Abstract
The copper-metabolism Murr1 domain (COMMD) protein family plays an essential role in tumours and the immune system. However, the multi-omics characterisation of COMMD family genes and their role in tumour patients has yet to be explored. We collected data from 33 types of cancer and conducted a comprehensive analysis of the expression abnormalities, diagnostic and prognostic roles, subcellular localisation, pathway enrichment, immune microenvironment and immune checkpoint associations of COMMD family genes in these diseases. We also predicted patient responses to immunotherapy using ICIs. Finally, we confirmed the role of COMMD7 in ccRCC and bladder cancer through in vivo and in vitro experiments. We found differential expression and diagnostic biomarker value of the COMMD family of proteins in pan-cancer, and also found that they play a key role in the tumour microenvironment. COMMD proteins are closely related to common immune checkpoints, TMBs and MSIs, and COMMD family proteins can predict immunotherapy response in patients with various cancers. Finally, knockdown of COMMD7 was found to inhibit the progression of ccRCC and bladder cancer cells, as verified by in vivo and in vitro experiments. Our study highlights the great potential of COMMDs as prognostic and immunotherapeutic response biomarkers, which could pave the way for further research into tumour infiltration mechanisms and the therapeutic potential of COMMDs in cancer.
Similar content being viewed by others
Introduction
Cancer is a major global public health problem as it is the leading cause of related deaths worldwide1. Most of the genomic alterations interacting with the tumour microenvironment and their role in immunotherapeutic response are currently unexplored, and there is a great deal of genetic variation across tumour types as well as between individual tumours2,3,4.
Copper-metabolism Murr1 domain proteins (COMMDs) consist of 10 members, that regulate cellular signalling through endosomal transport regulates cell signalling5. Members of the COMMDs family have been reported to be involved in the regulation of a variety of tumourigenesis and tumor microenvironment6,7,8. For example, COMMD1 protein is associated with the regulation of copper homeostasis and affects the immune response, cell growth and cell cycle in cancer cells9,10. COMMD3 and COMMD8 are essential for B-cell migration11. Currently, studies have shown that COMMDs are involved in the progression of multiple tumours and can be used as a prognostic biomarker for a variety of tumours, but their systematic studies in pan-cancer have not been elaborated. systematic studies have not been elaborated.
In this study, we explored the differential expression and diagnostic biomarker value of the COMMDs family in pan-cancer. COMMDs were also found to play a key role in pan-cancer in relation to the tumour microenvironment. Finally, it was verified by in vivo and in vitro experiments that knockdown of COMMD7 inhibited the proliferation and invasion of renal clear cell carcinoma and bladder cancer cells.
Materials and methods
Data sources
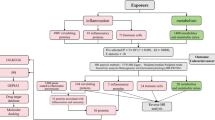
We used TCGA database and UCSC Xena public datasets to download data from normal and tumour tissue samples for 33 cancer types, which included transcript expression and clinical information.
Diagnostic and prognostic value analysis
ROC graphs can reflect the relationship between sensitivity and specificity. The diagnostic value of the COMMD family of genes was estimated using the ‘pROC’ package to draw ROC curves for visualisation.
Information related to the prognosis of pan-cancer patients was downloaded from the UCSC Xena database. We positioned risk factors with HR > 1 and protective factors with 0 < HR < 1 of the analysed results.
GSEA and immune infiltration analysis
We downloaded the gene sets of HALLMARK cancer-associated pathways and GO signalling pathways in the MSigDB database, which were used to assess the expression of each COMMD gene correlation with cancer-related pathways12.
The correlation between the expression of COMMD family genes and the level of immune cell infiltration was downloaded from the TISIDB database (http://cis.hku.hk/TISIDB/), and the correlation between COMMDs genes and immune cell infiltration.
Immunotherapy response
We calculated the association between each member of the COMMD family and the level of immune cell infiltration in each of the tumour microenvironments as well as common immunotherapy biomarkers.
Cell culture
Human renal clear cell carcinoma 786-O and ACHN and bladder cancer cell lines T24 and UM-UC3 were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences and used.786-O, ACHN, T24 and UM-UC3 cells were cultured in an incubator at 37 °C, 5% CO2 with medium containing 10% fetal bovine serum.
