Abstract
Geographic atrophy, the advanced form of age-related macular degeneration (AMD), is associated with increased oxidative stress and chronic inflammation. Pro-inflammatory genes, like TNF-α and IL-1β, are under the regulation of the transcription factor p65/RelA. We have previously shown that adeno-associated virus (AAV) delivery of the RelA inhibitory gene M013 blocks retinal inflammation in uveitis models. In this study, we evaluated the effects of RelA inhibition in an oxidative stress-driven geographic atrophy mouse model. We injected Sod2RPEcKO mice with rAAV, delivering either secreted GFP (sGFP control) or sGFP fused to a cell-penetrating version of the tagged M013 (sGFP-TatM013v5). Over nine months, we measured retinal function, structure, and morphological changes using electroretinography, optical coherence tomography, and fundoscopy. We quantified changes in inflammatory markers using multiplex ELISA, RT-qPCR, and immunofluorescence staining of the retinal tissue. Finally, we generated an NF-kB-luciferase reporter microglia cell line to study the impact of immune signaling changes on microglia. Mice injected with the rAAV delivering M013 had transient protection of their retinal function at 3 months. Based on ERG evaluations, the intravitreal injection of rAAV delivering sGFP-TatM013v5 significantly delayed the loss of retinal function. Furthermore, the rAAV-mediated expression of the sGFP-TatM013v5 protected photoreceptors’ outer and inner segments based on OCT and immunofluorescence analysis. Analysis of postmortem tissues showed decreased migration of immune cells towards the RPE. Retinas injected with the sGFP-M013v5 vector showed increased levels of IL-9, IL10 and LIF. Finally, adding LIF to our NF-kB reporter cell line showed decreased TNF-induced reporter expression and modulation of microglia-specific genes. Our results indicate that modulating retinal inflammation could significantly slow the degeneration associated with geographic atrophy. Specifically, inhibiting the RelA protein in the retina may offer protective effects against retinal degeneration. Additionally, we demonstrated that LIF can counteract the influence of TNF on microglial gene expression. Future research will explore the dynamic interactions between RelA and other transcription factors and the NF-kB signaling pathway in the retina as they relate to retinal diseases.
Similar content being viewed by others
Introduction
Age-related macular degeneration (AMD) is the principal cause of vision loss among individuals aged 60 years and older in developed countries1. Affected individuals accumulate lipid-rich deposits called drusen beneath the retinal pigment epithelium (RPE). Complement and complement-associated proteins in drusen and genome-wide association studies (GWAS) implicate an inflammatory process in AMD2. GWAS studies have identified variants of complement factor H and the ARMS2/HTRA1 locus strongly associated with AMD development3. More recently, the expression of complement factor-related genes has been associated with AMD, further strengthening the role of inflammation in this disease4. Plasma levels of TNFɑ are elevated in intermediate AMD and geographic atrophy, the advanced severe form of dry-AMD, and the NLRP3 inflammasome becomes activated in the RPE and inflammatory cells in the retina5,6.
There is no animal model that can recapitulate all the pathological features of AMD. However, there are many animal models that show phenotypic characteristics like the pathology of AMD. The models range from transgenic mouse models (e.g., mice carrying the CFH variant linked to increased AMD risk), knock-out mice (e.g., LXRα−/− mouse, and the Sod2RPEcKO mouse), and acute stress models (e.g., sodium iodate mouse model). Deletion of the Sod2 gene in the RPE of the Sod2RPEcKO mouse increases oxidative stress in these cells. Over time, the RPE degenerates and leads to a slow decline in retinal function and thinning of the RPE and outer nuclear layer5. We have previously observed that this model, similar to human geographic atrophy, has a retinal inflammatory response6. Therefore, we carried out our experiments using the Sod2RPEcKO mouse model of geographic atrophy.
Viruses have evolved to evade the host’s inflammatory response. Myxoma virus is a poxvirus belonging to the genus Leporipoxvirus. Deleting the M013 gene in this virus permits the detection of the virus by host immune cells and triggers an inflammatory response7. The M013 protein contains a pyrin domain that inhibits the NLRP3-inflammasome signaling pathway. Its C-terminal domain also inhibits the nuclear translocation of p65/RelA (an NF-κB subunit)8,9, thereby blocking the expression of potent pro-inflammatory genes, such as TNFɑ and interleukin-1 beta (IL-1β). These signaling pathways have been linked to AMD; however, their contribution to AMD pathology remains unclear.
Our group has developed an adeno-associated viral (AAV) vector that delivers a secretable and cell-penetrating form of the myxoma M013 gene10. This vector significantly decreased the number of infiltrative cells within the vitreous humor in an animal model of experimental autoimmune uveitis11. We also demonstrated that this vector strongly reduced the concentration of IL-1β in the vitreous humor of these animals.
