Abstract
Melanoma immunotherapy urgently requires approaches that can accurately predict drug responses to minimize unnecessary treatments. Deep learning models have emerged as powerful tools in this domain due to their robust predictive capabilities. Integrating functional characteristics with expression data from mRNA transcripts shows promise for enhancing prediction accuracy. We developed a deep learning model called AMU (Attention mechanism Model for melanoma immUnotherapy) that incorporates a self-attention mechanism to predict clinical responses to immune checkpoint inhibitors in melanoma patients based on mRNA expression profiles. We evaluated AMU’s performance against established machine learning approaches including Support Vector Machine (SVM), Random Forest, AdaBoost, XGBoost, and classical Convolutional Neural Networks (CNN). In the validation set (pre-treatment tissue samples), AMU exhibited outstanding performance, with an AUC of 0.941 and an mAP of 0.960. In the test set (post-treatment tissue samples), its AUC was 0.672, and the mAP was 0.800. Model interpretation revealed that the TNF-TNFRSF1A pathway was a crucial pathway influencing the efficacy of melanoma immunotherapy. Additionally, the expression levels of CD80 and CCR3 were closely correlated with the survival rate (hazard ratios of 0.761 and 0.134, respectively) and the response to immune checkpoint inhibitors in melanoma patients. The deep learning model integrated with the self-attention mechanism has demonstrated strong efficacy in processing mRNA expression data for melanoma immunotherapy response prediction. After rigorous evaluation, including batch effect correction and cross-validation, AMU achieved superior performance compared to traditional machine learning approaches. Beyond prediction accuracy, our model interpretation work identified the TNF-TNFRSF1A pathway as potentially crucial in determining melanoma ICI response, a finding aligned with recent experimental evidence. The embedding architecture’s ability to capture meaningful gene-gene relationships, partially consistent with established protein interaction networks, suggests broad potential for representation learning in transcriptomic analysis. While acknowledging the limitations of current sample sizes and the need for prospective validation, this work provides both methodological advances in applying transformer architectures to gene expression data and biological insights into immunotherapy response mechanisms. The integration of robust machine learning approaches with domain-specific biological knowledge represents a promising direction for developing clinically relevant biomarkers in precision oncology.
Similar content being viewed by others
Introduction
The 2012 publication of AlexNet rekindled widespread interest in neural networks among researchers1. In the decade since, deep learning (DL) has emerged as a leading scientific field and core artificial intelligence technology, driving significant advancements across various sectors. DL-powered AI has become pivotal in the current industrial revolution.
In image recognition and segmentation, DL has matured into a widely adopted industrial technology. For natural language processing (NLP), Google’s 2017 introduction of the self-attention mechanism in “Attention is All You Need“2 substantially enhanced predictive accuracy for tasks including speech recognition and machine translation. The transformer network architecture, which leverages self-attention, quickly established itself as a superior foundation for various applications. Its adaptation to computer vision (CV) through models such as Vision Transformer (ViT) and Swin Transformer3,4 significantly improved accuracy across CV tasks.
In medicine, transformer-based CV and NLP models have gained widespread adoption5, accelerating AI-assisted diagnosis and medical data extraction6. In scientific research, graph neural networks (GNNs) facilitate drug sensitivity and molecular affinity predictions4,7, while AlphaFold2’s breakthrough in protein structure prediction8 demonstrates the potential to revolutionize protein function studies. For gene multi-omics data analysis, traditional Convolutional Neural Network (CNN) approaches have shown limitations4,9,10. Therefore, we investigated the application of transformer encoders with self-attention mechanisms for gene expression data analysis, achieving promising results.
We named our DL model AMU, which means Attention mechanism Model for predicting melanoma iMMUnotherapy checkpoint inhibitor (ICI) response. In recent years, malignancy immunotherapy has made great progress and significantly improved patients’ overall survival, especially for melanoma, immunotherapy has already acquitted as the standard treatment in the advanced disease10,11. However, the clinical tumor response to immunotherapy is not satisfied, and the objective response rate (ORR), which is the standard assessment criteria for evaluating anti-tumor drug activation, is around 30%12, in some other tumors, the ORR is even lower, around 10%−20%13,14. So how to precisely identify which group of patients can be beneficial from immunotherapy has caused much attention15,16,17. Currently approved immunotherapy treatment includes PD-1/PD-L1 inhibitor and CTLA-4 inhibitors, the biomarkers for these drugs are usually PD-L1 expression level, tumor mutation burden (TMB) and MSI-H/dMMR status, but these biomarkers have low prediction accuracies and often contradict with each other18,19,20. The search for more precise methods has not stopped, we considered drug response is related to complex biological pathways and conducted this study using muti-gene mRNA expression values to predict ICI response.
