Abstract
Epidemiological evidence suggests an association between dioxin and dioxin-like compound (DLC) exposure and human liver disease. In rodents, the prototypical DLC, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), has been shown to induce the progression of reversible hepatic steatosis to steatohepatitis with periportal fibrosis and biliary hyperplasia. Although the effects of TCDD are mediated by aryl hydrocarbon receptor (AHR) activation, the underlying mechanisms of induced pathologies have not been resolved. In the present study, male C57BL/6NCrl mice were gavaged every 4 days for 28 days with 0.03–30 μg/kg TCDD or sesame oil vehicle and evaluated for liver histopathology and gene expression as well as complementary 1-dimensional proton magnetic resonance (1-D 1H NMR) urinary metabolic profiling. Urinary trimethylamine (TMA), trimethylamine N-oxide (TMAO), and 1-methylnicotinamide (1MN) levels were altered at doses of ≤ 3 μg/kg TCDD; other urinary metabolites, such as glycolate, urocanate, and 3-hydroxyisovalerate, were only altered following the induction of moderate to severe steatohepatitis. Hepatic differential gene expression of rate-limiting enzymes of choline, gloxylate, and amino acid metabolism coincided with the altered urinary metabolites. Published single-nuclear RNA-seq (snRNA-seq), AHR ChIP-seq, and AHR knockout gene expression datasets provided further support for hepatic cell-type and AHR-regulated disruption of the affected metabolic pathways.
Similar content being viewed by others
Introduction
Metabolic dysfunction-associated steatotic liver disease (MASLD) includes a spectrum of liver pathologies, from simple and reversible hepatic steatosis to steatohepatitis with fibrosis in the absence of alcohol consumption, viral infection, or lipodystrophy that increases the risk for diabetes, end-stage liver disease, and hepatocellular carcinoma (HCC)1,2,3,4. Genetics, lifestyle, and diet are commonly cited as causal factors. Yet, epidemiological studies have suggested MASLD and associated pathologies can also be linked to environmental pollutant exposures5,6. One such class of compounds, polychlorinated dibenzo-p-dioxins (PCDDs), such as the prototypical congener 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), and dioxin-like compounds (DLCs), are environmental pollutants, which have been associated with steatotic liver disease (SLD) and hepatotoxicity in humans7,8,9,10,11,12,13.
TCDD toxicity is mediated by the ligand-activated transcription factor, the aryl hydrocarbon receptor (AHR)14,15,16. The AHR is a member of the basic-helix-loop-helix (bHLH) Per-Arnt-Sim (PAS) transcription factor family14,15. Although it is promiscuous and binds many structurally diverse ligands with varying affinities, no single eminent physiological ligand has been identified17,18. Upon ligand binding, the AHR translocates to the nucleus where it heterodimerizes with the AHR nuclear translocator (ARNT). Canonically, the AHR-ARNT dimer binds to dioxin response elements (DREs) within the locus of target genes and recruits coactivators and RNA Polymerase II to elicit species, sex, age, tissue, and cell-specific differential gene expression14. However, AHR-mediated differential gene expression has also been shown to involve DRE-independent regulation5,19 and interactions with other transcription factors20.
In male mice, TCDD induces dose-dependent hepatic steatosis that can progress to steatohepatitis with periportal fibrosis and biliary hyperplasia13,21,22. Yet, the underlying pathways disrupted due to AHR-mediated primary and secondary gene expression effects following treatment with TCDD are poorly understood23,24,25. Moreover, in human populations, correlation of TCDD exposure with endogenous metabolite changes has identified various disrupted pathways but without the identification of the responsible tissues or cell type-specific gene expression responses26. Previous studies on tissue extracts have integrated complementary hepatic gene expression and metabolomic analyses to investigate SLD progression in relation to disrupted metal, carbohydrate, lipid, and amino acid metabolism21,22,23,27,28,29,30,31,32,33,34,35,36 but were limited by metabolite validation, method-dependent extraction, and chromatographic separation of specific metabolites within complex matrix mixtures. In this study, we tested the hypothesis that 1-dimensional proton magnetic resonance (1-D 1H NMR) could identify tissue-specific disrupted pathways responsible for TCDD elicited changes in urinary metabolite profiles. Although less sensitive than mass spectrometry, 1-D 1H NMR can simply, simultaneously, and confidently identify and measure hundreds of small molecules in complex matrices. Therefore, we collected urine samples from mice dose-dependently treated with TCDD at levels that spanned human accidental exposures to an intentional poisoning22,37. 1-D 1H NMR profiles were integrated with complementary liver histopathology and gene expression data to identify tissue-specific pathways disrupted by TCDD that contributed to changes in urinary metabolite levels.
Results
TCDD-elicited effects
The dose range and treatment regimen used in this study (Supplementary Fig. 1) have been previously shown to elicit dose- and time-dependent liver pathologies in mice, including the progression of steatosis to steatohepatitis with periportal fibrosis and biliary hyperplasia13,21,22,38,39. Chow consumption was not changed in any dose group (Supplementary Fig. 2) as previously reported34. Euthanasia and tissue collection occurred between ZT 0–3 to control for changes due to diurnal regulation including oscillating liver weight34,40,41. TCDD dose-dependently increased absolute and relative liver weights along with a decrease in body weight at 30 μg/kg21,34,35. There was also a dose-dependent increase in hepatic steatosis, hepatocellular hypertrophy, mixed inflammatory cell infiltration (mainly monocytes and lymphocytes with lesser numbers of neutrophils), and biliary hyperplasia, especially at ≥ 10 μg/kg TCDD (Supplementary Fig. 3 and Table 1). A slight increase in hepatic necrosis (Table 1) was consistent with the modest increase in serum ALT35. Overall, TCDD did not elicit overt systemic toxicity or hepatotoxicity but dose-dependently induced steatohepatitis with evidence of biliary hyperplasia at 30 μg/kg TCDD.
1-D 1H NMR urinary metabolite profiling
In addition to TCDD-induced hepatic pathologies, treatment is reported to cause AHR-mediated metabolic reprogramming and alter metabolite levels across various tissues. 1-D 1H NMR analysis of urine (n = 5 per dose) collected on post-natal day (PND) 55 (i.e., on day 25 of 28-day study; 1 day after the last gavage) between ZT 0–3 identified 107 unique metabolites in mouse urine samples (Supplementary Table 1).
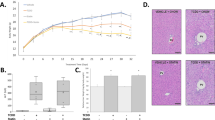
Creatinine normalization (CN) is commonly used for evaluating the relative level of urinary metabolites as urinary creatinine is assumed to be constant within individuals over time and across individuals, but normalizing to creatinine alone may under- or overestimate the levels of metabolites of interest42. Other methods for resolving the orders of magnitude difference in urinary metabolite levels, such as probabilistic quotient normalization (PQN) have also been developed for 1-D 1H NMR43. Principal component analysis (PCA) was used to evaluate creatinine normalization, PQN, or the combination to identify the best approach to resolve the dose-dependent separation of urine metabolites along PC1. Non-normalized urinary creatinine did not show a dose-dependent trend (Supplementary Fig. 4). PC1 from non-normalized urinary metabolite profile data was dominated by noise that accounted for 47.5 percent explained variance (PEV), while PC2 showed dose-dependent separation of urinary profiles at ≥ 10 μg/kg TCDD (PEV = 8.2%) (Supplementary Fig. 4). Similarly, PCA analyses of urinary profiles after CN (Supplementary Fig. 4) or PQN (Supplementary Fig. 4) were dominated by outliers for PC1 (PEVCN = 46.6% and PEVPQN = 24.4%) while PC2 showed dose-dependent separation. CN of urinary profiles followed by PQN (Supplementary Fig. 4) provided the best resolution along PC1 (PEVCPQN = 15.48%) with separation of samples occurring due to altered glycolate, pyruvate, succinate, urocanate, and 2-hydroxyisovalerate (2HIV) at ≥ 10 μg/kg (Supplementary Fig. 4). Therefore, statistical analyses of urinary metabolite levels were conducted on metabolite levels first normalized to creatinine and then subject to PQN. The levels of succinate, methionine, trimethylamine (TMA), trimethylamine N-oxide (TMAO), and 1-methylnicotinamide (1MN) were altered below 10 μg/kg TCDD (Fig. 1A). Other urinary metabolites were only altered at 10 or 30 μg/kg TCDD, including pyruvate, tyrosine, glutamic acid, 2HIV, 3-hydroxyisovalerate (3HIV), 2-hydroxyvalerate, methylsuccinate, methylamine (MA), butyrate, isobutyrate, formate, and glycolate (Fig. 1A). Of the altered metabolites, the median lower (BMDL10%) to upper bound (BMDU10%) benchmark dose responses ranged from 3.2–9.0 μg/kg. This included 1MN (BMDU10%: 2.3 μg/kg), N,N’-dimethylglycine (DMG) (BMDU10%: 6.8 μg/kg), isobutyrate (BMDU10%: 8.1 μg/kg), tyramine (BMDU10%: 4.70 μg/kg), butyrate (BMDU10%: 8.79 μg/kg), TMAO (BMDU10%: 8.72 μg/kg), glycolate (BMDU10%: 9.26 μg/kg), 2HIV (BMDU10%: 8.06 μg/kg), and urocanate (BMDU10%: 8.34 μg/kg) (Fig. 1B).
TCDD-elicited changes in urinary metabolites. Urine was collected from individual mice (n = 5) one day before euthanasia on PND 57 between ZT0-3. Metabolites were normalized by creatinine levels followed by probabilistic quotient normalization. (A) Heatmap of dose-dependent changes in metabolite levels. The black triangle (upper right hand corner) indicates a significant (p ≤ 0.05) change from controls following Benjamini-Hochberg (BH) and post hoc Dunnett’s tests relative to urine from sesame oil gavaged mice. Maximum concentrations are colored according to low (0.25 μM/μM creatinine) and high concentrations (2 μM/μM creatinine). (B) Benchmark dose–response values for metabolites in (A) calculated using BMDExpress2 (v2.3). The 10% benchmark dose (BMD10%) was estimated with a 95% confidence interval to calculate a lower (BMDL10%) and an upper (BMDU10%) bound. The median BMDL10%, BMD10%, and BMDU10% of all significant metabolites (p value ≤ 0.05).
Integration of hepatic gene expression
To identify metabolic pathways responsible for the altered urinary metabolite levels following TCDD treatment, liver samples (n = 5 per dose) from the same study were examined by bulk liver RNA-seq. TCDD caused a dose-dependent shift of the bulk hepatic transcriptome at ≥ 1 μg/kg TCDD (Fig. 2A). Approximately, 6,722 genes in total were differentially expressed (DEGs) by at least one dose (Fig. 2B) (Supplementary Table 2). 2,983 DEGs had a BMD10% upper bound (BMDU10%) < 10 μg/kg TCDD (Fig. 2C), suggesting widespread differential gene regulation occurring at the same doses as minimal to mild inflammation and cell death but below the dose at which TCDD elicited marked steatohepatitis, fibrosis, and biliary hyperplasia. Functional analysis of DEGs with a BMDU10% below 10 μg/kg TCDD showed enrichment for metabolic pathways associated with TCDD-altered urinary metabolite levels, including the metabolism of pyruvate (KEGG:00,620), choline (KEGG:05,231), branched-chain amino acids (BCAAs) (KEGG:00,280), glyoxylate (KEGG:00,630), and tryptophan (KEGG:00,380) (Fig. 2D and Supplementary Table 3). For DEGs with a BMDU10% greater than 10 μg/kg TCDD, functional analysis enriched for some of the same pathways, including glutathione metabolism (KEGG:00,480) and cofactor biosynthesis (KEGG:01,240) (Supplementary Fig. 5 and Supplementary Table 4).