Plate cloning
From 786-O, ACHN, T24 and UM-UC3 cells in logarithmic growth phase, cell suspensions were inoculated in 6-well plates with 800 cells per well. The plates were placed in an incubator at 37 °C with 5% CO2 for 14 days. Cells were fixed using 4% paraformaldehyde and then stained with 1% crystal violet stain.
Wound-healing experiment
Use a gun tip to make cell scratches, the direction of the scratches is perpendicular to the marking line, and choose the appropriate time point to take pictures, such as 0 h and 48 h after removing the cells to take pictures under the microscope.
Transwell experiment
Appropriate amount of cell suspension was taken and added into a small chamber, 600μL of complete medium containing 10% FBS was added into the lower chamber of a 24-well culture plate, and the plate was incubated in a CO2 incubator at 37℃ for 24 h, and the cells were fixed with paraformaldehyde after removing the small chamber, and then stained with crystal violet solution, and finally photographed under a microscope.
Xenograft experiments
Xenograft experiments in nude mice were conducted at the Experimental Animal Center of Hunan University. The nude mice were purchased from Jiangsu Huachuang Xinnuo Pharmaceutical Science and Technology Co Ltd. 10 mice were randomly divided into two groups. Each mouse in the control group was injected 786-O cells, and each mouse in the knockdown group was injected 786-O cells stably transfected with COMMD7 knockdown lentivirus. After 30 days of intervention, the nude mice were deeply anesthetized with inhaled isoflurane (1–2% volume) and euthanized by cervical dislocation. The xenograft experiments were approved by the Animal Care and Use Committee of the Ethics Committee of The Affiliated Hospital of Hunan University. All animal experiments and experimental procedures were conducted in accordance with the guidelines of the Ethics Committee of The Affiliated Hospital of Hunan University, and adhered to the recommendations of the National Institutes of Health as well as the ARRIVE Guidelines (https://arriveguidelines.org).
Statistical analysis
P-values less than 0.05 were considered statistically significant. Image J and GraphPad software were used for experimental data processing and plotting.
Results
Expression of COMMDs family genes in human normal tissues and cancers
By collecting 33 cancers from the TCGA database and the GTEx database as well as the corresponding normal tissues, we went on to analyse the differences in the expression of each member of the COMMDs family in pan-cancer. We found that the COMMD1, COMMD2, COMMD4, COMMD5, COMMD7, COMMD8, and COMMD9 genes were significantly highly expressed in most of the tumours compared to the corresponding normal tissues, whereas the expression of COMMD6 was significantly reduced in most of the tumours (Fig. 1).
Diagnostic and prognostic value of COMMD family genes in pan-cancer
The value of COMMD family genes as diagnostic biomarkers was further assessed by ROC curves in pan-cancer. For example, COMMD5 in BRCA, COMMD4 and COMM4 in CESC, each COMMD family member (COMMD1-10) in CHOL, COMMD2, COMMD4 and COMMD5 in ESCA, COMMD2 and COMMD4-10 in GBM, COMMD5 in HNSC, COMMD1-2, COMMD6-7 and COMMD9 in KICH and COMMD2-5, COMM7 and COMMD9 in LIHC had AUC values greater than 0.9, suggesting that these COMMD family genes have high diagnostic value for the above cancers (Fig. 2A). Therefore, it can be inferred that abnormally expressed COMMD family members can be used as diagnostic biomarkers for cancer.
Diagnostic and prognostic analysis of COMMD family genes. (A) Diagnostic ROC curves of COMMD family genes in pan-cancer. an AUC > 0.9 is considered a high diagnostic value, 0.9 ≥ AUC > 0.7 is an intermediate diagnostic value, and 0.7 ≥ AUC > 0.5 is a low diagnostic value. (B) Results of DFI, DSS, OS and PFI prognostic analysis of COMMD family genes in pan-cancer.
Prognostic analyses, including uniCox regression and Log-rank analysis, were performed by each COMMD family gene (COMMD1-10) in pan-cancer, which in turn assessed the value of each COMMD family gene as a prognostic biomarker in pan-cancer. The results showed that COMMD1 was protective in KIRC but risky in KICH, LGG, STAD and UVM, COMMD2 was protective in KIRC but risky in ACC, LIHC and SARC, COMMD3 was protective in BLCA and LUSC but risky in ACC and LIHC, COMMD4 is hazardous in ACC, COMMD5 is hazardous in MESO and UVM, COMMD6 is protective in LGG and UVM and hazardous in ACC and HNSC, COMMD7 is hazardous in ESCA, HNSC, LGG, LIHC, PAAD and UVM, COMMD8 is hazardous in KIRC and dangerous in HNSC, LIHC, and MESO, COMMD9 was protective in KIRC and dangerous in KICH, and COMMD10 was protective in KIRC and dangerous in ESCA, HNSC, LGG, and STAD (Fig. 2B).