We summarized our contributions as follows:
-
1)
In model building and developing level:
We provided reliable evidence to reveal the superiority of our model AMU achieving excellent performance in both validation dataset and independent testing dataset for melanoma ICI response prediction, highlighting the strong predictive power and generalization ability of our model.
We proved that the self-attention mechanism could work in 1-D vector data, even if the input data is not spatial positional type image or sequence sensitive type natural language.
We discovered the embedding architecture could be used for representation learning of gene features and combining mRNA expression quantitative information, the interactions of the learned representation vector had local consistence with the widely accepted Functional Protein Association Network (STRING, https://cn.string-db.org/)21, which proved the embedding architecture is suitable and promising for gene vector representation. Self-attention mechanism was superior and benefit for digitating data inner correlation. The interpretation of the embedding layer made the DL network becoming more convincing, which was especially important in the biomedical areas.
-
2)
In biological level:
According to the model interpretation work, we put forward an assumption that the TNF- TNFRSF1A pathway might be a key pathway to decide melanoma ICI response.
CD80 and CCR3 expressions were identified as related to both survival (hazard ratios were 0.761 and 0.134 respectively) and ICI response for melanoma.
Materials and methods
We compiled our datasets from publicly available sources. AMU’s performance was evaluated against five other machine learning models using both validation and testing datasets. Following model training, we conducted comprehensive interpretation analyses to explore gene function importance and interactions. We implemented AMU using the PaddlePaddle 2.3.2 deep learning framework and trained the model on Paddle AI Studio (https://aistudio.baidu.com/aistudio/index). All figures were generated using the matplotlib package.
Github code: https://github.com/1692775560/AMU.
Overview of AMU framework
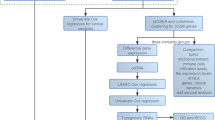
AMU is constructed by a transformer encoder followed by a convolutional network for an ICI clinical response binary classification task. The input data are 160 normalized mRNA expression values. As the same as other classification models, the output of AMU is a pair of probability values, which denotes non-response and response probability. The transformer encoder structure is classical as that in NLP, which will be described in detail in the following part. In convolutional network, we used ‘Convolution- Dropout - Batch Normalization - ReLU activation function - Adaptive Maximum Pool’ strategies. We used Adam algorithm as the optimizer for back-propagation process and two-step decay of learning rate for training. AMU takes the SoftMax activation function at the end of the net and cross-entropy as the loss function. A total of 83,462 parameters are trainable in AMU. For details see Fig. 1 and Supplement (Table S1).
mRNA embedding and transformer encoder layer
We set a 20-D gene embedding for gene feature learning, and the initialized embedding input is an integer “1” to ”160”, then we multiply the embedded values with mRNA expression values in order to add expression information to every embedding, this method was inspired by NLP process in which word position information is added to the embedding layer. We consider that gene features can be learned in the end-to-end training process just like words can be representation learned in a large text corpus. However, the genes is not a sequence data so that the position information is not necessary and expression information should be instead. Genes have interactions and association with each other, so self-attention mechanism will be work.
In the process of transplanting the transformer encoder layer, no structure needs to be changed, which includes Layer normalization, Dropout, Muti-head attention, and Multilayer Perceptron (MLP). In the model training experiment, we used eight Muti-head attentions and repeated the transformer encoder layer eight times to avoid underfitting.
mRNA data preprocessing Raw RNA-seq data underwent a systematic preprocessing pipeline
Quality Control: Raw sequencing reads were filtered using FastQC to remove low-quality reads (Phred score < 20) and adapter sequences.
Alignment and Quantification: Filtered reads were aligned to the human reference genome (GRCh38) using STAR aligner, followed by gene expression quantification with HTSeq-count.
Normalization: Raw counts were normalized to Transcripts Per Million (TPM) to account for differences in sequencing depth and gene length.