Given the association of inflammatory pathways (such as the inflammasome) with AMD12,13,14,15,16,17, we tested the effects of a single intravitreal injection of AAV delivering the secretable and cell-penetrating form of M013 in the RPESod2cKO mouse model of geographic atrophy (an advanced form of AMD)5,18,19. This model renders the retinal pigmented epithelium (RPE) more susceptible to oxidative damage and recapitulates some cardinal features of human disease, including increased lipofuscin autofluorescence, Bruch’s membrane disruption, sub-RPE accumulation deposits, retinal inflammation, and complement protein deposition. These changes can be documented in living mice by declines in electroretinography (ERG) responses, fundus abnormalities, and alterations in photoreceptors and RPE reflectance measured by spectral-domain optical coherence tomography, SD-OCT. The current study tested whether inhibiting the retinal inflammation by blocking the activity of the RelA/p65 transcription factor could protect the retina against degeneration.
Materials and methods
Chemicals and antibodies
Table 1 lists the antibodies used and their dilutions. The antibodies were stored at 4 °C. Primer sequences are provided in Supplementary Table 2. All primers synthesized by Eurofins Genomics were resuspended in RNAse- and DNAse-free water and stored at -20 °C.
Animals
The University of Florida IACUC approved the experiments described in this study. All procedures adhered to ARVO guidelines for the use of animals in biomedical research. This study is reported in accordance with ARRIVE guidelines. Our studies included equal numbers of male and female mice. Mice were reared in a 12-hour light and 12-hour darkness cycle. C57BL6J mice (8–19 weeks of age) were used in the experiments involving the endotoxin-induced uveitis model, as described below. We used the RPE-specific Sod2 conditional KO mouse model to study geographic atrophy. We fed the mice two weeks of doxycycline-containing chow (200 mg doxycycline/diet kg). We then switched to regular chow for the remainder of the experiment. Before each procedure, mice were anesthetized with an intraperitoneal injection of a ketamine/xylazine mixture (100 mg/kg ketamine, 4 mg/kg xylazine). After the procedure, the mice received an intraperitoneal injection of atipamezole (1 mg/kg) to reverse the effects of anesthesia. Animals were kept on a heated plate until fully ambulatory before returning to their racks. Mice were euthanized using CO2 asphyxiation followed by cervical dislocation.
Vector design
We created the sGFP-TatM013v5 vector by fusing the sequence of the cell-penetrating peptide of HIV Tat and the M013 gene of the myxoma virus containing a V5 epitope tag. We cloned this fusion downstream of the IgG kappa light-chain leader sequence linked in-frame to the coding sequence of green fluorescent protein (GFP). This gene cassette was cloned into the pTR-smCBA-AAV plasmid (Fig. 1A). The pTR-smCBA-Igκ-GFP- TatM013v5 plasmid was packaged, purified on iodixanol gradients, and titrated by the Ophthalmology Vector Core at the University of Florida according to published methods20,21.
Intravitreal injection of AAV
The eyes of mice were dilated using ophthalmic solutions of atropine (1%) and phenylephrine (2.5%). Next, the mice were anesthetized with a ketamine/xylazine mixture (100 mg/kg ketamine and 4 mg/kg xylazine). A drop of proparacaine hydrochloride ophthalmic solution (0.5%) was added to the eye as a topical anesthetic. An incision was made in the limbus region using a 27 G needle, then a Hamilton syringe with a 33 G needle was inserted. Once inside the vitreous, 1 µL of the appropriate AAV dose was injected, as observed using a Leica stereoscope. Subsequently, the needle was retracted, and the mice were given an intraperitoneal injection of Antisedan® (atipamezole, 1 mg/kg) and topical antibiotics at the injection site. The mice were placed on a warm pad until ambulatory before returning to their housing rack.
Endotoxin-induced uveitis (EIU) model
We used the EIU mouse model to validate the anti-inflammatory properties of the V5-tagged M013 gene as previously described10. Briefly, C57BL/6J mice were injected intravitreally with the corresponding AAV vector. Four weeks after vector injection, we injected the mice intravitreally with 125 ng of lipopolysaccharide (LPS) per eye.
Spectral domain optical coherence tomography (SD-OCT)
Mouse eyes were dilated with topical atropine and epinephrine solutions and anesthetized with a ketamine-xylazine mixture (100 mg/kg ketamine: 4 mg/kg xylazine) administered intraperitoneally (i.p.). The anesthetized mice were placed on a platform and restrained in place. A total of 250 B-scans centered on the optic nerve head were obtained using the Bioptigen spectral domain optical coherence tomography equipment. Afterward, the mice received an i.p. injection of atipamezole (0.25 mg/kg) and were kept on a heated pad until ambulatory. Scans were averaged into 25 B-scans subjected to auto-segmentation using Diver Software from Bioptigen.