Subcellular localisation and and functional enrichment analysis of COMMD family members
The HPA database was used to obtain immunofluorescence images of COMMD family member proteins to clarify the subcellular localisation of COMMD member proteins. The results showed that COMMD1, COMMD5 and COMMD8 proteins were located in Nucleoplasm and Cytosol, COMMD2 and COMMD7 proteins were located in Vesicles, COMMD4 was located in Vesicles, Plasma membrane and Cytosol, COMM9 was located in Nucleoplasm, Golgi apparatus and Cytosol, and COMMD10 is located in Nucleoplasm (Fig. 3A).
Protein localisation and functional enrichment analysis of COMMD family members. (A) Immunofluorescence images of COMMD family member proteins. (B) Network map of COMMD family members and other related role proteins. (C-D) Heatmap of enrichment of each COMMD family member in Hallmark signalling pathway.
We first constructed a network map through the COMMDs family members and other related role proteins (Fig. 3B). This was further followed by gene set enrichment analysis to elucidate the role of COMMDs in cancer progression. We found that gene sets were mainly enriched in the HIF-1 signalling pathway, Ubiquitin mediated proteolysis, NF-kappa B signalling pathway and cullin-RING ubiquitin ligase complex signalling pathway (Fig. 3C-D).
Association of COMMD family genes with tumour microenvironment
By correlating the COMMD family genes with the degree of infiltration of various immune cells in the pan-cancer dataset, it was found that COMMD1, COMMD8, and COMMD9 showed a positive correlation with the majority of immune cells in a variety of tumours, whereas COMMD2 and COMMD6 showed a negative correlation with the degree of infiltration of a variety of immune cells in a variety of tumours (Fig. 4A). By further investigating the correlation between the expression of common immune checkpoints and the COMMD family of genes in a variety of tumours, we found that COMMD2 showed a significant positive correlation with CTLA4, PD-1 and PD-L1 in most cancers, while COMMD6 showed a significant negative correlation with CTLA4, PD-1 and PD-L1. In addition, each COMMD family gene was significantly positively associated with common immune checkpoints in UVM and LGG (Fig. 4B-D). Suggesting that our COMMD family genes may have an important impact on immunotherapy.
Association between the COMMD family and immunotherapy
We assessed the correlation between each member of the COMMD family and TMB in pan-cancer and found that COMMD1 showed significant positive correlation with TMB in BRCA and LIHC, and significant negative correlation with TMB in SARC and THYM (Fig. 5A). COMMD7 was positively correlated with MSI in DLBC, HNSC, LIHC, PRAD, SKCM, and TGCT, whereas in COAD, GBM, and LUSC it was negatively correlated with with MSI.COMMD10 in ACC, READ, and UCEC was positively correlated with MSI in BLCA, BRCA, DLBC, GBM, HNSC, LUAD, LUSC, OV, PRAD and TGCT (Fig. 5B).
We subsequently evaluated the role of the COMMD family in immunotherapy by immunotherapy data, which showed that in Mammary cancer (EMT6) COMMD1 expression was significantly elevated in the anti-PD-L1 treatment response group compared to baseline levels. In addition, the expression level of COMMD3 was significantly elevated in Mammary cancer (EMT6) and Colorectal carcinoma (CT26) compared to baseline (Fig. 5 C).