Batch Effect Correction: To address potential batch effects from different data sources, we implemented ComBat, as verified by principal component analysis before and after correction (Supplementary Figure S1).
Feature Selection: From the initial gene set, we selected 160 genes that were consistently measured across all datasets and relevant to melanoma biology and immune response.
Final Normalization: Selected gene expression values were log-transformed and scaled to have zero mean and unit variance within each dataset, then integrated into the final analysis matrix.”
Batch effect assessment and correction since
Our data originated from multiple sources with potential technical variations, we implemented a robust batch effect correction strategy. First, we performed Principal Component Analysis (PCA) on the combined dataset before correction to visualize batch-specific clustering (Supplementary Figure S1A)40. We then applied the ComBat algorithm from the sva R package to remove batch effects while preserving biological variation. Post-correction PCA analysis confirmed substantial reduction in batch-specific clustering (Supplementary Figure S1B). All models were trained and evaluated using both raw and batch-corrected data to assess the impact of batch correction on prediction performance.
Building dataset
As shown in Table 1, all the cases fed into the models are collected from published data, including three independent datasets GSE78220, GSE91061, GSE165278 from GEO Datasets and one dataset from the paper of Liu (PMID:31792460)22,23,24,25,26. We collected total 206 patients diagnosed with advanced melanoma treated with immunotherapy checkpoint inhibitors, including Nivolumab, Pembrolizumab and Ipilimumab. The whole-transcriptome sequencing (RNA-seq) was conducted on pretreatment tumor tissues.
To evaluate our model’s generalization ability across different clinical scenarios, we included two testing datasets: (1) a primary testing set consisting of 58 pre-treatment tissue samples from an independent cohort, and (2) a secondary testing set built from 58 post-treatment tissue samples from GSE91061. While we acknowledge that pre-treatment samples provide the most clinically relevant prediction scenario, the inclusion of post-treatment samples tests the model’s robustness to treatment-induced expression changes.Clinical information is not available in most datasets and the patients’ characteristics cannot be described. Dataset details are shown in Table 1.
According to previous studies, 169 genes have been described potentially associated with melanoma, inflammation, immunity, the PD-L1/CTLA4 pathways and ICI response27,28,29. Finally, 160 genes were overlapped in the four datasets and selected. For response digital representation, we took records with “complete response (CR)” and “partial response (PR)” as responses (classed as numeric 1), “stable disease (SD)” and “progression disease (PD)” as non-response (classed as numeric 0). Supplement (Data file S1) lists the full 160 gene names.
We also conducted a heatmap analysis on the provided raw data to analyze and discuss the expression levels of different genes under different samples or conditions. The depth of the color indicates the level of expression, as shown in Fig. 2.
Figure 2 displays expression patterns of the 10 most influential genes identified through SHAP analysis, rather than randomly selected genes from the 160-gene set. These genes demonstrated the highest impact on model predictions across all evaluated models. The ‘Important Region’ and ‘Key Cluster’ annotations highlight expression patterns strongly associated with treatment response outcomes. Specifically, the Important Region contains gene expression profiles that consistently appear in responders, while the Key Cluster represents a signature pattern that distinguishes between response groups with high confidence. These patterns align with known immunological pathways, particularly the TNF signaling pathway and T-cell activation mechanisms, supporting the biological relevance of our model’s predictions.
We calculated the TPM-normalized expression values by the raw data provided by authors. And continued to normalize the TPM values to constituent ratios, then we selected the 160 values from their original datasets and convert to constituent ratios again. After this step, all the values from different datasets represent mRNA relative expression quantity and are comparable. At last, we logarithm them.
To address class imbalance in our dataset, we implemented the Synthetic Minority Over-sampling Technique (SMOTE) algorithm rather than simple duplication. SMOTE generates synthetic samples in the feature space by interpolating between existing minority class instances, creating more diverse synthetic samples compared to simple duplication. This approach helps prevent overfitting while improving model performance on minority classes. After SMOTE application, our training/validation dataset contained 148 positive samples and 132 negative samples. A total of 280 samples were in our training/validation dataset including 148 positive samples and 132 negative samples. We randomly divided the training/validation dataset into the training (224) and validation (56) sets, which corresponded to 80% and 20% of the total instances, respectively. In order to get reliable model performance, we randomly split the training and validation data five times, which will be mentioned as “5-fold cross validation” in the following part.