TCDD elicited dose-dependent bulk liver differential gene expression analysis. Mice gavaged every 4 days for 28 days with 0.03–30 μg/kg TCDD or sesame oil vehicle were euthanized on PND 58 between ZT0-3. Bulk liver RNA-seq was performed (n = 5). (A) Principal component analysis of all genes expressed in the liver. (B) Number of differentially expressed (IFold-ChangeI ≥ 1.5 and a P1(t) ≥ 0.8) induced and repressed genes at each dose. (C) The 10% benchmark dose (BMD10%) of all differentially expressed genes was estimated with a 95% confidence interval to calculate a lower (BMDL10%) and an upper (BMDU10%) bound using BMDExpress2 (v2.3). The red dashed line indicates 10 μg/kg TCDD. (D) KEGG pathways enriched for DEGs with a BMDU10% less than 10 μg/kg TCDD. The dashed red line indicates − log10(adjusted p value = 0.05).
Choline metabolism
Choline and carnitine are vitamin-like nutrients, which may be metabolized by gut microbiota to form TMA prior to their absorption44 (Fig. 3A and Supplementary Fig. 6). Urinary choline levels decreased 2.6-fold following TCDD treatment (Fig. 3B), while no change occurred in carnitine (Fig. 3C). No significant change was detected in mouse cecum in the copy number of relevant TMA-producing microbial enzymes, including cutC (EC: 4.3.99.4), grdB (EC: 1.21.4.4), or cntA (EC: 1.14.13.139) (Supplementary Fig. 6). Moreover, as with previous reports28,29, there was little evidence suggesting choline was redirected to phosphatidylcholine (PtdC) biosynthesis by the Kennedy pathway or the synthesis of betaine to support homocysteine remethylation to methionine (Supplementary Fig. 4). Phosphorylcholine levels were unaltered (Supplementary Fig. 6), while Chpt1, which encodes the final and rate-limiting step of PtdC biosynthesis, was repressed 2.5-fold (Supplementary Fig. 6). DMG is produced when betaine donates a methyl group for the regeneration of methionine catalyzed by BHMT and was reduced 1.7-fold by TCDD (Supplementary Fig. 6). Chdh and Aldh7a1, which respectively encode enzymes for the two consecutive oxidations of choline → DMG → betaine (undetected in urine), were induced 1.3-fold and decreased 2.0-fold, respectively (Supplementary Fig. 6). Considering AHR-binding at pDREs and the 20-fold reduction in Bhmt (Supplementary Fig. 6), the decrease in urinary DMG was likely due to hepatic AHR-mediated downregulation.
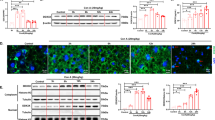
TCDD effect on urinary metabolites involved in choline metabolism. (A) The TMA pathway. Circles indicate metabolites. Arrows indicate enzymatic reaction. The numbers map to the genes in the heatmap (H), encoding the enzyme associated with the reaction. (B–G) Urinary metabolite levels as measured by 1-D 1HNMR (n = 5). An asterisk (*) indicates significance from a post hoc Dunnett test. Mean and standard deviation are shown. (H) Heatmap of hepatic TMA/TMAO pathway related genes. pDREs were determined by a position weight matrix with a Matrix Similarity Score (MSS) ≥ 0.856. Hepatic AHR ChIP-seq was detected in mouse 2 h after oral gavage with 30 μg/kg TCDD—green tiles indicate an FDR of ≤ 0.05 (GSE97634). In the dose response bulk RNA-seq gene expression, the black flags indicate a P1(t) ≥ 0.8. The Counts column refers to the maximum estimate of aligned reads of each gene where a lower level of expression (≤ 500 reads) is depicted in yellow and a higher level of expression (≥ 10,000) is depicted in pink. TMA Trimethylamine, TMAO Trimethylamine N-oxide, DMA Dimethylamine, MA Methylamine.
Metabolism of TMA in the gut and the liver leads to TMAO, dimethylamine (DMA), and methylamine with the possibility of TMA regeneration from TMAO (Fig. 3A). Urinary TMA decreased 8.0-fold (Fig. 3D), while TMAO increased as much as 5.0-fold at 3 μg/kg TCDD but fell to 2.0-fold at 30 μg/kg TCDD (Fig. 3E). Urinary DMA was unchanged (Fig. 3F), but methylamine was reduced 1.6-fold at 30 μg/kg TCDD (Fig. 3G). FMO3 exhibits the greatest TMA oxidizing activity and is not normally expressed in adult male mice45. However, 30 μg/kg TCDD induced Fmo3 5.3-fold at 8 h, 102.6-fold after 7 days, and > 600-fold after treatment every 4 days for 28 days (Fig. 3H). Moreover, AHR genomic binding was present at a putative dioxin response element (pDRE) at the Fmo3 transcription start site (TSS) 2 h after a bolus gavage of TCDD (Fig. 3H). Fmo3 induction was greatest in hepatocytes as observed with single nuclear (sn)RNA-seq (Supplementary Fig. 7) with FMO3 protein levels also exhibiting dose-dependent increases (Supplementary Fig. 7). Urinary TMA and TMAO levels were altered at doses as low as 3 μg/kg TCDD coincident with the dose required to initiate induction of FMO3. Bulk liver RNA-seq datasets from whole body Fmo3 knockout (KO) and wild-type (WT) mice treated with weekly injections of 25 μg/kg TCDD for 6 weeks concluded that Fmo3 induction was responsible for the increased plasma TMAO levels although whether FMO3 induction is relevant to TCDD hepatotoxicity is questionable (Supplementary Fig. 7)46,47. Furthermore, whole-body Ahr KO mice treated with a bolus of TCDD confirmed Fmo3 induction is AHR dependent (Supplementary Fig. 7)48. Basal expression of Fmo3 in adult male mice is minimal in all tissues except for lung. TCDD did not strongly induce Fmo3 in gonadal white adipose tissue, kidney, bone, or the intestinal tract (Supplementary Fig. 5). Moreover, in female, ovariectomized C57BL/6NCrl mice gavaged with TCDD every 4 days for 28 days, control mice had relatively high Fmo3 expression relative to males (Supplementary Table 5). Yet, TCDD dose-dependently induced Fmo3 4.4-fold in female liver (Supplementary Table 5), suggesting AHR-mediated changes in urinary TMA and TMAO are not specific to male mice. Therefore, increased FMO3 levels by TCDD in hepatocytes is likely the primary source of increased urinary TMAO and decreased urinary TMA levels with similar gene expression profiles in both sexes.
Because TMAO is a charged compound, several potential transporters including organic cation transporters and ATP-binding cassettes have been reported to export or import TMAO across cell membranes49. In TCDD-exposed mice, the most likely TMAO exporter from hepatocytes is ABCC4, which, located on the basolateral membrane, exports various molecules into the bloodstream. Its mRNA levels were induced 46.9-fold after oral gavage with 30 μg/kg TCDD every 4 days for 28 days (Fig. 3H), and the Abcc4 locus exhibited AHR genomic binding in the presence of a pDRE (Fig. 3H). It exhibited similar gene expression to Fmo3 in the time course with induction after a bolus TCDD dose as early as 8 h post-gavage (Fig. 3H).
TMAO has been reported to be a chemical chaperone that can alleviate fatty acid induced unfolded protein response (UPR) via ERN1 (also known as IRE1)50. TMAO can also activate EIF2AK3 (also known as PERK)51 to induce UPR. Excessive activation of the UPR leads to cell death52. Functional analysis of DEGs did not enrich for pathways associated with the UPR (Supplementary Tables 3–4). TCDD did not affect the gene transcription of Eif2ak3 or Atf4, a transcription factor associated with UPR, and did not affect the expression of Ddit353, an established marker of UPR (Supplementary Table 2), suggesting UPR was not activated. In addition, Ern1, a sensor of unfolded proteins in the endoplasmic reticulum (ER) that triggers UPR, was repressed 2.5-fold along with negative regulators of the ERN1-mediated UPR, such as Hspa5, Dnajb9, Pdia6, Tmbim6, Ddrgk1, and Ufl1 (1.9-, 1.7-, 2.0-, 2.1-, 1.9-, and 1.6-fold, respectively) (Supplementary Table 2). Although the bulk liver gene expression did not reflect a TCDD-induced UPR, the downregulation of Ern1 coinciding with increased TMAO suggests a potential interaction with TMAO and the ERN1 branch of the UPR.
Hydroxyproline and glyoxal metabolism
Urinary glycolate, a hydroxyproline and glyoxal metabolism product (Fig. 4A), was dose-dependently increased 7.4-fold by TCDD (Fig. 4B), while the related compound, serine, was increased in the urine 1.6-fold (Fig. 4C).
Hydroxyproline and glyoxal metabolism. (A) Hydroxyproline and glyoxal metabolism. (B, C) Urinary glycolate and L-serine levels measured by 1-D 1HNMR (n = 5). Data was normalized by creatinine and subsequently probabilistic quotient normalization. p values were calculated using a Kruskal–Wallis test and adjusted by the Benjamini–Hochberg method. An asterisk (*) indicates significance from a post hoc Dunnett test (p value ≤ 0.05). Mean and standard deviation are shown. (D, E) Heatmaps of hydroxyproline and glyoxal metabolism, respectively. pDREs were determined by a position weight matrix with a Matrix Similarity Score (MSS) cut off of ≥ 0.856 based on the sequence of characterized functional DREs. Hepatic AHR ChIP-seq was detected in mice 2 h after oral gavage with 30 μg/kg TCDD (GSE97634). The green tiles indicate an FDR of ≤ 0.05. In the dose response bulk RNA-seq gene expression, the black flags indicate a P1(t) ≥ 0.8. The Counts column refers to the maximum estimate of aligned reads of each gene where a lower level of expression (≤ 500 reads) is depicted in yellow and a higher level of expression (≥ 10,000) is depicted in pink. (F–H) Hepatic metabolite levels measured by GC–MS (n = 5). p values were calculated using a Kruskal–Wallis test. An asterisk (*) indicates significance from a post hoc Dunnett test. Mean and standard deviation are shown.
Hydroxyproline catabolism involves glyoxylate as an intermediate with subsequent metabolism to glycolate and oxalate (Fig. 4A). In mice gavaged every 4 days for 28 days with TCDD, Prodh2, which encodes the first catalytic step of hydroxyproline metabolism, was downregulated 5.0-fold (Fig. 4D). Subsequent reactions, encoded by Got2 and Hoga1, which produce glyoxylate, were repressed 2.1 and 4.0-fold, respectively (Fig. 4D). Grhpr, which converts glyoxylate to glycolate, was also downregulated 2.7-fold (Fig. 4D). In peroxisomes, HAO1 and AGXT convert glycolate into glycine. In the bulk liver RNA-seq data, Hao1 and Agxt were downregulated 222.7- and 6.5-fold respectively (Fig. 4D). Decreased expression of HAO1 and AGXT were confirmed by Western blot (Supplementary Fig. 8). Interestingly, no pDREs or AHR binding 2 h after a bolus gavage of TCDD were detected at the Hao1 locus (Fig. 4D). snRNA-seq data suggests the dose-dependent repression of Hao1 and Agxt primarily occurred in hepatocytes (Supplementary Fig. 8). Furthermore, Hao1 was not affected in a TCDD-treated whole-body Ahr KO model (Supplementary Fig. 8)48. Hepatic Hao1 in female, ovariectomized mice was also dose-dependently repressed 142.9-fold (Supplementary Table 5).