Knockdown of COMMD7 inhibits the proliferation and migration of renal clear cell carcinoma and bladder cancer cells
To investigate the role of COMMD7 in renal clear cell carcinoma and bladder cancer cells, we designed two shRNAs (COMMD7-1, COMMD7-2) to silence COMMD7 expression in renal clear cell carcinoma (786-O and ACHN) and bladder cancer cells (T24 and UM-UC3). We performed plate cloning, Transwell and wound healing assays on renal clear cell carcinoma (786-O and ACHN) and bladder cancer cells (T24 and UM-UC3) transfected with COMMD7 lentivirus, respectively. The results of plate cloning showed that the proliferative capacity of renal clear cell carcinoma (786-O and ACHN) and bladder cancer cells (T24 and UM-UC3) after transfection with COMMD7 lentivirus was significantly lower than that of the NC group (Fig. 6A-B).The results of the Transwell assay and wound healing assay indicated that the knockdown of COMMD7 significantly reduced the proliferative capacity of renal clear cell carcinoma (786-O and ACHN) and bladder cancer cells (T24 and UM-UC3) in the NC group, as compared with that of the COMMD7 lentivirus. O and ACHN) and bladder cancer cells (T24 and UM-UC3) had significantly reduced migratory ability after knockdown of COMMD7 (Fig. 6C-F). The above results indicated that COMMD7 could promote the proliferation and migration of renal clear cell carcinoma and bladder cancer cells in vitro. To further explore the oncogenic role of COMMD7 in vivo, a subcutaneous tumour model was established. Knockdown of COMMD7 significantly delayed tumour progression (Fig. 6G-I).
COMMD7 promotes growth and metastasis of ccrcc and bladder cancer cells in vivo and in vitro. Plate cloning assay (A-B), transwell cell migration ability (C-D), and wound healing ability assay (E–F) of renal clear cell carcinoma (786-O and ACHN) and bladder cancer cells (T24 and UM-UC3) transfected with COMMD7 lentivirus. (G-I) Knockdown of COMMD7 significantly inhibited tumour growth, such as tumour growth rate, tumour weight. Note * p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
The current study found that various members of the COMMD family play key roles in cancer13. We comprehensively analysed the expression abnormalities, diagnostic and prognostic roles, correlation with cancer-related pathways, association with the immune microenvironment and immune checkpoints, and prediction of patient immunotherapeutic response to ICI and targeted small molecule drugs in cancer by using multi-omics pan-cancer data for each gene of the COMMD family. We found that COMMD family gene expression levels are specific and vary across many cancer types. For example, COMMD1, COMMD2, COMMD4, COMMD5, COMMD7, COMMD8, and COMMD9 genes are significantly highly expressed in most tumours, whereas COMMD6 is significantly less expressed in most tumours.Various members of the COMMD family (COMMD1-10) have high diagnostic value in CHOL.Various COMMD family members have significant correlation with immune checkpoints, implying that the COMMD family has important research value in the direction of immunotherapy. Finally, we verified that knockdown of COMMD7 inhibited the proliferation and invasion of renal clear cell carcinoma and bladder cancer cells by in vivo and in vitro experiments.
Major breakthroughs in tumour therapy by immunotherapy have led to new directions in tumour treatment, however, the complexity of the tumour microenvironment responds to immunotherapy differently from tumour to tumour14. Understanding the tumour microenvironment is more helpful in identifying new effective immunotherapy targets6,7,8,15. Researchers have found that hypoxia-induced COMMD1 regulates signalling and metabolism in human macrophages and thus inhibits osteoclastogenesis16. COMMD8 is a signalling articulator for chemoreceptors, and COMMD3 and 8 form a protein complex that can affect humoral immune responses and autoimmunity13.
COMMD family members can serve as therapeutic targets as well as diagnostic and prognostic indicators, according to a comprehensive analysis conducted in this work. Our study does, however, still have certain shortcomings. First, there are a number of biases, including the retrospective design, the limited number of patients, and the mixed usage of tumor specimens that had preoperative chemotherapy and those that did not. To further confirm the veracity of our findings, a sizable cohort of clinical cases must yet be gathered and evaluated. In this study, we used one-way cox regression analysis to determine whether COMMD family members were associated with survival in pan-cancer patients, but one-way cox regression analysis can only assess the effect of a single factor and cannot consider the effects of multiple factors at the same time and the use of multifactorial cox regression analysis is limited by factors such as data availability or study design. Ultimately, further in vivo and in vitro investigations are required in subsequent research to confirm the role of COMMD family member genes in cancer and to methodically identify the mechanism by which COMMD family members operate as therapeutic targets and cancer biomarkers.