Competing methods
We chose four machine learning models and designed one simple CNN as competing methods. All competing models input data are 160-D vector of mRNA expression values we have been described.
-
SVM (Support Vector Machine) is a first-class classification machine learning model. We employed the grid search strategy to find the optimal model hyperparameter. ‘kernel’ included ‘linear’, ‘poly’ and ‘rbf’, ‘C’ was in the list [1, 10, 100], ‘gamma’ was in the list [1, 0.1, 0.001].
-
Random Forest is a tree-based ensemble learning method that can be used for both classification and regression tasks. In this study, we implemented RandomForestClassifier for the binary classification of ICI response, despite its common description as a regression model in literature. The classifier version constructs multiple decision trees during training and outputs the class that is the mode of the classes output by individual trees. We set the number of trees in the forest from range (2,10) and ‘n_estimators’ from arrange (10,300,10). The best hyperparameters were chosen in the comparing experiments.
-
AdaBoostClassifier is a tree-based ensemble model and the best hyperparameters were chosen as the same as Random Forest.
-
XGBoost (eXtreme Gradient Boosting) is a scalable tree boosting system. It implements machine learning algorithms under the gradient boosting framework. One of the advantages of XGBoostClassifier is the convenience for model interpretation30.
-
CNN, we built a simple CNN to represent the traditional DL model without self-attention mechanism. The model included three Conv1D layers and total 737 trainable parameters. Details see Supplement (Table S2).
Model evaluation
In the classification experiments, AUC and PR curves, two commonly used measurements, were chosen as our classification metrics. To further evaluate the performance of our model, we demonstrated the results under validation dataset and testing dataset, respectively. We also used several common metrics in five-fold cross validation, including accuracy, precision, recall and f1 score.
Model interpretation
We selected SVM, XGBoost and AMU model to explore the model interpretation works.
For SVM and XGBoost models, we applied SHAP (SHapley Additive Explanations) which is a game theoretic approach to estimate the gene feature importance, then we used GO pathway enrichment analysis and overall survival COX analysis to describe the important gene features.
For AMU model, Shap also can identify the gene importance, but more information can be analysis through mRNA embedding layer. Just be inspired by NLP word embedding, we toke mRNA embedding layer 20-D trainable parameters as gene features. We tried the cluster analysis and calculated the Euclidean distance, cosine similarity and t-distributed Stochastic Neighbor Embedding (t-SNE) among gene feature vectors to describe the gene association and interaction. Then we compare the gene correlations with Functional Protein Interaction Network (STRING, https://cn.string-db.org/) to evaluate the gene features learned from AMU.
For model interpretation analysis, we focused on SVM, XGBoost, and AMU models based on three key considerations: (1) these models demonstrated the highest performance metrics in our evaluation, making their interpretations most relevant; (2) they represent diverse machine learning paradigms (traditional statistical learning, ensemble learning, and deep learning), providing complementary perspectives on feature importance; and (3) these models offer established interpretation methods like SHAP values that allow for consistent cross-model comparison. Other models were excluded from detailed interpretation due to either inferior performance or limitations in applicable interpretation techniques.
Results
Impact of batch effect correction on model performance
Batch effect correction significantly improved model performance across all evaluated algorithms. For AMU, the AUC on the validation dataset increased from 0.941 to 0.953 after correction. This improvement validates the importance of addressing technical variation in multi-source transcriptomic data. Supplementary Figure S1 illustrates the PCA visualization before and after batch correction, demonstrating clear reduction in dataset-specific clustering while maintaining biological signal relevant to treatment response.
AMU accurately predicted melanoma immunotherapy response
We identified the performance of our model on validation and testing datasets compared with currently advanced machine learning models in five-fold cross validation. Supplement Table S3 shows the binary classification reports of the validation dataset predicted by the original training data. DL models were not preferred, and SVM had the best performance according to the accuracy score (0.633) and recall score (0.633). All models had unsatisfactory performance. XGBoost model got the highest f1-score (0.567) followed by AMU (0.55), the CNN model had the lowest f1-score of 0.45.