Glycolate is also formed by the detoxification of glyoxal which is associated with advanced glycation end-products (AGEs)54 unless metabolized into glycolate by Glo1 and Hagh, which were downregulated 5.0- and 1.9-fold, respectively (Fig. 4). Reductions in GLO1 protein and activity were confirmed by western blot (Supplementary Fig. 8) and an enzymatic assay (Supplementary Fig. 8). Glo1 and Hagh are primarily expressed in hepatocytes (Supplementary Fig. 8).
Although glycolate dose-dependently increased in the urine, hepatic glycolate and hydroxyproline levels were unchanged (Fig. 4F, G), despite a slight reduction in hepatic oxalic acid (Fig. 4H).
Vitamin B3 metabolism
Urinary 1MN, a product of nicotinamide adenine dinucleotide (NAD+) and NAD phosphate (NADP+) metabolism (Fig. 5A), is a biomarker of de novo NAD+ and NADP+ biosynthesis from tryptophan55,56. Although TCDD did not alter hepatic33, serum33, or urinary tryptophan levels (Fig. 5B), 1MN was dose-dependently deceased 10.5-fold at 30 μg/kg TCDD (Fig. 5C). Trigonelline also exhibited a decrease but did not achieve significance (Fig. 5D). Indoleamine 2,3-dioxygenase 2 (Ido2) and tryptophan 2,3-dioxygenase (Tdo2) catalyze the first and rate-limiting step in the conversion of tryptophan to nicotinamide. Hepatic Ido2 and Tdo2 were repressed 4.0- and 1.6-fold, while Ido1 expression was not detected (Fig. 5E). The expression of genes encoding subsequent steps including Afmid, Kmo, Kynu, Haao, and Qprt were reduced 10.0-, 5.0-, 4.8-, 8.3-, and 6.3-fold, respectively (Fig. 5E). Ido2, Afmid, Kmo, Haao, and Qprt all had AHR genomic binding in the presence of a pDRE (Fig. 5E) suggesting repression may be AHR-mediated. Nnmt, which methylates nicotinamide to form 1-methylnicotinamide, was not repressed (Fig. 5E). Cd38, which catalyzes the hydrolysis of NAD+57, was induced 3.4-fold (Supplementary Table 2). Nadk2, which encodes the mitochondrial enzyme that converts NAD+ to NADP+, was repressed 1.8-fold (Supplementary Table 2). In female, ovariectomized mice, hepatic Afmid, Kmo, Haao, and Qprt was repressed 4.6-, 1.5-, 2.0-, and 1.7-fold, respectively, following TCDD treatment (Supplementary Table 5).
Effect of TCDD on nicotinamide biosynthesis. (A) Nicotinamide biosynthesis pathway. (B–D) Urinary tryptophan, trigonelline, and 1-methylnicotinamide levels as measured by 1-D 1H NMR (n = 5). Data was normalized by creatinine and subsequently probabilistic quotient normalization. An asterisk (*) indicates significance from a post hoc Dunnett test (p ≤ 0.05). Mean and standard deviation are shown. (E) Heatmap of dose dependent effects of TCDD on gene expression associated with nicotinamide biosynthesis. pDREs were determined by a position weight matrix with a Matrix Similarity Score (MSS) cut off of ≥ 0.856 based on the sequence of characterized functional DREs. ChIP-seq analysis detected AHR genomic binding in mouse liver 2 h after oral gavage with 30 μg/kg TCDD (GSE97634). The green tiles indicate an FDR of ≤ 0.05. In the dose response bulk RNA-seq gene expression, the black flags indicate a P1(t) ≥ 0.8. The Counts column refers to the maximum estimate of aligned reads of each gene where a lower level of expression (≤ 500 reads) is depicted in yellow and a higher level of expression (≥ 10,000) is depicted in pink. ACMS Aminocarboxymuconate semialdehyde, AMS Aminomuconate semialdehyde, NAMN Nicotinic acid mononucleotide, NA Nicotinic acid, NAAD Nicotinic acid dinucleotide, NAD Nicotinamide dinucleotide, NMN Nicotinamide mononucleotide, 2PY N-methyl-2-pyridone-5-carboxamide, 4PY N-methyl-4-pyridone-3-carboxamide.
Branched-chain amino acid catabolism
At homeostasis, catabolism of the branched-chain amino acids (BCAA) isoleucine, leucine, and valine share the first two enzymatic steps (Fig. 6A). Cytosolic BCAT1 or mitochondrial BCAT2 reversibly convert BCAA and α-ketoglutarate into branched-chain α-keto acids (BCKA) and glutamate (Fig. 6A). Next, an irreversible oxidative decarboxylation by the branched-chain alpha-keto acid dehydrogenase (BCKDH) converts the BCKA into a coenzyme A (CoA)-activated branched hydrocarbon (Fig. 6A). Although liver, serum, and urinary valine, leucine, and isoleucine were unaltered by TCDD (Fig. 6B–D)58, dose-dependent increases in the urinary levels of 2HIV, 3-hydroxyisovalerate (3HIV), and methylsuccinate (MSA) of 2.1-, 3.6-, and 3.6-fold, respectively, suggested BCAA metabolism was disrupted (Fig. 6E–G), specifically within the BCKDH complex. In rodents and humans, the initial step of BCAA metabolism is extrahepatic59, and therefore the modest effects of TCDD on hepatic Bcat1 and Bcat2 expression are negligible (Fig. 6H). Moreover, TCDD had minor effects on Bcat1 and Bcat2 expression in kidney, gonadal white adipose tissue, and intestinal data (Supplementary Fig. 9). The second step of BCAA metabolism is catalyzed by BCKDH, an inner mitochondrial membrane multienzyme complex consisting of three enzymatic catalytic components: E1, encoded by Bckdha and Bckdhb; E2, encoded by Dbt; and E3, encoded by Dld. Bckdha was repressed 2.5-fold at 30 μg/kg TCDD in the absence of AHR genomic binding (Fig. 6H). Furthermore, Bckdhb exhibited dose-dependent repression and AHR genomic binding in the presence of a pDRE (Supplementary Fig. 10). E2 and E3, encoded by Dbt and Dld, respectively, were unaltered by treatment (Fig. 6H). Bckdha and Bckdhb repression occurred mainly in the liver (Supplementary Fig. 9). BCKDH is inhibited by phosphorylation from the kinase BCKDK and is activated by the phosphatase PPM1K (Fig. 6A). Ppm1k expression was dose-dependently reduced 2.9-fold, while Bckdk expression was unaltered. If BCKDH activity is inhibited, alternative metabolism of the BCKA occurs via BCKA-specific enzymes (Fig. 6A). Like oxalic acid, 2HIV is produced from Ldha60, the expression of which was unaltered by TCDD (Fig. 6H). Hpd expression, which converts α-ketoisocaproate into 3HIV61, was reduced 2.6-fold (Fig. 6H). Although methylsuccinate is likely produced by the transformation of α-ketoisovalerate, the responsible enzyme is not known62. Bcat2, Bckdha, Bckdhb, Ppm1k, and Bckdk were mainly expressed in hepatocytes and immune cells (Supplementary Fig. 9). KLF15, was repressed 2.3-fold (Supplementary Table 2), is a transcriptional regulator of BCAA metabolism63. At homeostasis, KLF15 binds to the Bckdha, Bckdhb, and Ppm1k transcription start sites (TSS) (Supplementary Fig. 10). AHR genomic binding occurs at the TSS in the presence of a pDRE following TCDD treatment (Supplementary Fig. 10). Consistent with the repression of BCKDH, dose-dependent decreases in the hepatic levels of α-isobutyryl-CoA were identified in a complementary untargeted metabolomic dataset35. In the same dataset, isovaleryl-CoA and α-methylbutyryl-CoA (also known as 2-methylbutyryl CoA) also exhibited dose-dependent decreases, but they could not be resolved from isomers (Supplementary Table 6). This untargeted mass spectrometry data was not verified by targeted analysis. In female, ovariectomized mice, hepatic Bckdhb and Ppmk1 were repressed 1.5- and 1.6-fold, respectively, by TCDD.
Alternative branched-chain amino acid catabolism. (A) The first two steps in the canonical branched-chain amino acid (BCAA) catabolic pathway (bold lines). An alternative pathway (dashed lines) is also depicted when step 2 is inhibited. Step 2 may be inhibited by phosphorylation and activated by dephosphorylation (steps 3 and 4, respectively). The numbers map to the genes in the heatmap (H), encoding the enzyme associated with the reaction. (B–G) Urinary metabolite levels as measured by 1-D 1H NMR (n = 5). An asterisk (*) indicates significance from a post hoc Dunnett test. Mean and standard deviation are shown. (H) Heatmap of hepatic BCAA catabolism related genes. pDREs were determined by a position weight matrix with a Matrix Similarity Score (MSS) ≥ 0.856. AHR ChIP-seq was detected in mouse liver 2 h after oral gavage with 30 μg/kg TCDD—green tiles indicate an FDR of ≤ 0.05 (GSE97634). In the dose response bulk RNA-seq gene expression, the black flags indicate a P1(t) ≥ 0.8. The Counts column refers to the maximum estimate of aligned reads of each gene where a lower level of expression (≤ 500 reads) is depicted in yellow and a higher level of expression (≥ 10,000) is depicted in pink. Ile, Isoleucine; Leu, Leucine; Valine, Valine; αKG, α-Ketoglutarate; Glu, Glutamate; KMV, α-Keto-β-methylvalerate; KIC, α-Ketoisocaproate; KIV, α-Ketoisovalerate; Pi, Phosphate; IV-CoA, Isovaleryl-CoA; α-MB-CoA, Alpha-methyl-butyryl-CoA; IB-CoA, Isobutyryl-CoA; 2HIV, 2-Hydroxyisovalerate; 3HIV, 3-Hydroxyisovalerate; MSA, Methylsuccinate.
Histidine metabolism
Histidine is metabolized by three possible pathways in mammalian liver, leading to the formation of histamine, imidazole pyruvate, or glutamate (Supplementary Fig. 11). Urinary urocanate, an intermediate in histidine catabolism, was induced 2.8-fold (Supplementary Fig. 11). Although urinary histidine was unchanged (Supplementary Fig. 9) glutamate increased 1.7-fold (Supplementary Fig. 11). TCDD has been shown to increase the levels of hepatic and serum histidine as well as hepatic glutamate33. The first step in histidine metabolism is a nonoxidative deamination catalyzed by Hal (repressed 25.0-fold), which forms urocanate and ammonia (Supplementary Fig. 11). In contrast, histidine decarboxylation by Hdc or conversion to imidazole propionate by Hat1 were induced 1.6- and 1.8-fold, respectively (Supplementary Fig. 11). Following Hal, Uroc1, which converts urocanate to imidazole propionate, was downregulated 10.0-fold (Supplementary Fig. 11). Then, Amdhd1, which converts imidazole propionate to formiminoglutamate, and Ftcd, which converts formiminoglutamate to glutamate with the cofactor tetrahydrofolate (THF), were downregulated 7.7- and 5.3-fold, respectively (Supplementary Fig. 11). Alternatively, histidine bioavailability may be modulated by the gut flora hut system64. The hut genes detected by metagenomic analysis, hutH, hutU, and hutI, were not altered (Supplementary Fig. 11), suggesting host metabolism was primarily responsible for altered histidine and urocanate levels.
