K. Phenolic compounds as antioxidants and chemopreventive drugs from Streptomyces cellulosae strain TES17 isolated from the rhizosphere of Camellia sinensis. BMC Complement. Altern. Med. 8, 1–15 (2018).
Thenmozhi, M. & Kannabiran, K. Studies on isolation, classification and phylogenetic characterization of novel antifungal Streptomyces sp. VITSTK7 in India. Curr. Res. J. Biol. Sci. 2(5), 306–312 (2010).
Karthik, L., Kumar, G. & Rao, K. V. B. Antioxidant activity of newly discovered lineage of marine actinobacteria. Asian Pac. J. Trop. Med. 6, 325–332 (2013).
Taechowisan, T., Samsawat, T., Puckdee, W. & Phutdhawong, W. S. Cytotoxicity and antibacterial activities of crude extract of Streptomyces sp. W08, an endophyte of Amomum krervanh Pierre. J. Appl. Pharm. Sci. 11, 134–138 (2021).
Saraswathi, K. et al. In vitro biological properties of Streptomyces cangkringensis isolated from the floral rhizosphere regions. Saudi J. Biol. Sci 27, 3249–3257 (2020).
Bhat, M. P., Nayaka, S. & Kumar, R. S. A swamp forest Streptomyces sp. strain KF15 with broad spectrum antifungal activity against chilli pathogens exhibits anticancer activity on HeLa cells. Arch. Microbiol. 204, 540 (2022).
Bhat, M. P. & Nayaka, S. Cave soil Streptomyces sp. strain YC69 antagonistic to chilli fungal pathogens exhibits in vitro anticancer activity against human cervical cancer cells. Appl. Biochem. Biotechnol. 1–24 (2023).
Acknowledgements
This work was financially supported by RUSA 2.0 Rashtriya Uchchatar Shiksha Abhiyan, MHRD, India.
Funding
The funding was provided by Indian Council of Medical Research (No. AMR/Fellowship/18/2019-ECD-II), University Grants Commission (No. 43-468/2014(SR) dated 24/9/15).
Author information
Authors and Affiliations
Contributions
R.K.M. designed and planned the research work, analyzed and interpreted the data, and drafted and critically edited the manuscript for intellectual content. J.V. planned and executed the research work, analyzed and interpreted the data, and wrote the manuscript under the guidance of the research supervisor. M.S. analyzed data related to pure compound characterization and assisted in drafting the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in the order of the Figures. Figure 2 was published as figure 3, figure 3 was published as figure 4, figure 4 was published as figure 5, figure 5 was published as figure 6, figure 6 was published as figure 7 and figure 7 was published as figure 2.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Verma, J., Sharma, M. & Manhas, R.K. Purification and characterization of an antimicrobial compound against drug-resistant MRSA and VRE produced by Streptomyces levis strain HFM-2. Sci Rep 15, 26016 (2025). https://doi.org/10.1038/s41598-025-10572-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10572-3