When evaluated solely on pre-treatment samples, AMU achieved superior performance with an AUC of 0.953 and mAP of 0.972, compared to its performance on post-treatment samples (AUC of 0.672, mAP of 0.800). This difference highlights the importance of using pre-treatment data for clinical prediction applications, while demonstrating AMU’s ability to identify key predictive signals even in treatment-modified expression profiles.
However, after data enhancement, all model performances were significantly improved except CNN (Table 2), which was hard to converge. AMU model showed the best performance with f1-score 0.93, the area under the curve (AUC) 0.941 and mean average precision (mAP) 0.960, respectively. In the testing dataset, AMU also demonstrated superior predictive perform as shown in Table 3 and achieved the highest AUC (0.672) and mAP (0.800) respectively. The receiver operating characteristic curve (ROC) and Precision-Recall (PR) curve were shown in Fig. 3.
ROC and PR in the validation dataset. ROC and PR curves for model comparison across datasets. (A) ROC curves of six models on the validation dataset, demonstrating AMU’s superior discrimination ability (AUC = 0.953) compared to other methods. (B) Precision-Recall curves on the validation dataset showing AMU’s excellent performance (mAP = 0.972) in balancing precision and recall. (C) ROC curves on the testing dataset of post-treatment samples, where AMU maintains relatively robust performance (AUC = 0.672) despite the challenging transfer scenario. (D) PR curves on the testing dataset highlighting AMU’s ability to maintain precision (mAP = 0.800) even with treatment-modified expression profiles. The diagonal line in ROC curves represents random prediction (AUC = 0.5), while higher curves indicate better performance. For PR curves, higher curves represent superior precision-recall balance across threshold values.
Model interpretation
We listed the top-10 Shap value genes of SVM, XGBoost and AMU (Supplement Table S4)31. The top genes were quite different among models. The intersection of these top-10 genes was including TNF and its receptor TNFRSF1A. TNF encodes a multifunctional proinflammatory cytokine. TNFRSF1A is a member of the TNF receptor superfamily of proteins. The details of Shap values were in Supplementary (Fig S1−3).
Gene Ontology (GO) analysis was performed on the top-50 genes prioritized by each model, yielding 112 unique genes for pathway enrichment analysis (Supplementary Fig S4)32,33,34. To distinguish model-selected pathways from biases inherent in our initial gene selection, we performed differential enrichment analysis comparing these 112 genes against the background of all 160 input genes. While the initial gene set was indeed enriched for inflammatory processes, our models specifically emphasized genes in the lymphocyte proliferation pathway (adjusted p-value = 0.003, enrichment ratio = 2.7), suggesting that this biological process is particularly predictive of ICI response beyond the baseline inflammatory signature. This differential enrichment approach highlights the models’ ability to identify biologically relevant predictive signatures rather than merely reflecting the composition of the input gene set.Then overall survival cox analysis of these 112 genes was conducted (Supplementary Fig S5), 17 genes showed statistical significance, most genes showed protecting effects, and only 2 genes (CD 80 and CCR3) had noteworthy hazard ratios (HRs) (0.761 and 0.134 respectively). CD 80 protein is activated by the binding of CD28 or CTLA-4 and then induces T-cell proliferation and cytokine production. CCR3 protein is a receptor for C-C type chemokines.
Finally, but most importantly, the genes features learned by AMU showed biological significance. We found that the mRNA embedding matrix was hard to perform a desirable cluster analysis, also, the Euclidean distance and cosine similarity algorithm both revealed the gene features distributed uniformly and no aggregation. However, the genes association and interaction calculated with t-SNE algorithm showed locally similar with STRING. Four cases were visualization in Fig. 4. For CD4-MAPK14-PTPRC-SOCS1 subgroup, both mRNA embedding and STRING indicated an inner close association, and NEDD9 relatively isolated to them. For PDE3B-ELANE-CXCL8, mRNA embedding successfully mapped the close distance. In NRAS-LAGLS3-IL10-FCGR2B-CDKN1A-HMGB1 subgroup, most links were accurately figured out with a local difference that STRING showed CDKN1A -NRAS, but mRNA embedding showed CDKN1A- FCGR2B association. Another case was in CASP1-TLR9-CXCR3-ITGAL-TXNRD1 subgroup, most links were consistent except STRING described an interaction with CXCR3-ITGAL, but mRNA embedding didn’t figure it out.