Quantification of infiltrative cells from SD-OCT images
SD-OCT b-scans images were opened with Image J software using the OCT reader plugin. Images were then inverted and flipped. The background was subtracted using the software function (using a 50-pixel setting) to improve the signal-to-noise ratio. The image was then scaled by using a 1024 pixels/1400 microns ratio. The brightness was adjusted using the auto-adjust brightness/ contrast option, followed by adjusting the threshold to 11–12% depending on the image. Using the binary option under the process tab, we selected the convert to mask option. Clusters of cells in the vitreous humor are separated digitally using the watershed option in Image J. The vitreous humor was selected, and the area outside was cropped using the polygon option. Finally, the infiltrative cells were counted using the count particles function with the following settings: 50–300 microns2, exclude on edges. This process was repeated for 3 images from each eye, and the average number of cells was determined.
Fundoscopy
The eyes of the mice were dilated with drops of phenylephrine and atropine ophthalmic solutions. The mice were anesthetized with a mixture of ketamine and xylazine, as described previously. Next, the mice were placed on a platform, and their eyes were covered with a drop of Genteal to avoid dehydration. A Micron III fundus camera was focused on the retina, centered on the optic nerve. Images were captured using bright-field and fluorescent filters. The same exposure time and light intensity were used for all images.
Electroretinography (ERG)
Mice were dark-adapted for 16 h overnight. The following day, the eyes of the mice were dilated with atropine and epinephrine eye drops, as described earlier. Next, the mice were placed on a platform, and electrodes were placed on their mouth, tails, and corneas. The corneal electrodes were adjusted until similar impedance values (< 10 µV) were obtained for both eyes.
In comparison, the mouth and tail impedances are < 16 µV. Mice were then subjected to three flashes of 20 cds/m2 separated by 2-minute intervals using a full-field dome. After the last recording, the values were averaged, and the a-, b-, and c-wave amplitudes were determined. Finally, the mice received an i.p. injection of atipamezole, as previously described.
Retina RNA extraction
Mice were humanely euthanized, and their retinas were extracted and placed in 0.4 mL of TRIzol reagent. An equal volume of 100% ethanol was then added to each sample and mixed. We used the Direct Zol miniprep kit from Zymo Research (Irvine, CA, USA) to isolate RNA according to the manufacturer’s protocol. RNA concentration was quantified using Qubit 2.0, as per the manufacturer’s protocol. The samples were stored at -80 °C.
Real-time quantitative PCR (RT-qPCR)
Following the manufacturer’s protocol, a cDNA library was generated using 500 ng of total RNA and the iScript cDNA synthesis kit from Bio-Rad. The concentration of the cDNA was then quantified using the Qubit 2.0, assay for oligos, per the manufacturer’s protocol. The samples were then diluted to a concentration of 2 ng/µL. qPCR was performed using 3 µL of each cDNA sample (6 ng), 5 µL of Sosofast Eva Green (Bio-Rad) 2X master mix, and 2 µL of forward and reverse pre-mixed primers (10µM each). The reactions were duplicated for each sample and gene using a hard-shell 96-well PCR plate. The primer sequences are given in Table 2. The PCR conditions were as follows: 95 °C for 30 s, 98 °C for 10 s, 60 °C for 30 s, repeating steps 2 and 3 another 39 times, and melt curve from 65 to 95 °C with increments of 0.5 °C every 5 s. The actin gene primer set was used as the constitutive gene to standardize the Ct values of each sample. Fold changes were determined using the ΔΔCt method22,23. Values were plotted as Log2 of fold change.
Immunofluorescence
Mice were euthanized using CO2, followed by cervical dislocation. Eyes were enucleated, placed in 4% paraformaldehyde in PBS 1X, and kept on ice for 20 min. Eyes were then incubated in 10% sucrose in PBS 1X solution for 1 h, followed by another hour of incubation in 20% sucrose in phosphate-buffered saline (PBS) solution. The samples were then transferred to 30% sucrose in PBS and stored at 4 °C overnight. The eyes were rinsed once in Optimum Cutting Temperature (OCT) medium and then placed in a plastic mold containing OCT medium. The eyes were snap-frozen in liquid nitrogen. We collected 14-micron-thick sections using a Leica cryostat and fixed them on a glass slide using cold acetone. The slides were stained using a Shandon Sequenza slide rack and permeabilized with 1% Triton-X100 in PBS supplemented with 5% normal horse or goat serum, depending on the antibodies used. Primary and secondary antibodies were used at the dilutions listed in Table 1. DAPI was used at a 1:1,000 dilution as a nuclear counterstain. Finally, the slides were imaged using a wide-field Leica fluorescence microscope and the LAS-X software from the same company.