Discussion
Although TCDD and DLCs are associated with human and rodent liver dysfunction, the mechanism of TCDD hepatotoxicity via AHR-mediated metabolic reprogramming is not well understood23. A recently published study using LC–MS/MS examined DLCs’ effects on serum metabolites in Dutch laborers and identified changes in levels of pyruvate, BCAA, histidine, methionine, and vitamin B3 metabolites26. Our study demonstrated that TCDD elicited changes in mouse urinary metabolites levels that correlated with liver histopathology and differential gene expression. In addition to being used to monitor effects on systemic metabolism65, changes in urinary metabolite may precede histological responses. For example, urinary levels of 1MN, TMA, and TMAO were altered at doses below those required to induce moderate to severe steatohepatitis (≤ 3 μg/kg), while changes in the levels of glycolate, pyruvate, BCAA catabolites, urocanate, or methionine were only observed at ≥ 10 μg/kg TCDD.
AHR activation by TCDD mediated the induction of FMO3 and increased urinary TMAO levels as previously reported46,66,67,68,69. The present study further demonstrated an inverse association between TMAO and TMA with no effect on cutC, the primary microbial enzyme responsible for the metabolism of choline to TMAO. Changes in urinary TMA and TMAO levels occurred at doses as low as 3 μg/kg TCDD with Fmo3 induction occurring at as low as 0.3 μg/kg TCDD. The nominal bile acid and glutathione transporter ABCC4 is likely responsible for TMAO export from hepatocytes into the systemic circulation. Upstream of TMA and TMAO metabolism, TCDD also reduced urinary choline. In humans, increased TMAO or TMA levels are reported to increase the risk of cardiovascular disease70 while there is a positive correlation between TMAO and atherosclerosis in rodents, with DLCs accelerating lesion development66. Furthermore, TMAO is associated with steatosis and all-cause mortality in steatotic patients71,72. This suggests TMAO may indicate TCDD-induced steatosis, although the role TMAO plays in hepatic fat accumulation is unclear. A modest level of cell death was also observed in mice gavaged with doses of TCDD as low as 0.3 μg/kg, coinciding with FMO3 induction. In vitro studies using rodent or human hepatocytes have implicated that TMAO attenuates UPR by acting as a “chemical chaperone” and dampens palmitate-induced ERN1 activation50. Other studies have implicated TMAO activation of the EIF2AK3 branch of the UPR51. Significantly, persistent activation of the UPR leads to the death of hepatocytes52. In the present study, TCDD dose-dependently repressed Ern1 and negative regulators of ERN1 signaling potentially by TMAO. However, bulk liver gene expression data does not support the hypothesis that TCDD induced the UPR. Hypothetically, TMAO from AHR-mediated FMO3 induction may protect hepatocytes from UPR. Overall, the modest hepatic necrosis induced by TCDD was likely not related to UPR or TMAO, and therefore its potential roles in TCDD hepatotoxicity warrant further investigation.
Glycolate is a two-carbon metabolite at the nexus of hydroxyproline catabolism, glyoxal detoxification, and glyoxylate detoxification. Hydroxyproline is a significant component of collagen fibers and is elevated in fibrotic liver samples73,74,75,76. Although bulk liver RNA-seq and AHR ChIP-seq data suggested AHR-mediated downregulation of hydroxyproline catabolism, GC–MS analysis of aqueous liver extracts showed no effect on hydroxyproline levels. A previous study using the same dosing regimen and study design reported mild periportal fibrosis, which may not be sufficient to detect increased hydroxyproline levels despite the induction of several collagen genes22,34. Disruption of glyoxylate metabolism can lead to kidney stones due to calcium oxalate accumulation77,78. Glyoxylate is reduced to glycolate by cytosolic or mitochondrial GRHPR. The glycolate may then be further metabolized by the peroxisomal HAO1 and AGXT to prevent calcium oxalate accumulation and minimize the risk of kidney stone formation. In this study, Hao1 and Agxt were repressed with corresponding reductions in protein levels. Hao1 knockouts in rodents and dysfunction in humans are reported to increase glycolate levels compared to wild-type animals79, implicating Hao1 repression in elevated urinary glycolate levels. Another possible source of glycolate, is glyoxal, an electrophilic dialdehyde that covalently binds to macromolecules, including lipids, proteins, and DNA. This is referred to as glycation, resulting in the formation of advanced glycation end-products (AGEs), a biomarker for diabetes80. Glyoxal can be detoxified into glycolate by the glyoxalase system (GLO1 and HAGH) using glutathione as a cofactor. Glo1 and Hagh were both dose-dependently repressed by TCDD, with GLO1 activity also reduced. Overall, elevated urinary glycolate levels were most likely due to AHR-mediated repression of hepatic HAO1 expression. HAO1 repression may reflect peroxisomal metabolic reprogramming, including the loss of peroxisomal fatty acid metabolism, and even lower peroxisome levels, which may contribute to hepatic lipid accumulation30. More likely, HAO1 repression is an adaptive response to TCDD-induced oxidative stress36,81, because the metabolism of glycolate to glyoxylate releases hydrogen peroxide.
Niacin (vitamin B3) is a nutrient that can be replenished by the transformation of tryptophan. Furthermore, diets deficient in tryptophan or niacin can be rescued with tryptophan administration82. 1MN is an intermediate of de novo niacin synthesis that correlates with tryptophan load55,56. Although estimates differ regarding the exact percentage, most free tryptophan after a bolus dose is oxidized83,84 and circulates as kynurenine produced by liver-specific TDO285,86,87. TCDD did not affect liver, serum, or urinary tryptophan levels. However, 1MN was dose-dependently reduced at doses as low as 3 μg/kg TCDD88. Although hepatic Tdo2, the rate limiting enzyme in kynurenine formation, was modestly repressed at a high TCDD dose, the negligible effects on tryptophan levels suggest that Tdo2 repression did not lead to reduced 1MN levels. Yet, downregulation of Afmid, Kmo, and Kynu may reduce kynurenine levels and de novo niacin synthesis while sparing tryptophan oxidative metabolism88. Additionally, TCDD has been shown to reduce hepatic levels of NAD+ and NADP+34. The TEKLAD 8940 chow contained 63 mg/kg nicotinic acid, which is converted to NAD+, NADP+, and NAM. NADP+ levels are reduced in NADK2 deficiency89. Using a mouse knockout model, Cd38, which hydrolyzes NAD+, was strongly associated with hepatic NAD+ levels90. Although dose-dependent Nadk2 repression and Cd38 induction by TCDD may partly explain the decreases in NAD+ and NADP+, they do not adequately explain 1MN reductions. 1MN formation also requires S-adenosylmethionine (SAM), which was dose-dependently decreased by TCDD29. Therefore, the cumulative burden of repressed gene expression associated with de novo niacin biosynthesis and lower hepatic SAM levels may contribute to reduced urinary 1MN levels. Interestingly, exogenous 1MN reduces immune cell-mediated liver necrosis in a concanavalin A mouse model91. Additional studies are needed to elucidate the role of de novo niacin production, NAD+ salvage, and SAM on 1MN, NAD+, NADP+ synthesis and how changes in these may affect TCDD-induced hepatotoxicity92.
Valine, leucine, and isoleucine are BCAAs that constitute a disproportionately large percentage of proteins93. BCAA metabolism is initially catalyzed by branched-chain aminotransferases (BCAT), which are expressed in all tissues59. The subsequent irreversible step by branched-chain keto acid dehydrogenase (BCKDH) primarily occurs in the liver and commits BCCAs to degradation for energy production59. Although urinary BCAA levels were not altered by TCDD, urinary levels of catabolic intermediates, 2HIV, 3HIV, and MSA, were increased. Each one results from a different enzyme acting on the respective BCKAs formed by BCAT. A published metabolomics dataset for acyl-coenzyme A intermediates in liver extracts from mice gavaged every 4 days for 28 days with TCDD showed a reciprocal decrease in α-isobutyryl-CoA as well as the unresolvable peak belonging to either isovaleryl-CoA or α-methylbutyryl-CoA, suggesting inhibition of BCKDH activity. Ppm1k, a gene encoding a phosphatase, which prevents kinase-induced inhibition encoded by Bckdk, was repressed. Bckdk and Ppm1k are regulated by KLF15, which was also repressed by TCDD. The repression of Bckdha and Ppm1k may lead to a decrease in total BCKDH and reduce the un-phosphorylated to phosphorylated BCKDH ratio, resulting in further inhibition of BCKDH, and increased formation of 2HIV, 3HIV, and MSA. Considering BCKDH subunit repression occurred specifically in the liver, the increase in BCAA intermediates was likely liver-dependent.
In addition to oxidative stress from Phase I enzyme induction contributing to AHR-mediated liver toxicity94, accumulating evidence suggests TCDD-induced hepatotoxicity is due to the cumulative burden of the disruption of multiple metabolic pathways23. The present study sought to further elucidate two questions concerning the effects of TCDD: (1) is the liver the source of the TCDD-induced changes in metabolite profiles and (2) are the changes in the urinary metabolite profiles related to the development of TCDD-induced pathology. This study showed compelling evidence for a significant hepatic origin of altered urinary metabolites. However, it was less clear what direct effects these altered metabolites and disrupted pathways had on the progression of steatosis to steatohepatitis with fibrosis. Nevertheless, TMA, TMAO or 1MN may serve as urinary biomarkers of TCDD exposure or TCDD-induced liver inflammation and cell death, while others, such as glycolate, may serve as biomarkers of TCDD-induced steatohepatitis. This study provides further evidence of the disruption of several hepatic pathways in response to persistent AHR activation by TCDD.
Materials and methods
Study design
TCDD elicits similar effects of steatohepatitis and hepatotoxicity in both sexes of C57BL/6NCrl mice as previously reported21. However, male C57BL/6NCrl mice exhibit heightened sensitivity to TCDD-elicited liver toxicity compared to female C57BL/6NCrl mice (i.e., the TCDD dose–response curve of female C57BL/6NCrl liver toxicity is right-shifted). To observe the range of histological, gene expression, and metabolic differences elicited by TCDD, C57BL/6NCrl males were prioritized. Post-natal day (PND) 25 C57BL/6NCrl males from Charles River Breeding Laboratories (Kingston, NY) were acclimatized for five days prior to treatment. Mice were housed in Innovive Innocages (San Diego, CA) containing ALPHA-dri bedding (Shepherd Specialty Papers, Chicago) at an ambient temperature of 21 °C with 30–40% humidity and a 12 h/12 light/dark cycle. All mice were fed the TEKLAD diet 8940 (Madison, WI) ad libitum. At PND 30, mice were orally gavaged with 0.1 ml sesame oil vehicle (Sigma-Aldrich, St. Louis, MO) or 0.03, 0.1, 0.3, 1, 3, 10, or 30 μg/kg TCDD (AccuStandard, New Haven, CT) every 4 days for 28 days from Zeitgeber (ZT) 0–3 for a total of 7 administered doses to induce steady-state levels of TCDD. Moreover, the doses used in the current study were chosen to account for the relatively short study duration as opposite to lifelong cumulative human exposure from diverse AhR ligands, the bioaccumulative nature of halogenated AhR ligands, and different half-lives of TCDD in humans (1–11 years)37,95 and mice (8 − 12 days)96. The same dose range and treatment regimen has been used in previous studies where we found that mouse hepatic tissue levels were comparable to serum levels following the intentional poisoning of Viktor Yushchenko (1–30 μg/kg) and following the accidental exposure from a 1976 chemical plant accident of Seveso women with and without chloracne (0.03–0.3 μg/kg)22. On PND 55, urine and feces were collected from individual mice from ZT 0–3, snap-frozen in liquid nitrogen, and stored at − 80 °C. Three days later, PND 58 mice were euthanized by CO2. Blood, liver, kidney, and epididymal adipose tissues were collected. Whole livers were weighed, snap-frozen in liquid nitrogen, and stored at − 80 °C. Mice were monitored every day for fluctuations in body weight and changes in chow weight from ZT 0–3. Food consumption per day was calculated by dividing the difference in chow weight from the previous day by the number of mice per cage. This study was conducted in accordance with relevant guidelines and regulations. All animal procedures were approved by the Michigan State University Institutional Animal Care and Use Committee (IACUC; PROTO 202,100,219) and meet the ARRIVE guidelines.