Discussion
In industrial 4.0 age, DL has been the most advanced model algorithm. Since the Alexnet proposed in 2012, the convolutional networks have renewed and a new wave of artificial intelligence research and applications have begun. Then the transformer has been the most advanced deep learning technique and exhibited powerful performers in CV and NLP areas for its strong feature extraction ability of sequential and spatial interactions of data. AMU is a model connecting the transformer encoder with a convolutional network, it’s a successful trial of proving that the transformer structure is also feasible and superior for 1-D gene expression data (just like NLP) prediction task and splendid for gene feature learning. AMU also showed superior performance in testing dataset which tissue biopsies were post ICI and some of the features and data distribution had to be different from that of pre-ICI, which further proving AMU learned some essential features. Previously, several works have been done in using mRNA expression and clinical data to predict melanoma ICI response. Noam Auslander etc. reported an AUC of 0.83 for their IMPRES predictor35,. Another algorithm proposed by Philip Friedlander etc. was validated in the validation set with AUCs of 0.6227. By this cross-experiment comparison, AMU exhibited its advantage.
Furthermore, features abstracted from the embedding layer showed local similarity with laboratory results or curated databases, indicating the strong gene presentation abilities of the transformer encoder should be fully researched and utilized for more gene related downstream tasks. DL studies should sufficiently take advantage of the power of the transformer. Moreover, model interpretation is quite important for medical studies and clearly gene embedding can facilitate this work.
Additionally, in the model interpretation part, SVM, XGBoost and AMU consistently indicated that TNF- TNFRSF1A axes possessed the most important genes related to melanoma ICI response process. Previous mouse experiments published in Nature showed that anti-PD-1 and anti-CTLA-4 (NIVO + IPI) combined with TNF-α inhibitors could improve the course of colitis in a mouse model and enhance the anti-tumor effect36,. Phase Ib clinical trial showed the promising effect of combining Nivolumab and Ipilimumab with TNF-α inhibitor in advanced melanoma37. These facts indicate that machine learning models not only can be applied in predictive scenarios but also can provide suggestive information for further investigation.
Our work has several limitations that should be considered when interpreting the results:
-
1)
Sample Size and Representativeness: The total sample size (206 patients) remains modest for deep learning applications, potentially limiting the model’s ability to capture the full spectrum of biological variation. The heterogeneity of melanoma and treatment responses likely requires larger cohorts to develop truly robust predictors. Future work should aim to incorporate additional independent cohorts to validate and refine the model.
-
2)
Feature Selection: While our 160-gene set provides good interpretability and computational efficiency, it represents a pre-selected subset of the transcriptome based on prior knowledge. This approach may miss novel predictive markers not previously associated with melanoma or immune response. An unbiased whole-transcriptome approach might identify additional predictive signatures, though at the cost of interpretability and increased risk of overfitting given the sample size constraints38.
-
3)
Data Modality Limitations: Our model relies solely on mRNA expression data, while immunotherapy response is influenced by multiple biological factors including genomic alterations, protein expression, tumor microenvironment, and host factors. Integration of multi-omics data (including DNA mutations, methylation, proteomics) and clinical variables would likely enhance prediction accuracy, though such comprehensive datasets remain scarce.
-
4)
Post-treatment Testing: While we included post-treatment samples to test model robustness, the ideal clinical application requires accurate pre-treatment prediction. The performance difference between pre- and post-treatment samples highlights the biological reality that treatment itself alters the expression landscape.
-
5)
Biological Validation: Although our model identifies biologically plausible gene signatures, experimental validation of the causal relationship between these signatures and treatment response remains necessary. Functional studies in cell lines or animal models would strengthen the biological foundations of our predictions.
Despite these limitations, our approach demonstrates the value of transformer-based architectures for transcriptomic data analysis and provides a foundation for future refinement as larger, more comprehensive datasets become available.”
Data availability
All data and code have been made publicly available in an English-language GitHub repository: https://github.com/1692775560/AMU. This repository contains: Complete preprocessing scripts for all datasetsImplementation of the AMU model architectureTraining and evaluation code for all modelsModel interpretation scriptsCode to reproduce all figures and tablesProcessed datasets ready for model trainingDetailed documentation and usage instructions in EnglishFor users preferring the original Chinese platform, the project is also available at: https://aistudio.baidu.com/aistudio/projectdetail/4298990.