Conclusion
In this study, we explored the differential expression and diagnostic biomarker value of the COMMDs family in pan-cancer. COMMDs were also found to play a key role in pan-cancer in relation to the tumour microenvironment.COMMDs are closely related to immunotherapy. Small molecule compounds that can target COMMD9, COMMD3 and COMMD8 protein complex proteins were found by molecular docking. We verified that knockdown of COMMD7 inhibited the proliferation and invasion of renal clear cell carcinoma and bladder cancer cells by in vivo and in vitro experiments. In this study, we analysed the role and feasibility of COMMD family members as diagnostic and prognostic biomarkers and therapeutic targets through a multi-omics system, providing new targets for future cancer drug therapy.
Data availability
All data utilized in this study are included in this article and all data supporting the findings of this study are available on reasonable request from the corresponding author.
References
Sanchez-Vega, F. et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 173(2), 321–37.e10 (2018).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502(7471), 333–339 (2013).
Xu, Y. et al. The role of circular RNA during the urological cancer metastasis: Exploring regulatory mechanisms and potential therapeutic targets. Cancer Metastasis Rev. 43(3), 1055–1074 (2024).
Xu, Y. et al. Identification of PANoptosis-related signature reveals immune infiltration characteristics and immunotherapy responses for renal cell carcinoma. BMC Cancer 24(1), 292 (2024).
Laulumaa S, Varjosalo M. Commander Complex-A multifaceted operator in intracellular signaling and cargo. Cells. 10(12) 2021
Ye, B. et al. Navigating the immune landscape with plasma cells: A pan-cancer signature for precision immunotherapy. BioFactors (Oxford, England). 51(1), e2142 (2025).
Sun, W. et al. Systemic immune-inflammation index predicts survival in patients with resected lung invasive mucinous adenocarcinoma. Transl. Oncol. 40, 101865 (2024).
Ye, B. et al. iMLGAM: Integrated machine learning and genetic algorithm-driven multiomics analysis for pan-cancer immunotherapy response prediction. iMeta. 4(2), e70011 (2025).
Tai, P. et al. Multi-omics analysis of the oncogenic value of copper Metabolism-Related protein COMMD2 in human cancers. Cancer Med. 12(10), 11941–11959 (2023).
Devlin, A. M. et al. HCaRG is a novel regulator of renal epithelial cell growth and differentiation causing G2M arrest. Am. J. Physiol. Renal Physiol. 284(4), F753–F762 (2003).
Nakai, A. et al. The COMMD3/8 complex determines GRK6 specificity for chemoattractant receptors. J. Exp. Med. 216(7), 1630–1647 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 53(D1), D672–D677 (2025).
Shirai, T. et al. Celastrol suppresses humoral immune responses and autoimmunity by targeting the COMMD3/8 complex. Sci. Immunol. 8(81), eadc9324 (2023).
Li, H., Xu, Y., Shan, J. & Lun, Y. Pan-cancer landscape of DCTPP1 and preliminary exploration of DCTPP1 in renal clear cell carcinoma. Sci. Rep. 14(1), 27710 (2024).
Xu, Y. et al. Integration of multi-omics and clinical treatment data reveals bladder cancer therapeutic vulnerability gene combinations and prognostic risks. Front. Immunol. 14, 1301157 (2023).
Murata, K. et al. Hypoxia-sensitive COMMD1 integrates signaling and cellular metabolism in human macrophages and suppresses osteoclastogenesis. Immunity 55(11), 2209 (2022).
Funding
This work was supported by the Scientific research project of Xiangtan Medical Association (2024-xtyx-33), Natural Science Foundation of Hunan Province (2024JJ9559) and Supporting project of China Medical and Health Care Development Foundation (chmdf2024-xrzx08-13).
Author information
Authors and Affiliations
Contributions
SHW: formal analysis, data curation, conceptualization, writing—original draft. JLC: formal analysis, visualization, writing—original draft. ZYH: software, investigation, writing—original draft. HJZ: formal analysis, visualization, software, investigation. LFY: software, investigation, writing—original draft. JYL: formal analysis, visualization, software, investigation. YYD: formal analysis, visualization, software, investigation. YH: writing—review and editing, supervision, project administration, funding acquisition. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, S., Cheng, J., Hu, Z. et al. Bioinformatics analysis of COMMD family in pan-cancer reveals potential biomarkers and therapeutic targets. Sci Rep 15, 25490 (2025). https://doi.org/10.1038/s41598-025-11118-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-11118-3