Histopathology
A liver sample was fixed in 10% formaldehyde for 24 h before being transferred to a 30% ethanol solution. Liver tissue was sectioned at 5 microns and stained with hematoxylin and eoson (H&E). Whole glass histology slides were digitally scanned using an Olympus VS200 Research Slide Scanner. Liver sections were microscopically examined by a board-certified veterinary pathologist for TCDD-associated histopathology and semi-qualitatively scored for severity of inflammation, apoptosis or necrosis, lipid vacuolation, bile duct hyperplasia, and hepatocellular hypertrophy (0 = 0% of liver tissue area ─ minimal (1) = 1 < 10% of liver tissue area; mild (2) = 10–25% of liver tissue area; moderate (3) = 26–50% of liver tissue area; marked (4) = 51–75% of liver tissue area; severe (5) = 76–100% of liver tissue area affected). The median severity score for each dose was calculated. Severity scores were statistically tested by a Kruskal–Wallis followed by a Dunnett test in R (v4.2.1).
1H NMR preparation, acquisition, analysis
Urine samples (500 μl) in a 1.5 mL Eppendorf tube were centrifuged at 12,000 g for 10 min at 4 °C to remove debris. The supernatant (300 μL) was transferred to a clean 1.5 mL Eppendorf tube containing 35 μL of D2O and 15 μL of buffer (11.667 mM DSS [disodium-2,2-dimethyl-2-silapentane-5-sulphonate], 730 mM imidazole, and 0.47% NaN3 in H2O). Samples (350 μL) were then transferred to a standard 3 mm thin-walled glass NMR tube for 1H NMR spectral analysis. All 1H NMR spectra were randomly collected on a Bruker Ascend HD 600 MHz spectrometer equipped with a 5 mm TCI cryoprobe and acquired at 25 °C using the modified version of the first transient of the Bruker noesy-presaturation pulse sequence providing a high degree of quantitative accuracy. Spectra were collected with 128 transients and 16 steady-state scans using a 5-s acquisition time and a 5.1-s recycle delay. Prior to spectral analysis, all FIDs were zero-filled to 128 K data points, and line broadened by 0.5 Hz. The methyl singlet produced by a known quantity of DSS (1000 μM) was used as an internal standard for chemical shift referencing (set to 0 ppm) and for quantification. All 1H NMR spectra were processed and analyzed using Chenomx NMR Suite Professional software package version 8.3 (Chenomx Inc., Edmonton, CA). Prior to statistical analysis, all NMR spectra were manually inspected for technical faults. Analysis identified 107 unique metabolites in the mouse urine samples. The concentration of all metabolites were normalized to creatinine signal followed by probabilistic quotient normalization (PQN). Each metabolite distribution was tested for differences across the dose response by a Kruskal–Wallis test. p values were adjusted by the Benjamini–Hochberg method. Sixteen metabolites were determined to be significant (adjusted p value ≤ 0.05). Post hoc, each metabolite was subject to a Dunnett test. The raw files were uploaded to Metabolomics Workbench97 (ST003845).
RNA extraction
RNA was extracted as previously described13,34. Briefly, ~ 100 mg of frozen tissues was submerged in 1.3 mL TRIzol and disrupted with a Mixer Mill 300 and a 3 mm stainless steel ball. Following the addition of chloroform, samples were manually shaken and left at room temperature for 3 min before centrifugation (12,000 × g) at 4 °C for 15 min. The supernatant was removed to another Eppendorf tube. Then, 125:24:1 phenol:chloroform:isoamylalcohol was added, and the mixture was centrifuged at 4 °C for 10 min. The supernatant was removed, and equal parts isopropanol was added and left at − 20 °C overnight. The next day, the samples were centrifuged at 4 °C for 15 min (12,000 × g) and the isopropanol was removed. The pellet was washed with 70% ethanol and centrifuged again for 10 min at 4 °C (12,000 × g). The ethanol was removed, and the pellet was broken up, dissolved in RNA Storage Solution (Thermo Scientific, Waltham, MA), heated at 70 °C for 1 min, and stored at − 80 °C. Total RNA was assayed for concentration and purity by a Nanodrop and examined for integrity using an Agilent 2100 Bioanalyzer.
Bulk RNA sequencing, processing, and analysis
Bulk liver (n = 5 per dose) and kidney (n = 4–5 per dose) RNA-seq were performed by Novogene on a NovaSeq 6000 for a total of 20 M 150 base-pair un-stranded paired-end reads per sample. Alignment to GRCm39 (release 104) was performed with STAR (v2.7.3a)98 after trimming adaptors with Trimmomatic (v0.39-Java-11)99. Quality control of sequencing and alignment data was respectively performed with fastqc (v0.11.7-Java-1.8.0_162) and qualimap100 using R (v4.0.3). Separately, gene counts from liver and kidney were variance stabilized using DESeq2101 and modeled using Wolfinger’s mixed linear approach102. Then, posterior probabilities (P1(t)’s) were calculated from the derived t-values using a Bayesian approach to reduce false discoveries. Genes were considered differentially expressed when the absolute fold-change was ≥ 1.5 and the P1(t) was ≥ 0.8. Data wrangling and transformation were done with R (v4.2.1). Principal component analyses were performed using the method `prcomp`. Plots were generated through ggplot2 and formatted using Adobe Illustrator (v25.2.3). Raw and processed data for the liver and kidney are available at GSE203302 and GSE272652, respectively31. Functional enrichment of KEGG103 pathways was performed in R using g:Profiler104.
Benchmark dose (BMD) analysis
BMD response values were calculated for bulk liver gene expression and urinary metabolites using BMDExpress2 (v2.3)58. Each metabolite was fit to parameterized models: Exponential 2, Exponential 3, Linear, Polynomial 2, Polynomial 3, Hill, and Power. Fitting converged over a maximum of 250 iterations to calculate a 10% BMD response value with a 95% confidence level, consisting of a BMD upper (BMDU) and lower (BMDL) bound. Constant variance was assumed. Power was not restricted. BMDL and BMDU were utilized in the best model selection. A nested likelihood ratio test determined the best linear or polynomial model with a p value cutoff of 0.05. Subsequently, the Akaike information criterion (AIC) was compared among the best linear or polynomial model and the remaining models. Hill models were flagged with less than 1/3rd of the lowest positive dose. If a Hill model was flagged, the next best model according to AIC was selected if the p value was greater than 0.05.
Putative dioxin response element (pDRE) identification
pDREs were identified as previously described27,34,105. Briefly, the mouse genome (mm10 GRCm38 build) was computationally searched for the DRE core consensus core 5’-GCGTG-3’. Each core was extended by 7 base pairs (bp) upstream and downstream. The resulting 19 bp sequences were scored using a position weight matrix constructed from bona fide functional DREs. For annotation at the gene level, pDRE locations were compared against the regulatory region (10 kb upstream of the TSS together with 5’ and 3’ UTRs) and the coding sequence of each mouse gene obtained from the UCSC genome browser. The raw bedGraph file for the mouse pDRE analysis is available on Harvard Dataverse106.
Hepatic AHR ChIP-sequencing
ChIP-seq was performed as previously described27 and is available on GEO (GSE97634). Briefly, liver samples were collected from C57BL/6NCrl adult male mice 2 h after a single oral gavage with 30 μg/kg TCDD. Cross-linked DNA was immunoprecipitated with either rabbit IgG or rabbit IgG and anti-AHR as previously described105,107. Libraries prepared using the MicroPlex kit (Diagenode) were pooled and sequenced at a depth of approximately 30 M on an Illumina HiSeq 2500 at the Michigan State University (MSU) Research and Technology Support Facility Core. Read processing and analysis were performed using the MSU High-Performance Computing Center. Quality was determined using FASTQC v0.11.2 and adaptor sequences were removed using Cutadapt v1.4.1 while low-complexity reads were cleaned using FASTX v0.0.14. Reads were mapped to GRCm38 (release 76) using Bowtie v2.0.0 and alignments were converted to SAM format using SAMTools v0.1.19. Normalization and peak calling were performed using CisGenome108 determined by comparison of IgG control and AHR enriched samples (n = 5) using a bin size (b) of 25 and boundary refinement resolution (bw) of 1 with default parameters.
Metagenomics
Metagenomic analysis was performed as previously described109. Briefly, microbial DNA from cecum contents (~ 25 mg) of C57BL/6NCrl male mice repeatedly gavaged every 4 days for 28 days was extracted using the FastDNA spin kit for soil and sequenced on a NovaSeq 6000 by Novogene. Reads aligning to the C57BL/6NCrl genome were removed. The HUMAnN 3.0 bioinformatic pipeline was used with default settings to classify reads to UniRef90 protein identifications, which were mapped to enzyme commission (EC) number entries. Read abundance was normalized to gene copies per million reads (CPM). Multivariate association between dose and EC number relative abundance was established using Maaslin2 with the following settings: normalization (total sum scaling), analysis method (general linear model), and Benjamini–Hochberg multiple test correction. Raw sequencing files and processed data can be found at the NCBI Sequence Read Archive under the accession ID PRJNA719224.
Western blotting
Lysates (20 μg) and the PageRuler Prestained Protein Ladder (Thermo Scientific, Waltham, MA) were resolved via 10% SDS-PAGE gels (Bio-Rad, San Diego, CA) and transferred to nitrocellulose membranes (GE Healthcare, Chicago, IL) using the Mini Trans-Blot Cell Unit (Bio-Rad, San Diego, CA) by wet electroblotting (100 V, 45 min). The membranes were then blocked with 5% nonfat milk (in Tris-buffered saline [TBS] + 0.01% Tween) for 1 h and incubated with primary antibodies: anti-FMO3 (1:5000; ab126711; Abcam, Waltham, MA), anti-GLO1 (1:1000; MA531148, Life Technologies Corporation, Carlsbad, CA), anti-HAO1 (1:5000; ab194790; Abcam, Waltham, MA), anti-AGXT (1:1000; ab178708; Abcam, Waltham, MA), and anti-ACTB (1:1000; #4970; Cell Signaling, Danvers, MA) overnight at 4 °C. Blots were visualized using horseradish peroxidase (HRP)-linked secondary antibodies of goat anti-mouse (1:10,000; Elabscience, Houston, TX) and anti-rabbit (1:1000, Cell Signaling, Danvers, MA) and an ECL kit (Millipore Corporation, Billerica, MA). Membranes were scanned on a Sapphire Biomolecular Imager (Azure Biosystem, Dublin, CA). Protein density values were assessed and calculated using ImageJ (v1.53). Protein expression was standardized to ACTB levels per sample. Raw images are available at the Data Dryad data repository, including raw images, raw images with appropriate labeling of samples by lane and the protein size ladder110.
Bulk TCDD exposure RNA-seq
Time-course liver (GSE109863), dose–response duodenum (GSE87542), dose–response jejunum (GSE90097), dose–response proximal (GSE171942) and distal (GSE89430) ileum, dose–response colon (GSE171941), dose–response femur (GSE104551), and female dose–response gonadal white adipose tissue (GSE272683) and liver (GSE62902) RNA-seq data were processed as previously described21,111. Liver (n = 5) data were from C57BL/6NCrl males orally gavaged with sesame oil vehicle or 30 μg/kg TCDD and euthanized 2, 4, 8, 12, 24, 72, or 168 h post-gavage. Duodenum (n = 3), jejunum (n = 3), proximal (n = 3) and distal (n = 3) ileum, femur (n = 3), colon (n = 3), and kidney (n = 4–5) data were from C57BL/6NCrl males orally gavaged every 4 days for 28 days with TCDD or sesame oil vehicle. Gonadal white adipose tissue (GWAT) data were from C57BL/6NCrl females gavaged every 4 days for 92 days with TCDD or sesame oil vehicle. Unlike liver samples, male bone, male intestinal and GWAT tissue and liver collected from C57BL/6NCrl females were not collected within a narrow ZT (0–3) window.