References
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Commun. ACM. 60 (6), 84–90 (2017).
Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929, (2020).
Liu, Z. et al. Swin transformer: Hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF International Conference on Computer Vision 10012-10022. (2021).
Devlin, J. et al. Bert: Pre-training of deep bidirectional Transformers for Language Understanding. arXiv preprint arXiv:1810.04805. (2018).
Li, J. et al. Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspective. arXiv preprint arXiv:2206.01136. (2022).
Zuo, Z. et al. SWnet: a deep learning model for drug response prediction from cancer genomic signatures and compound chemical structures. BMC Bioinform. 22(1), 434. https://doi.org/10.1186/s12859-021-04352-9 (2021).
Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 28 (9), 704–705. https://doi.org/10.1038/s41594-021-00650-1 (2021).
Liu, Q., Hu, Z., Jiang, R. & Zhou, M. DeepCDR: a hybrid graph convolutional network for predicting cancer drug response. Bioinformatics 36 (Suppl_2), i911–i918. https://doi.org/10.1093/bioinformatics/btaa822 (2020).
He D, Xie L. A Cross-level information transmission network for hierarchical omics data integration and phenotype prediction from a new genotype. Bioinformatics. 38(1):204–210. https://doi.org/10.1093/bioinformatics/btab580. (2021)
Dummer, R. et al. Five-Year analysis of adjuvant Dabrafenib plus Trametinib in stage III melanoma. N Engl. J. Med. 383 (12), 1139–1148. https://doi.org/10.1056/NEJMoa2005493 (2020).
Eggermont, A. M. M. et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma (EORTC 1325-MG/KEYNOTE-054): distant metastasis-free survival results from a double-blind, randomised, controlled, phase 3 trial. Lancet Oncol. 22(5), 643–654 (2021).
Weber, J. S. et al. Sequential administration of nivolumab and ipilimumab with a planned switch in patients with advanced melanoma (CheckMate 064): an open-label, randomised, phase 2 trial [published correction appears in Lancet Oncol.17 (7):e270]. Lancet Oncol. 17(7):943–955. https://doi.org/10.1016/S1470-2045(16)30126-7 (2016).
Janjigian, Y. Y. et al. CheckMate-032 Study: Efficacy and Safety of Nivolumab and Nivolumab Plus Ipilimumab in Patients With Metastatic Esophagogastric Cancer. J Clin Oncol 36(28), 2836–2844. https://doi.org/10.1200/JCO.2017.76.6212 (2019).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl. J. Med. 366 (26), 2455–2465. https://doi.org/10.1056/NEJMoa1200694 (2012).
Rozeman, E. A. et al. Survival and biomarker analyses from the opacin-neo and opacin neoadjuvant immunotherapy trials in stage III melanoma. Nat. Med. 27 (2), 256–263. https://doi.org/10.1038/s41591-020-01211-7 (2021).
Zeng, D. et al. Tumor microenvironment evaluation promotes precise checkpoint immunotherapy of advanced gastric cancer. J. Immunother Cancer. 9 (8), e002467. https://doi.org/10.1136/jitc-2021-002467 (2021).
Marabelle, A. et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 21 (10), 1353–1365. https://doi.org/10.1016/S1470-2045(20)30445-9 (2020).
Janjigian, Y. Y. et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet 398 (10294), 27–40. https://doi.org/10.1016/S0140-6736(21)00797-2 (2021).
Diaz, L. A. Jr et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 23 (5), 659–670. https://doi.org/10.1016/S1470-2045(22)00197-8 (2022).
Hellmann, M. D. et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl. J. Med. 378 (22), 2093–2104. https://doi.org/10.1056/NEJMoa1801946 (2018).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets [published correction appears in Nucleic Acids Res. 49(18):10800]. Nucleic Acids Res. 49(D1):D605-D612. https://doi.org/10.1093/nar/gkaa1074 (2021).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma [published correction appears in Nat Med. 26(7):1147]. Nat Med. 25(12):1916–1927. https://doi.org/10.1038/s41591-019-0654-5 (2020).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41 (Database issue), D991–D995. https://doi.org/10.1093/nar/gks1193 (2013).
Hugo, W. et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 165(1), 35–44. https://doi.org/10.1016/j.cell.2016.02.065 (2017).