Retinal protein extraction
Mice were humanely euthanized by CO2 inhalation, followed by cervical dislocation to ensure death. A horizontal incision was made through the cornea using 10 scalpels. The aqueous humor was blotted onto the tissue. The cornea was removed using micro-scissors, and the lens, vitreous, and retina were squeezed by applying pressure on the back of the globe with a pair of forceps. The lens and vitreous were separated from the retina, which was then placed in 100 µL of NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl (pH 8.0), 150 mM NaCl) supplemented with Halt’s protease inhibitor cocktail (Sigma-Aldrich). The retinas were then homogenized using 30 firm strokes of a motorized pestle. The samples were centrifuged at 10,000 × g for 5 min at 4 °C to remove non-solubilized material. Retina homogenates were collected and transferred into a new and sterile 1.5 mL tube. Finally, the protein concentration was determined using the DC Assay (Bio-Rad) according to the manufacturer’s protocol. The samples were then diluted to 1 µg/µL using NP-40 lysis buffer.
Protein concentration assay
protein concentration was determined using the Bradford assay (Bio-Rad). Briefly, the BSA concentration was determined using a nano-spectrophotometer. Standard was diluted into 1.5, 1.2, 0.9, 0.6, and 0.3 µg/µL using NP-40 lysis buffer. Reagent A from the kit was mixed with reagent S as indicated by the manufacturer’s protocol to prepare reagent A’. A total of 25 µL of A’ was added to each well, followed by 5 µL of the sample. Next, 200 µL of Reagent B was added to each well using a multichannel pipette, followed by a 15-minute incubation at room temperature with constant shaking. The absorbance at 750 nm was measured using a plate reader. The absorbance of the blank wells was subtracted from that of all the standards and samples. The sample concentrations were extrapolated from linear regression generated using BSA standards.
Multiplex ELISA
Twenty-five micrograms of protein lysate were used to set up a multiplex ELISA plate. According to the manufacturer’s instructions, the MILLIPLEX Map Mouse cytokine/chemokine Magnetic Bead Panel-Pre-mixed Plex-Immunology Multiplex Assay (MCYTMAG-70 K-PX32) was used. The Mouse Cytokine/Chemokine Magnetic Bead Panel plate was prepared by washing with 200 µL of Wash Buffer and shaking vigorously for 10 min. The Wash Buffer was decanted, and the residual buffer was removed by lightly tapping the plate on an absorbent towel. Standards and Quality Controls (25 µl) were added to the appropriate wells. The assay buffer was used as the blank standard. Assay Buffer (25 µL) was added to all wells, corresponding to the samples, standards, and quality controls. Each sample (25 µL) was added in duplicate to the appropriate wells. Antibody-Immobilized Pre-mixed Beads were prepared by sonicating the bottle for 30 s and then mixing the contents using a vortex mixer for 1 min. After mixing, 25 µL of pre-mixed beads was added to each well. Beads were mixed intermittently by vortexing after each column. The plate was sealed with a foil plate sealer and incubated overnight at 4 °C with agitation on a plate shaker.
The next day, the contents of the wells were removed by placing the plate on a magnetic 96-well plate separator (Millipore, Burlington, MA, US) for 1 min to allow the magnetic particles to remain bound. Wash Buffer (200 µL) was added to the plate, shaken vigorously for 30 s, placed on the magnetic plate for 1 min, and then decanted. This step is repeated twice. After the detection antibodies were warmed to room temperature, 25 µL was added to each well containing the Standards, the Quality Controls, and samples. The plate was sealed with a foil plate sealer and incubated at room temperature with agitation on a plate shaker for 1 h. Streptavidin-phycoerythrin (25 µL) was added to each well containing 25 µL of detection antibody. The plate was sealed with a foil plate sealer and incubated at room temperature with agitation on a plate shaker for 30 min. The plate was placed on a magnetic plate for 1 min to keep the magnetic particles bound, and then the plate was decanted. Wash Buffer (200 µL) was added to the plate, shaken vigorously for 30 s, placed on the magnetic plate for 1 min, and then decanted. This step is repeated twice. Next, 150 µL of Wash Buffer was added to all wells containing the Standards, the Quality Controls, and samples. The plate was run on MAGPIX with Xponent Software (Millipore, Burlington, MA, US). A total of 150 beads were acquired per well.