Single nuclear RNA-sequencing (snRNA-seq) analysis
Available at a public repository (GSE184506), TCDD dose–response snRNA-seq of liver cell types was processed as previously described112. Male C57BL/6NCrl mice were gavaged with TCDD (0.03–30 μg/kg) or sesame oil vehicle every 4 days for 28 days. 11 Cell types were identified in line with previous studies113,114,115 by cluster analysis and semi-automated annotation. Dot plots of average cell type-specific gene expression at a certain dose of TCDD or sesame oil vehicle were generated in R using Seurat.
Publicly available bulk liver gene expression data analysis
Verification of TCDD-induced FMO3-induction-related gene expression was performed by pulling down bulk liver RNA-seq data from wild-type (WT) and Fmo3 knockout C57BL/6 mice, exposed to TCDD (GSE191138). Mice were treated with weekly injections of 25 μg/kg TCDD for 6 weeks46. The data was aligned and transformed identically to the TCDD dose–response bulk liver RNA-seq (See Method Bulk RNA sequencing, processing, and analysis). Verification of TCDD-induced, AHR-mediated gene regulation was performed by pulling down bulk liver microarray data from WT and Ahr knockout C57BL/6 mice, orally gavaged with a bolus of TCDD or corn oil vehicle (GSE15858). Data was processed in R as previously described48.
GC–MS
Extraction, derivatization, and measurement of hepatic metabolites by gas-chromatography mass spectrometry (GC–MS) were performed using protocols adapted from the publicly available Michigan State University Mass Spectrometry and Metabolomics Core website (https://rtsf.natsci.msu.edu/sites/_rtsf/assets/File/MSU_MSMC_004_v1_2_Two_phase_extraction_of_metabolites_from_animal_tissues.pdf). Briefly, ~ 50 mg of frozen liver tissue (n = 5 per dose) was supplemented with 6 nmol of 13C and 15N labeled amino acid internal standards (767,964, Sigma-Aldrich) and lysed with a Mixer Mill 300 and a 3 mm stainless steel ball in a solution of methanol/chloroform (1:2 v/v), 1% formic acid, and 0.01% BHT. Subsequently, the samples were sonicated for 15 min. The mixture was twice supplemented with Milli-Q water, vortexed, centrifuged, and the resulting aqueous supernatant was transferred to a separate Eppendorf tube and made basic by the addition of NaOH. The samples were evaporated to dryness by a SpeedVac without heat and stored at − 20 °C. For derivatization, the dried samples were set at 60 °C for 12 h with 100 μL of 40 mg/mL of methoxyamine HCl dissolved in pyridine and then incubated again at 60 °C for another 12 h with an additional 100 μL of N-Methyl-N-tert-butyldimethylsilyltrifluoroacetamide (MTBSTFA) containing 1% tert-butyldimethylsilyl chloride. Derivatized samples (1 μL) were injected with a 1:10 split at 230 °C and analyzed on an Agilent 7890A GC/ single quadrupole mass spectrometer with 5975C inert XL MSD. The carrier gas was helium, which flowed at 1.0 mL/min. Separation was achieved on an Agilent J&W VF-5 ms (30 m × 0.25 mm × 0.25 μm) (Agilent, Santa Clara, CA) using the following temperature profile: initially set at 40 °C, ramped to 80 °C at 20 °C/min, immediately ramped to 250 °C at 5 °C/min and held for 1 min, and ramped to 320 °C at 50 °C/min and held for 5 min. The mass spectrometer was operated at an electron ionization of 70 eV and a scan range of 50–600 m/z. Glycolic acid (PHR1427, Sigma), hydroxyproline (PHR1939, Sigma), oxalic acid (PHR3291, Sigma), and urea (PHR1406, Sigma) were used as external standards to calculate metabolite levels per sample. Quantification of the derivatized metabolites and internal standard were calculated using the peak area from the extracted ion chromatogram. The SIM ion used for derivatized glycolic acid, hydroxyproline, oxalic acid, and urea were 247, 416, and 261 m/z, respectively. Derivatized glycolic acid and hydroxyproline were normalized to labeled glycine, while oxalic acid was normalized to labeled alanine. Oxalic acid and alanine were quantified by separate runs in the SIM mode, while the other compounds were quantified from runs in the scan mode. Additionally, hydroxyproline was normalized to labeled proline. After calculating concentrations via a linear calibration curve, concentrations were normalized to the liver weight per sample from which metabolites were extracted. The raw and processed files were uploaded to Metabolomics Workbench (ST003476).
Activity assay
GLO1 activity was assayed using a commercial colorimetric kit (ab241019; Abcam, Waltham, MA) according to the manufacturer’s instructions. Liver tissue was disrupted with a Mixer Mill 300 and a 3 mm stainless steel ball.
Data availability
All pDRE annotations, RNA-seq, Western blot, 1-D 1H NMR, GC-MS, and LC-MS data presented in this manuscript are available on public repositories. The 1-D 1H NMR data was deposited to Metabolomics Workbench under the accession identifier ST003845. RNA-seq data for the 28-day repeated-dose dose-response studies for the liver, kidney, gonadal white adipose, femur, duodenum, jejunum, proximal ileum, distal ileum, and colon were deposited to GEO under the respective accession identifiers GSE203302, GSE272652, GSE104551, GSE272683, GSE87542, GSE90097, GSE171942, and GSE89430. Female liver RNA-seq data for the 28-day repeated-dose dose-response studies were deposited to GEO under the accession identifier GSE62902. Raw sequencing files and processed data for the snRNA-seq data can be found at GEO under the accession identifier GSE184506. Raw sequencing files and processed data for the metagenomic data can be found at the NCBI Sequence Read Archive under the accession ID PRJNA719224. Time-course liver RNA-seq data and ChIP-seq data were deposited to GEO under the accession identifiers GSE109863 and GSE97634. The raw and processed files for GC-MS were uploaded to the Metabolomics Workbench under the accession identifier ST003476. The Western blot data were uploaded to the Dryad repository (https://doi.org/10.5061/dryad.mpg4f4r8f). The pDRE annotations are available at Dataverse (https://doi.org/10.7910/DVN/JASCVZ).
References
Fouad, Y. et al. What’s in a name? Renaming ‘NAFLD’ to ‘MAFLD’. Liver Int. 40, 1254–1261. https://doi.org/10.1111/liv.14478 (2020).
Geh, D., Anstee, Q. M. & Reeves, H. L. NAFLD-associated HCC: Progress and opportunities. J. Hepatocell. Carcinoma 8, 223–239. https://doi.org/10.2147/JHC.S272213 (2021).
Gofton, C., Upendran, Y., Zheng, M. H. & George, J. MAFLD: How is it different from NAFLD?. Clin. Mol. Hepatol. 29, S17–S31. https://doi.org/10.3350/cmh.2022.0367 (2023).
Loomba, R., Friedman, S. L. & Shulman, G. I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564. https://doi.org/10.1016/j.cell.2021.04.015 (2021).
Taylor, K. W. et al. Evaluation of the association between persistent organic pollutants (POPs) and diabetes in epidemiological studies: a national toxicology program workshop review. Environ. Health Perspect. 121, 774–783. https://doi.org/10.1289/ehp.1205502 (2013).
Casals-Casas, C. & Desvergne, B. Endocrine disruptors: from endocrine to metabolic disruption. Annu. Rev. Physiol. 73, 135–162. https://doi.org/10.1146/annurev-physiol-012110-142200 (2011).
Al-Eryani, L. et al. Identification of environmental chemicals associated with the development of toxicant-associated fatty liver disease in rodents. Toxicol. Pathol. 43, 482–497. https://doi.org/10.1177/0192623314549960 (2015).
Cave, M. et al. Polychlorinated biphenyls, lead, and mercury are associated with liver disease in American adults: NHANES 2003–2004. Environ.. Health Perspect.. 118, 1735–1742. https://doi.org/10.1289/ehp.1002720 (2010).
Uemura, H. et al. Prevalence of metabolic syndrome associated with body burden levels of dioxin and related compounds among Japan’s general population. Environ.. Health Perspect.. 117, 568–573. https://doi.org/10.1289/ehp.0800012 (2009).
Li, H. et al. Review of the toxicity and potential molecular mechanisms of parental or successive exposure to environmental pollutants in the model organism Caenorhabditis elegans. Environ. Pollut. 311, 119927. https://doi.org/10.1016/j.envpol.2022.119927 (2022).
Wahlang, B. et al. Toxicant-associated steatohepatitis. Toxicol. Pathol. 41, 343–360. https://doi.org/10.1177/0192623312468517 (2013).
Gao, J. et al. A review of food contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin and its toxicity associated with metabolic disorders. Curr. Res. Food Sci. 7, 100617. https://doi.org/10.1016/j.crfs.2023.100617 (2023).
Boverhof, D. R. et al. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol. Sci. 85, 1048–1063. https://doi.org/10.1093/toxsci/kfi162 (2005).
Denison, M. S. & Nagy, S. R. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334. https://doi.org/10.1146/annurev.pharmtox.43.100901.135828 (2003).
Avilla, M. N., Malecki, K. M. C., Hahn, M. E., Wilson, R. H. & Bradfield, C. A. The Ah receptor: Adaptive metabolism, ligand diversity, and the xenokine model. Chem. Res. Toxicol. 33, 860–879. https://doi.org/10.1021/acs.chemrestox.9b00476 (2020).
Wilson, R. H. & Bradfield, C. A. Rodent genetic models of Ah receptor signaling. Drug Metab. Rev. 53, 350–374. https://doi.org/10.1080/03602532.2021.1955916 (2021).
Denison, M. S., Soshilov, A. A., He, G., DeGroot, D. E. & Zhao, B. Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 124, 1–22. https://doi.org/10.1093/toxsci/kfr218 (2011).
Hubbard, T. D., Murray, I. A. & Perdew, G. H. Indole and tryptophan metabolism: Endogenous and dietary routes to Ah receptor activation. Drug Metab. Dispos. 43, 1522–1535. https://doi.org/10.1124/dmd.115.064246 (2015).
Huang, G. & Elferink, C. J. A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol. Pharmacol. 81, 338–347. https://doi.org/10.1124/mol.111.075952 (2012).
Yeager, R. L., Reisman, S. A., Aleksunes, L. M. & Klaassen, C. D. Introducing the “TCDD-inducible AhR-Nrf2 gene battery”. Toxicol. Sci. 111, 238–246. https://doi.org/10.1093/toxsci/kfp115 (2009).
Fader, K. A. et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-elicited effects on bile acid homeostasis: Alterations in biosynthesis, enterohepatic circulation, and microbial metabolism. Sci. Rep. 7, 5921. https://doi.org/10.1038/s41598-017-05656-8 (2017).
Nault, R. et al. Dose-dependent metabolic reprogramming and differential gene expression in TCDD-elicited hepatic fibrosis. Toxicol. Sci. 154, 253–266. https://doi.org/10.1093/toxsci/kfw163 (2016).