Zappasodi, R. et al. CTLA-4 Blockade drives loss of Treg stability in glycolysis-low tumours. Nature 591 (7851), 652–658. https://doi.org/10.1038/s41586-021-03326-4 (2021).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171 (4), 934–949e16. https://doi.org/10.1016/j.cell.2017.09.028 (2017).
Friedlander, P. et al. Whole-blood RNA transcript-based models can predict clinical response in two large independent clinical studies of patients with advanced melanoma treated with the checkpoint inhibitor, tremelimumab. J. Immunother. Cancer 5(1), 67. https://doi.org/10.1186/s40425-017-0272-z (2017).
Luo, Y. et al. Transcriptome profiling of whole blood cells identifies PLEK2 and C1QB in human melanoma. PLoS One. 6 (6), e20971. https://doi.org/10.1371/journal.pone.0020971 (2011).
Saenger, Y. et al. Blood mRNA expression profiling predicts survival in patients treated with Tremelimumab. Clin. Cancer Res. 20 (12), 3310–3318. https://doi.org/10.1158/1078-0432.CCR-13-2906 (2014).
Chen, T. & Guestrin, C. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, (13–17 August, 2016) 785–794. https://doi.org/10.1145/2939672.2939785 (2016).
Scott, M., Lundberg, S. I. & Lee A Unified Approach to Interpreting Model Predictions. Advances in Neural Information Processing Systems 30 (NIPS). 30 (2017).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25 (1), 25–29. https://doi.org/10.1038/75556 (2000).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47 (D1), D419–D426. https://doi.org/10.1093/nar/gky1038 (2019).
Gene Ontology Consortium. The gene ontology resource: enriching a gold mine. Nucleic Acids Res. 49 (D1), D325–D334. https://doi.org/10.1093/nar/gkaa1113 (2021).
Auslander, N. et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma published correction appears in Nat Med. 24(12):1942. Nat Med. 24(10):1545–1549. https://doi.org/10.1038/s41591-018-0157-9 (2018).
Perez-Ruiz, E. et al. Prophylactic TNF Blockade uncouples efficacy and toxicity in dual CTLA-4 and PD-1 immunotherapy. Nature 569 (7756), 428–432. https://doi.org/10.1038/s41586-019-1162-y (2019).
Montfort, A. et al. Combining nivolumab and ipilimumab with Infliximab or certolizumab in patients with advanced melanoma: first results of a phase Ib clinical trial. Clin. Cancer Res. 27 (4), 1037–1047. https://doi.org/10.1158/1078-0432.CCR-20-3449 (2021).
Ben, S. Robert Geirhos,Shashank Shekhar,Surya Ganguli. Beyond neural scaling laws: beating power law scaling via data pruning. arXiv:2206.14486 (2022).
Yin, Y. et al. AMU: Using mRNA Embedding in Transformer Network to Predict Melanoma Immune Checkpoint Inhibitor Response.. medRxiv https://doi.org/10.1101/2022.10.01.22280593 (2022).
Gschwind, A. & Ossowski, S. .AI model for predicting Anti-PD1 response in melanoma using. Multi-Omics Biomarkers Cancers. 17 https://doi.org/10.3390/cancers17050714 (2025).
Acknowledgements
The authors thank Yu Kang, Xiaohua Zhang, Cunxi Li for helpful discussion, Tianxuan Qi for coding enlightenment, Jianfeng Wang and Yunjian Huang for work incentives, and thanks to Baidu Paddle team for providing free online GPU and training courses to us. A preprint has previously been published as a preprint39.
Funding
We would like to thank the Joint Funds for the innovation of science and Technology, Fujian province (Grant number: 2024Y9499) for this work.
Author information
Authors and Affiliations
Contributions
Conceptualization: Yi Yin Methodology: Tao Zhang Visualization: Yi Yin, Ziming Wang Project administration: Yi Yin, Tao Zhang Supervision: Writing – original draft: Yi Ying Writing – review & editing: Yi Yin, Tao Zhang.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yin, Y., Zhang, T. & Wang, Z. An attention-based mRNA transformer network for accurate prediction of melanoma response to immune checkpoint inhibitors. Sci Rep 15, 31908 (2025). https://doi.org/10.1038/s41598-025-15830-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-15830-y