Western blot
Protein lysates were diluted to 1 µg/µL in NP-40 lysis buffer and 5X Laemmli Buffer24 containing 100 mM DTT. The samples were boiled for 5 min to denature the proteins. A total of 25 µg of protein was loaded onto a 4–12% SDS-PAGE gel and run at 200 volts for 22 min. Proteins were transferred onto a nitrocellulose membrane using iBind 2. The membrane was then incubated with anti-V5, anti-beta tubulin, donkey anti-mouse CW680, and donkey anti-rabbit CW800 antibodies using the iBind Flex system from Invitrogen, according to the manufacturer’s protocol. Finally, the membrane was scanned using the Odyssey CLx scanner.
Stable Sim A9 NFKBRE-luc cell line
A total of 3 × 105 Sim A925 cells were seeded into two wells of a 6-well plate in standard growth medium a day before viral transduction. The following day, the medium was changed with 1mL of nil medium (no serum or antibiotics), and the cells were transduced with an MOI of 10 of the lentiviral vectors at 10^8Tu/mL concentration (Vector Builder). Additionally, the medium was supplemented with 5ug/uL (Vector Builder) of polybrene to increase transduction efficiency. One well was left untreated as a control. The medium containing the virus and polybrene was removed 6 h post-transduction and deactivated according to the EH&S guidelines. The cells were supplemented with a growth medium and incubated for 48 h before antibiotic selection. Cells were selected using 10 µg/mL puromycin diluted in growth medium. Cells were selected using puromycin for 72 h. After 72 h, the cells were observed for GFP expression using a wide-field fluorescence microscope (Leica). Cells were moved to a 10 cm dish to expand the line and supplemented with a maintenance dose of 5ug/mL puromycin in growth media. Once the cells were confluent in the 10 cm dish, they were moved to a T75 flask.
Luciferase assay
A total of 40,000 Sim N9 NFKBRE-luc cells were seeded in a 96-well plate a day prior. The cells were treated with the appropriate concentrations of cytokines diluted in Nil medium for 24 h. The medium containing the cytokines was removed, and 30 µL of lysis buffer was added to the cells. The plate was incubated at -80 °C for 1 h. The plate was thawed for 5 min at room temperature. Afterward, the cell lysates were collected, stored in a 1.5 mL tube, and kept on ice. The protein concentration of the lysates was determined using the Bio-Rad Dc Assay according to the manufacturer’s protocol with the luciferase standard supplied in the Luciferase Assay System (Promega) A 50uL aliquot of substrate equilibrated to room temperature was added to each sample, gently mixed by flicking, and read immediately using a luminometer (Turner Biosystems).
Statistical analysis
Values are reported as the mean ± standard deviation. Data were analyzed using one-way or two-way ANOVA, followed by a post-hoc Holm-Sidak test using GraphPad Prism 9 (La Jolla, CA, USA). Statistical significance was considered when the p-value or adjusted p-value was ≤ 0.05.
Results
Tagged M013 retains its anti-inflammatory properties
We previously demonstrated that gene delivery of the myxoma virus M013 gene protects the retina in endotoxin-induced uveitis (EIU) and autoimmune uveitis mouse models10,11. However, there is currently no antibody to detect the expression of M013. Thus, we added a V5 epitope tag (GKPIPNPLLGLDST) to the C-terminus of the M013 cDNA (Fig. 1A). To confirm that adding a V5 tag did not affect the function of M013, we tested the sGFP-TatM013v5 AAV vector in the EIU mouse model, which was generated by injecting E. coli lipopolysaccharide (LPS) into the vitreous 24 h before SD-OCT analysis and sacrifice of the mice. All mice were injected intravitreally with AAV, delivering either secreted GFP (sGFP) or sGFP-TatM013v5 one month before EIU induction with LPS. We quantified the recruitment of inflammatory cells to the vitreous humor and retinal thickness using SD-OCT (Fig. 1B).
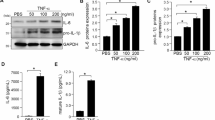
When the number of infiltrative cells was quantified before and after intravitreal injection of endotoxin (125 ng LPS), we observed a 28-fold increase in these cells in sGFP-treated retinas. In contrast, in retinas treated with TatM013v5, the increase in the number of infiltrating cells was not statistically significant. Furthermore, when we compared retinas treated with LPS only, treatment with TatM013v5 significantly decreased the number of infiltrating cells (Fig. 1C).
The total retinal thickness was also measured using SD-OCT images. The retinas treated with sGFP showed an average 73% increase in thickness when injected with LPS due to retinal edema and structural damage. In contrast, TatM013v5-treated retinas showed no significant increase in total thickness when treated with the same dose of LPS (Fig. 1D). Our results demonstrated that our M013 cDNA with a V5 tag retained robust anti-inflammatory properties.