Fader, K. A. & Zacharewski, T. R. Beyond the aryl hydrocarbon receptor: Pathway interactions in the hepatotoxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin and related compounds. Curr. Opin. Toxicol. 2, 36–41. https://doi.org/10.1016/j.cotox.2017.01.010 (2017).
Jeanneret, F. et al. Human urinary biomarkers of dioxin exposure: Analysis by metabolomics and biologically driven data dimensionality reduction. Toxicol. Lett. 230, 234–243. https://doi.org/10.1016/j.toxlet.2013.10.031 (2014).
Lee, C. C., Yao, Y. J., Chen, H. L., Guo, Y. L. & Su, H. J. Fatty liver and hepatic function for residents with markedly high serum PCDD/Fs levels in Taiwan. J. Toxicol. Environ. Health A 69, 367–380. https://doi.org/10.1080/15287390500244972 (2006).
Zhao, Y. et al. Dioxin(-like)-related biological effects through integrated chemical-wide and metabolome-wide analyses. Environ. Sci. Technol. 58, 258–268. https://doi.org/10.1021/acs.est.3c07588 (2024).
Nault, R. et al. Pyruvate kinase isoform switching and hepatic metabolic reprogramming by the environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 149, 358–371. https://doi.org/10.1093/toxsci/kfv245 (2016).
Nault, R., Fader, K. A., Lydic, T. A. & Zacharewski, T. R. Lipidomic evaluation of aryl hydrocarbon receptor-mediated hepatic steatosis in male and female mice elicited by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Chem. Res. Toxicol. 30, 1060–1075. https://doi.org/10.1021/acs.chemrestox.6b00430 (2017).
Fling, R. R., Doskey, C. M., Fader, K. A., Nault, R. & Zacharewski, T. R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) dysregulates hepatic one carbon metabolism during the progression of steatosis to steatohepatitis with fibrosis in mice. Sci. Rep. 10, 14831. https://doi.org/10.1038/s41598-020-71795-0 (2020).
Cholico, G. N. et al. Thioesterase induction by 2,3,7,8-tetrachlorodibenzo-p-dioxin results in a futile cycle that inhibits hepatic beta-oxidation. Sci. Rep. 11, 15689. https://doi.org/10.1038/s41598-021-95214-0 (2021).
Orlowska, K. et al. Dioxin-elicited decrease in cobalamin redirects propionyl-CoA metabolism to the beta-oxidation-like pathway resulting in acrylyl-CoA conjugate buildup. J. Biol. Chem. 298, 102301. https://doi.org/10.1016/j.jbc.2022.102301 (2022).
Orlowska, K., Fling, R. R., Nault, R., Schilmiller, A. L. & Zacharewski, T. R. Cystine/glutamate Xc(-) antiporter induction compensates for transsulfuration pathway repression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) to ensure cysteine for hepatic glutathione biosynthesis. Chem. Res. Toxicol. 36, 900–915. https://doi.org/10.1021/acs.chemrestox.3c00017 (2023).
Cholico, G. N., Fling, R. R., Sink, W. J., Nault, R. & Zacharewski, T. Inhibition of the urea cycle by the environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin increases serum ammonia levels in mice. J. Biol. Chem. 300, 105500. https://doi.org/10.1016/j.jbc.2023.105500 (2024).
Fader, K. A., Nault, R., Doskey, C. M., Fling, R. R. & Zacharewski, T. R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin abolishes circadian regulation of hepatic metabolic activity in mice. Sci. Rep. 9, 6514. https://doi.org/10.1038/s41598-019-42760-3 (2019).
Cholico, G. N. et al. Consequences of reprogramming acetyl-CoA metabolism by 2,3,7,8-tetrachlorodibenzo-p-dioxin in the mouse liver. Sci. Rep. 13, 4138. https://doi.org/10.1038/s41598-023-31087-9 (2023).
Fader, K. A. et al. Convergence of hepcidin deficiency, systemic iron overloading, heme accumulation, and REV-ERBalpha/beta activation in aryl hydrocarbon receptor-elicited hepatotoxicity. Toxicol. Appl. Pharmacol. 321, 1–17. https://doi.org/10.1016/j.taap.2017.02.006 (2017).
Sorg, O. et al. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) poisoning in Victor Yushchenko: Identification and measurement of TCDD metabolites. Lancet 374, 1179–1185. https://doi.org/10.1016/S0140-6736(09)60912-0 (2009).
Pierre, S. et al. Aryl hydrocarbon receptor-dependent induction of liver fibrosis by dioxin. Toxicol. Sci. 137, 114–124. https://doi.org/10.1093/toxsci/kft236 (2014).
Nault, R., Colbry, D., Brandenberger, C., Harkema, J. R. & Zacharewski, T. R. Development of a computational high-throughput tool for the quantitative examination of dose-dependent histological features. Toxicol. Pathol. 43, 366–375. https://doi.org/10.1177/0192623314544379 (2015).
Kast, A., Nishikawa, J., Yabe, T., Nanri, H. & Albert, H. Circadian rhythm of liver parameters (cellular structures, mitotic activity, glycogen and lipids in liver and serum) during three consecutive cycles in phenobarbital-treated rats. Chronobiol. Int. 5, 363–385. https://doi.org/10.3109/07420528809067782 (1988).
Sinturel, F. et al. Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169, 651–663. https://doi.org/10.1016/j.cell.2017.04.015 (2017).
Waikar, S. S., Sabbisetti, V. S. & Bonventre, J. V. Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney Int. 78, 486–494. https://doi.org/10.1038/ki.2010.165 (2010).
Dieterle, F., Ross, A., Schlotterbeck, G. & Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290. https://doi.org/10.1021/ac051632c (2006).
Romano, K. A., Vivas, E. I., Amador-Noguez, D. & Rey, F. E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio 6, e02481. https://doi.org/10.1128/mBio.02481-14 (2015).
Bennett, B. J. et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 17, 49–60. https://doi.org/10.1016/j.cmet.2012.12.011 (2013).
Massey, W. et al. Flavin-containing monooxygenase 3 (FMO3) is critical for dioxin-induced reorganization of the gut microbiome and host insulin sensitivity. Metabolites https://doi.org/10.3390/metabo12040364 (2022).
Agarwal, M. et al. Loss of flavin-containing monooxygenase 3 modulates dioxin-like polychlorinated biphenyl 126-induced oxidative stress and hepatotoxicity. Environ. Res. 250, 118492. https://doi.org/10.1016/j.envres.2024.118492 (2024).
Boutros, P. C., Bielefeld, K. A., Pohjanvirta, R. & Harper, P. A. Dioxin-dependent and dioxin-independent gene batteries: Comparison of liver and kidney in AHR-null mice. Toxicol. Sci. 112, 245–256. https://doi.org/10.1093/toxsci/kfp191 (2009).
Teft, W. A. et al. Identification and characterization of trimethylamine-n-oxide uptake and efflux transporters. Mol. Pharm. 14, 310–318. https://doi.org/10.1021/acs.molpharmaceut.6b00937 (2017).
Achard, C. S. & Laybutt, D. R. Lipid-induced endoplasmic reticulum stress in liver cells results in two distinct outcomes: Adaptation with enhanced insulin signaling or insulin resistance. Endocrinology 153, 2164–2177. https://doi.org/10.1210/en.2011-1881 (2012).
Chen, S. et al. Trimethylamine N-oxide binds and activates PERK to promote metabolic dysfunction. Cell Metab. 30, 1141–1151. https://doi.org/10.1016/j.cmet.2019.08.021 (2019).
Pfaffenbach, K. T. et al. Linking endoplasmic reticulum stress to cell death in hepatocytes: Roles of C/EBP homologous protein and chemical chaperones in palmitate-mediated cell death. Am. J. Physiol. Endocrinol. Metab. 298, E1027-1035. https://doi.org/10.1152/ajpendo.00642.2009 (2010).
Fusakio, M. E. et al. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol. Biol. Cell 27, 1536–1551. https://doi.org/10.1091/mbc.E16-01-0039 (2016).
Lange, J. N., Wood, K. D., Knight, J., Assimos, D. G. & Holmes, R. P. Glyoxal formation and its role in endogenous oxalate synthesis. Adv. Urol. 2012, 819202. https://doi.org/10.1155/2012/819202 (2012).
Fukuwatari, T. & Shibata, K. Nutritional aspect of tryptophan metabolism. Int. J. Tryptophan Res. 6, 3–8. https://doi.org/10.4137/IJTR.S11588 (2013).
Shibata, K. & Matsuo, H. Effect of dietary tryptophan levels on the urinary excretion of nicotinamide and its metabolites in rats fed a niacin-free diet or a constant total protein level. J. Nutr. 120, 1191–1197. https://doi.org/10.1093/jn/120.10.1191 (1990).
Chini, E. N., Chini, C. C., Kato, I., Takasawa, S. & Okamoto, H. CD38 is the major enzyme responsible for synthesis of nicotinic acid-adenine dinucleotide phosphate in mammalian tissues. Biochem J 362, 125–130. https://doi.org/10.1042/0264-6021:3620125 (2002).
Phillips, J. R. et al. BMDExpress 2: Enhanced transcriptomic dose-response analysis workflow. Bioinformatics 35, 1780–1782. https://doi.org/10.1093/bioinformatics/bty878 (2019).
Mann, G., Mora, S., Madu, G. & Adegoke, O. A. J. Branched-chain amino acids: Catabolism in skeletal muscle and implications for muscle and whole-body metabolism. Front. Physiol. 12, 702826. https://doi.org/10.3389/fphys.2021.702826 (2021).
Heemskerk, M. M., van Harmelen, V. J., van Dijk, K. W. & van Klinken, J. B. Reanalysis of mGWAS results and in vitro validation show that lactate dehydrogenase interacts with branched-chain amino acid metabolism. Eur. J. Hum. Genet. 24, 142–145. https://doi.org/10.1038/ejhg.2015.106 (2016).
Van Koevering, M. & Nissen, S. Oxidation of leucine and alpha-ketoisocaproate to beta-hydroxy-beta-methylbutyrate in vivo. Am. J. Physiol. 262, E27-31. https://doi.org/10.1152/ajpendo.1992.262.1.E27 (1992).
Nowaczyk, M. J. et al. Ethylmalonic and methylsuccinic aciduria in ethylmalonic encephalopathy arise from abnormal isoleucine metabolism. Metabolism 47, 836–839. https://doi.org/10.1016/s0026-0495(98)90122-6 (1998).
Fan, L., Hsieh, P. N., Sweet, D. R. & Jain, M. K. Kruppel-like factor 15: Regulator of BCAA metabolism and circadian protein rhythmicity. Pharmacol. Res. 130, 123–126. https://doi.org/10.1016/j.phrs.2017.12.018 (2018).
Quesada-Vazquez, S. et al. Potential therapeutic implications of histidine catabolism by the gut microbiota in NAFLD patients with morbid obesity. Cell Rep. Med. 4, 101341. https://doi.org/10.1016/j.xcrm.2023.101341 (2023).
Nagana Gowda, G. A. & Raftery, D. NMR-based metabolomics. Adv. Exp. Med. Biol. 1280, 19–37. https://doi.org/10.1007/978-3-030-51652-9_2 (2021).
Petriello, M. C. et al. Relationship between serum trimethylamine N-oxide and exposure to dioxin-like pollutants. Environ. Res. 162, 211–218. https://doi.org/10.1016/j.envres.2018.01.007 (2018).
Tijet, N. et al. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 69, 140–153. https://doi.org/10.1124/mol.105.018705 (2006).
Celius, T. et al. Aryl hydrocarbon receptor-dependent induction of flavin-containing monooxygenase mRNAs in mouse liver. Drug Metab. Dispos. 36, 2499–2505. https://doi.org/10.1124/dmd.108.023457 (2008).
Petriello, M. C. et al. Dioxin-like pollutants increase hepatic flavin containing monooxygenase (FMO3) expression to promote synthesis of the pro-atherogenic nutrient biomarker trimethylamine N-oxide from dietary precursors. J. Nutr. Biochem. 33, 145–153. https://doi.org/10.1016/j.jnutbio.2016.03.016 (2016).
Ufnal, M., Zadlo, A. & Ostaszewski, R. TMAO: A small molecule of great expectations. Nutrition 31, 1317–1323. https://doi.org/10.1016/j.nut.2015.05.006 (2015).
Theofilis, P., Vordoni, A. & Kalaitzidis, R. G. Trimethylamine N-oxide levels in non-alcoholic fatty liver disease: A systematic review and meta-analysis. Metabolites https://doi.org/10.3390/metabo12121243 (2022).
Flores-Guerrero, J. L. et al. Circulating trimethylamine-N-oxide is associated with all-cause mortality in subjects with nonalcoholic fatty liver disease. Liver Int. 41, 2371–2382. https://doi.org/10.1111/liv.14963 (2021).
Bissoondial, T. L. et al. Liver biopsy hydroxyproline content is a diagnostic for hepatocellular carcinoma in murine models of nonalcoholic steatohepatitis. Diagnostics (Basel) https://doi.org/10.3390/diagnostics10100784 (2020).
Gianmoena, K. et al. Epigenomic and transcriptional profiling identifies impaired glyoxylate detoxification in NAFLD as a risk factor for hyperoxaluria. Cell Rep. 36, 109526. https://doi.org/10.1016/j.celrep.2021.109526 (2021).
Mata, J. M., Kershenobich, D., Villarreal, E. & Rojkind, M. Serum free proline and free hydroxyproline in patients with chronic liver disease. Gastroenterology 68, 1265–1269 (1975).
Pellicano, A. J., Spahn, K., Zhou, P., Goldberg, I. D. & Narayan, P. Collagen characterization in a model of nonalcoholic steatohepatitis with fibrosis; A call for development of targeted therapeutics. Molecules https://doi.org/10.3390/molecules26113316 (2021).
Dutta, A. K. et al. Recurrent truncating mutations in alanine-glyoxylate aminotransferase gene in two South Indian families with primary hyperoxaluria type 1 causing later onset end-stage kidney disease. Indian J. Nephrol. 26, 288–290. https://doi.org/10.4103/0971-4065.171244 (2016).
Zabaleta, N. et al. CRISPR/Cas9-mediated glycolate oxidase disruption is an efficacious and safe treatment for primary hyperoxaluria type I. Nat. Commun. 9, 5454. https://doi.org/10.1038/s41467-018-07827-1 (2018).
McGregor, T. L. et al. Characterising a healthy adult with a rare HAO1 knockout to support a therapeutic strategy for primary hyperoxaluria. Elife https://doi.org/10.7554/eLife.54363 (2020).
Goh, S. Y. & Cooper, M. E. Clinical review: The role of advanced glycation end products in progression and complications of diabetes. J. Clin. Endocrinol. Metab. 93, 1143–1152. https://doi.org/10.1210/jc.2007-1817 (2008).
Recalcati, S., Tacchini, L., Alberghini, A., Conte, D. & Cairo, G. Oxidative stress-mediated down-regulation of rat hydroxyacid oxidase 1, a liver-specific peroxisomal enzyme. Hepatology 38, 1159–1166. https://doi.org/10.1053/jhep.2003.50417 (2003).
Pellagra treated with tryptophan. Nutr. Rev. 45, 142-148, https://doi.org/10.1111/j.1753-4887.1987.tb06348.x (1987).
Michael, A. F., Drummond, K. N., Doeden, D., Anderson, J. A. & Good, R. A. Tryptophan metabolism in man. J. Clin. Investig. 43, 1730–1746. https://doi.org/10.1172/JCI105048 (1964).
Patterson, J. I., Brown, R. R., Linkswiler, H. & Harper, A. E. Excretion of tryptophan-niacin metabolites by young men: Effects of tryptophan, leucine, and vitamin B6 intakes. Am. J. Clin. Nutr. 33, 2157–2167. https://doi.org/10.1093/ajcn/33.10.2157 (1980).
Maeta, A. et al. Contributions of tryptophan 2,3-dioxygenase and indoleamine 2,3-dioxygenase to the conversion of D-tryptophan to nicotinamide analyzed by using tryptophan 2,3-dioxygenase-knockout mice. Biosci. Biotechnol. Biochem. 78, 878–881. https://doi.org/10.1080/09168451.2014.905185 (2014).
Larkin, P. B. et al. Tryptophan 2,3-dioxygenase and indoleamine 2,3-dioxygenase 1 make separate, tissue-specific contributions to basal and inflammation-induced kynurenine pathway metabolism in mice. Biochim. Biophys. Acta 2345–2354, 2016. https://doi.org/10.1016/j.bbagen.2016.07.002 (1860).
Ferreira, P. et al. Hypertryptophanemia due to tryptophan 2,3-dioxygenase deficiency. Mol. Genet. Metab. 120, 317–324. https://doi.org/10.1016/j.ymgme.2017.02.009 (2017).
Kaiser, H., Parker, E. & Hamrick, M. W. Kynurenine signaling through the aryl hydrocarbon receptor: Implications for aging and healthspan. Exp. Gerontol. 130, 110797. https://doi.org/10.1016/j.exger.2019.110797 (2020).
Kim, H. et al. The mitochondrial NAD kinase functions as a major metabolic regulator upon increased energy demand. Mol. Metab. 64, 101562. https://doi.org/10.1016/j.molmet.2022.101562 (2022).
Young, G. S., Choleris, E., Lund, F. E. & Kirkland, J. B. Decreased cADPR and increased NAD+ in the Cd38-/- mouse. Biochem. Biophys. Res.. Commun. 346, 188–192. https://doi.org/10.1016/j.bbrc.2006.05.100 (2006).
Jakubowski, A. et al. 1-Methylnicotinamide protects against liver injury induced by concanavalin A via a prostacyclin-dependent mechanism: A possible involvement of IL-4 and TNF-alpha. Int. Immunopharmacol. 31, 98–104. https://doi.org/10.1016/j.intimp.2015.11.032 (2016).
Hayes, A. J. et al. Kynurenine monooxygenase regulates inflammation during critical illness and recovery in experimental acute pancreatitis. Cell Rep. 42, 112763. https://doi.org/10.1016/j.celrep.2023.112763 (2023).
Harper, A. E., Miller, R. H. & Block, K. P. Branched-chain amino acid metabolism. Annu. Rev. Nutr. 4, 409–454. https://doi.org/10.1146/annurev.nu.04.070184.002205 (1984).
Uno, S. et al. Cyp1a1(-/-) male mice: Protection against high-dose TCDD-induced lethality and wasting syndrome, and resistance to intrahepatocyte lipid accumulation and uroporphyria. Toxicol. Appl. Pharmacol. 196, 410–421. https://doi.org/10.1016/j.taap.2004.01.014 (2004).
Wolfe, W. H. et al. Determinants of TCDD half-life in veterans of operation ranch hand. J. Toxicol. Environ. Health 41, 481–488. https://doi.org/10.1080/15287399409531858 (1994).
Gasiewicz, T. A., Geiger, L. E., Rucci, G. & Neal, R. A. Distribution, excretion, and metabolism of 2,3,7,8-tetrachlorodibenzo-p-dioxin in C57BL/6J, DBA/2J, and B6D2F1/J mice. Drug Metab. Dispos. 11, 397–403 (1983).
Sud, M. et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463-470. https://doi.org/10.1093/nar/gkv1042 (2016).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Garcia-Alcalde, F. et al. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678–2679. https://doi.org/10.1093/bioinformatics/bts503 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Eckel, J. E., Gennings, C., Chinchilli, V. M., Burgoon, L. D. & Zacharewski, T. R. Empirical bayes gene screening tool for time-course or dose-response microarray data. J. Biopharm. Stat. 14, 647–670. https://doi.org/10.1081/BIP-200025656 (2004).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. https://doi.org/10.1093/nar/gkv1070 (2016).
Kolberg, L. et al. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 51, W207–W212. https://doi.org/10.1093/nar/gkad347 (2023).
Dere, E., Lo, R., Celius, T., Matthews, J. & Zacharewski, T. R. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics 12, 365. https://doi.org/10.1186/1471-2164-12-365 (2011).
Nault, R. & Zacharewski, T. (Harvard Dataverse, 2019).
Lo, R. & Matthews, J. High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicol. Sci. 130, 349–361. https://doi.org/10.1093/toxsci/kfs253 (2012).
Ji, H. et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26, 1293–1300. https://doi.org/10.1038/nbt.1505 (2008).
Fling, R. R. & Zacharewski, T. R. Aryl hydrocarbon receptor (AhR) activation by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) dose-dependently shifts the gut microbiome consistent with the progression of non-alcoholic fatty liver disease. Int. J. Mol. Sci. https://doi.org/10.3390/ijms222212431 (2021).
Sink, W. J. Western blots of liver-expressed genes in C57BL/NCrl mice gavaged every 4 days for 28 days with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Dryad Digit. Repos. https://doi.org/10.5061/dryad.mpg4f4r8f (2024).
Fader, K. A., Nault, R., Raehtz, S., McCabe, L. R. & Zacharewski, T. R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin dose-dependently increases bone mass and decreases marrow adiposity in juvenile mice. Toxicol. Appl. Pharmacol. 348, 85–98. https://doi.org/10.1016/j.taap.2018.04.013 (2018).
Nault, R. et al. Single-cell transcriptomics shows dose-dependent disruption of hepatic zonation by TCDD in mice. Toxicol. Sci. 191, 135–148. https://doi.org/10.1093/toxsci/kfac109 (2023).
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356. https://doi.org/10.1038/nature21065 (2017).
Nault, R., Fader, K. A., Bhattacharya, S. & Zacharewski, T. R. Single-nuclei RNA sequencing assessment of the hepatic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Cell Mol. Gastroenterol. Hepatol. 11, 147–159. https://doi.org/10.1016/j.jcmgh.2020.07.012 (2021).
Xiong, X. et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75, 644–660. https://doi.org/10.1016/j.molcel.2019.07.028 (2019).
Acknowledgements
GC-MS method development and instrumentation was performed in collaboration with the MSU mass spectrometry and metabolomics core, especially Dr. Cassandra Johnny. This project was supported by the National Institute of Environmental Health Sciences (NIEHS) Superfund Research Program [NIEHS SRP P42ES004911] and the NIEHS Research Project Grant Program [NIEHS R01ES029541] to T. Z. W. J. S. and R. R. F. were supported by NIEHS Multidisciplinary Training in Environmental Toxicology [NIEHS T32ES007255]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
W. J. S., S. F. G., T. Z. conceptualization; W. J. S., R. R. F., W. J. S., A. Y., D. G., and R. N. investigation; W. J. S. visualization; W. J. S., writing-original draft; J. R. H. formal analysis; W. J. S., R. R. F., A. Y., R. N., J. R. H., S. F. G. and T. Z. writing-review and editing; R. R. F., R. N., W. J. S., and A. Y. data curation; W. J. S. project administration; W. J. S. and A. Y. methodology; T. Z. and S.F.G. funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sink, W.J., Fling, R., Yilmaz, A. et al. Shift in the urinary metabolome associated with 2,3,7,8-tetrachlorodibenzo-p-dioxin activation of the hepatic aryl hydrocarbon receptor. Sci Rep 15, 26035 (2025). https://doi.org/10.1038/s41598-025-11772-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-11772-7