Gene delivery of M013 delays retinal loss-of-function in a GA mouse model
There are multiple mouse models of geographic atrophy; however, none can recapitulate all clinical features of human disease. We used the RPE-specific Sod2 KO (Sod2RPEcKO) mouse model developed by Mao et al.5. The RPESod2cKO mouse is a genetic model in which the Sod2 gene is deleted exclusively within RPE cells, thus leading to increased oxidative damage and a slow structural and functional decline of the retina26,27. We previously demonstrated that the RPESod2cKO mouse model develops slow retinal degeneration and chronic retinal inflammation6. Because of the well-characterized phenotype of the RPESod2cKO model5,6,18, we decided to use a secretable GFP-delivering AAV vector to compare transgene expression. We injected two-month-old mice of the correct genotype intravitreally with 1010 vector genome copies of AAV2quad(Y-F) + T495V vector delivering either secreted GFP (sGFP) or sGFP-TatM013v5 under the control of the constitutively active small CBA promoter. We evaluated mice one month after vector injection using fluorescence fundoscopy to validate transgene expression. Mice showed diffused GFP expression in both eyes, thus demonstrating that our AAV vectors express secreted GFP encoded in their genetic cargo (Fig. 2A–D). Furthermore, we detected secreted and cleaved TatM013v5 in retinal protein lysates as a 15 KDa band (Fig. 2E, and Supplementary Fig. 1) 9 months post-injection.
TatM013v5 decreases ocular inflammation in the EIU mouse model. (A) Genetic map of the pTR-smCBA-sGFP-FCS-TatM013v5 vector. The control vector has the same sequence except with the TatM013v5 insert. (B) Representative SD-OCT B-scans of mice treated with AAV that delivered either sGFP or sGFP-TatM013v5, twenty-four hours after endotoxin (LPS) injection. (C) Two-masked individuals quantified infiltrative cells in the vitreous humor using Image J software. LPS injection only significantly increased infiltrative cells in sGFP-treated eyes. (D) Total retinal thickness measurements were done before and after the LPS injection. LPS injection caused a significant increase in retina thickness only in sGFP-treated eyes. Values represent average ± standard deviation (n = 5–6 mice). ** = adjusted p-value ≤ 0.01, *** = adj. p-val ≤ 0.001, **** = adj. p-val ≤ 0.0001 (pTR – plasmid with terminal repeats, smCBA – small chicken-beta actin promoter, sGFP – secretable green fluorescent protein, FCS – furin cleavage site, SD-OCT – spectral domain optical coherence tomography, LPS – lipopolysaccharide).
To determine whether the TatM013v5 vector protected retinal function in our mouse model, we used electroretinography (ERG) to evaluate retinal sensitivity to light (Fig. 2F). At three months of age, one month after treatment, the amplitudes associated with the activity of the photoreceptors (a-wave), second-order neurons (b-wave), and even the RPE (c-wave) were significantly higher in eyes treated with the TatM013v5 expressing vector when compared with those treated with the sGFP-expressing vector. In addition, animals treated with the TatM013v5 vector had higher a, b-, and c-wave amplitudes at three and six months of age (Fig. 2G–I). Moreover, the average amplitude of all waves at nine months in TatM013v5-treated animals was like that of sGFP-treated animals at three months. These observations suggest that, although TatM013v5 expression does not stop retinal loss of function, it significantly delays retinal degeneration.
Retinal delivery of TatM013v5 protects the retina function of the RPESod2cKO mouse model. (A–D) Two-month-old mice of the correct genotype were injected intravitreally with 1010 vgc of AAV2quad(Y-F) + T491V, delivering either sGFP (A,B) or sGFP-TatM013v5 (C,D). One month later, fluorescent fundoscopy using a Micron III camera confirmed transgene expression. Representative images demonstrate the presence of GFP fluorescence in the retina of either sGFP or sGFP-TatM013-treated eyes. (E) Western blot showing the retinal expression of TatM013v5 in sGFP-TatM013v5-treated mice. Mice were evaluated with ERG at 3-, 6-, and 9-months of age. (F) Representative ERG recording at six months of age. The average amplitudes of their (G) a-wave, (H) b-wave, and (I) c-wave at each time point are graphed as a function of time in months. Eyes treated with the AAV vector delivering sGFP-TatM013 had significantly higher amplitudes when compared to sGFP-treated eyes. Values are reported as average ± standard deviation. (n = 5 mice per group). * = adj. p-val ≤ 0.05 (ERG – electroretinogram).
TatM013v5 treatment preserves photoreceptors’ inner and outer segments
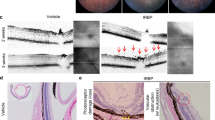
The effects of retinal expression of TatM013v5 on retinal structure in RPESod2cKO mice were studied using spectral-domain optical coherence tomography (SD-OCT). We evaluated mice at 3, 6, and 9 months of age to determine whether the functional effects observed by ERG correlated with the structural effects on the retina. No significant difference was observed between the thickness of the outer nuclear layer (ONL) of sGFP- and M013-treated eyes (data not shown), suggesting similar survival of photoreceptors regardless of treatment. However, the eyes treated with the M013 vector had longer inner and outer segments of photoreceptors (inner segments (IS) and outer segments (OS), respectively) than sGFP-treated eyes (Fig. 3A, B). Furthermore, when retinal cryosections from both groups were stained with peanut agglutinin (PNA) (a lectin that stains cone glycoproteins28,29, we observed that TatM013v5-treated retinas had more intact inner and outer segments (Fig. 3C, white arrows). These results indicate that TatM013v5 retinal expression can protect the inner and outer segments of photoreceptors, which explains the functional effects described earlier.
Effects of TatM013v5 on photoreceptor’s inner and outer segments of the RPESod2cKO mouse model. Two hundred and fifty B-scans of mice retinas were acquired using a Bioptigen spectral-domain optical coherence tomography. We averaged every 10 B-scans to obtain 25 high-resolution images. Bioptigen Diver auto-segmentation software was used to measure the thickness of each retina layer. The thickness of the (A) inner segments (IS), and (B) outer segments (OS) of retinas treated with either sGFP (green) or sGFP-TatM013v5 (red) were plotted as a function of the animal age. (C) Retina cross sections were stained with peanut agglutinin (PNA) to detect the structure of IS and OS. sGFP-TatM013v5 treated retinas had more intact IS and OS based on the PNA staining (red channel). Values were reported as average +/- standard deviation (n = 5 mice). * = adj. p-val ≤ 0.05, *** = adj. p-val. ≤ 0.001.
TatM013v5 retinal treatment decreases inflammation-associated genes
M013 blocks nuclear translocation of the RelA/p65 subunit of the NF-kB transcription factor. In addition, M013 interacts with ASC-1 through a different domain and prevents its incorporation into the NLRP3 inflammasome9. We hypothesized that the retinal expression of M013 would decrease the inflammatory response in our mouse model of GA. Therefore, frozen sections from animals treated with either the sGFP or sGFP-TatM013v5 vector were sectioned and stained with anti-CD45 antibody to detect the presence of immune cells within the retina. In sections of three-month-old mice, the CD45-positive cells were detected in the ONL and subretinal space in sGFP-treated eyes (Fig. 4A). In TatM013v5-treated eyes, CD45 cells were detected only in the inner retina (Fig. 4B), characteristic of the distribution of microglia and astrocytes in the absence of inflammation. CD45-positive cells interacted with the RPE layer only in sGFP-treated eyes.
We also studied the activation of Müller glia by immunostaining with anti-glial fibrillary acidic protein (GFAP). Based on this staining, Sod2 knockout eyes treated with the sGFP vector showed significant activation of Müller glia (Fig. 4C). This fluorescent pattern differs from that observed in TatM013v5-treated retinas, which had minor GFAP staining, especially in the transverse orientation, indicative of reduced Müller glial activation (Fig. 4D). We confirmed the specificity of our staining in sections stained without any of the primary antibodies (Supplementary File 2).
Mononuclear phagocytes (e.g., microglia, macrophages, and monocytes) express genes involved in either an active inflammatory response (M1 phenotype) or pro-angiogenesis response (M2 phenotype), which are considered anti-inflammatory30. To determine whether retinal M013 expression affects the levels of either M1- or M2-associated genes, we used RT-qPCR in retinal RNA samples to quantify gene expression changes. Expression of M1-related genes was decreased in TatM013v5-treated retinas in comparison to sGFP-treated retinas (Fig. 4E). Specifically, we found potent inhibition of nitric oxide synthase 2 (Nos2), signaling lymphocytic activation molecule 1 (Slamf1), and suppressor of cytokine signaling 3 (Socs3). However, no significant effect was observed on M2-associated genes, including arginase 1 (Arg1) and peroxisome proliferator-activated receptor gamma (Ppar-γ). These results suggest that the retinal expression of TatM013v5 can significantly suppress the pro-inflammatory response associated with our GA mouse model, thus explaining the functional protection of the retina.
TatM013v5 retinal expression decreases reactive gliosis and retinal inflammation. Cryosections from a 3-month-old Sod2RPEcKO mouse model were stained with antibodies against CD45 and Iba-1 or GFAP and Iba-1. (A) CD45-positive cells were detected through all the retinal layers in retinas treated with the sGFP vector, especially in the RPE layer (white arrows). (B) In contrast, retinas treated with the sGFP-TatM013v5 vector showed CD45 positive cells only between the RNFL and the IPL or below the RPE layer (white arrows). (C) Increased expression of GFAP through all retinal layers was observed in sGFP-treated retinas, indicating Müller glial activation. (D) The GFAP staining pattern in sGFP-TatM013v5 treated retinas was mainly limited to the RNFL, showing astrocyte expression and minimal staining in Müller glial cells. (E) Total RNA was isolated from retinas treated with an AAV vector, delivering either sGFP or sGFP-TatM013v5. Gene expression of M1-associated genes (e.g., STAT1 and Nos2) and M2-associated genes (e.g., PPar-γ and Arg1) were quantified using RT-qPCR. Beta Actin was used as the reference housekeeping gene for all samples, and the ΔΔCt method was used to determine the fold-change. Values are plotted as the Log2 of the fold-change (average ± standard deviation) to better illustrate the decrease in M1-gene expression induced by the expression of sGFP-TatM013v5. No significant changes were identified within the M2 genes. (n = 3 retina samples). * = adjusted p-value ≤ 0.05 (CD45 – cluster of differentiation 45, Iba-1 – ionized calcium-binding adaptor molecule 1, GFAP – glial fibrillary acidic protein, RNFL – retina nerve fiber layer, IPL – inner plexiform layer, RPE – retinal pigmented epithelium, RT-qPCR – reverse-transcribed quantitative polymerase chain reaction).
TatM013v5 retinal expression increases Stat3-activating cytokine expression
To determine the impact of TatM013v5 retinal expression on the concentrations of cytokines and chemokines in the retina, we quantified 32 analytes using multiplex ELISA. Retinal protein extracts were prepared, and equal amounts of protein were assayed. We compared all analyte concentrations to their respective concentrations at three months of age (Fig. 5A). IL-10, IL-9, and leukemia inhibitory factor (LIF) concentrations were differentially expressed among the 32 analytes. However, the concentration of these analytes decreased to the same levels as the sGFP control by nine months (Fig. 5B–D). Although not statistically significant, we also identified a trend towards an increase in IL-13 in TatM013v5-treated retinas at three months (Fig. 5E). Interestingly, IL-9, IL-10, LIF, and IL-13 signaling pathways activate STAT3 as part of their signaling pathway, which has been reported to have retinal neuroprotective effects31,33,33,34. Together, these results suggest that TatM013v5 inhibition of NF-kB activation in the retina induces retinal neuroprotection, potentially through STAT3 activation.
Retinal expression of TatM013v5 increases retinal concentration of IL-10, IL-9, and LIF. Retinal protein extracts from RPESod2cKO-treated mice were evaluated by multiplex ELISA using a pre-mixed kit for 32 cytokines and chemokines. Four of these analytes were significantly different, at least at a one-time point in the assay. (A) At three months of age, the anti-inflammatory cytokine IL-10 was increased in the sGFP-TatM013v5-treated retinas compared to sGFP-treated retinas (values relative to sGFP). (B) IL-9 was increased dramatically in sGFP-TatM013v5-treated retinas at three months of age. Likewise, IL-10 (C) and LIF (D) were increased substantially in sGFP-TatM013v5-treated retinas at three months. Finally, the cytokine IL-13 concentration increased in the sGFP-treated retinas by nine months and not in the sGFP-TatM013v5 (E). Values represent average ± standard deviation (n = 3–5 retina lysates). * = adj. p-val ≤ 0.05 (IL-10 – interleukin 10, IL-9 – interleukin 9, LIF – leukemia inhibitory factor, ELISA – enzyme-linked immunosorbent assay).
LIF antagonizes pro-inflammatory signals in microglia
The leukemia inhibitory factor (LIF) is a pleiotropic cytokine with retina neuroprotective effects35,36,37. It activates Stat3 by binding to its receptors, Lifr, and gp130. Although the effects of LIF on multiple retinal cells have been studied, its impact on microglial cells remains unknown. Because microglia are the resident immune cells of the retina, we tested whether changes in LIF concentration can inhibit the pro-inflammatory activity of microglia. To test our hypothesis, we generated a microglial cell line expressing a luciferase reporter under the control of the NF-κB response element (NF-kBRE-luc) (Fig. 6A). When incubated with TNF, the luciferase activity of these cells increased by approximately 60%. However, the addition of LIF in the presence of TNF significantly decreased luciferase activity in a dose-dependent manner (Fig. 6B). Similarly, luciferase activity was significantly reduced when incubated with LIF in the absence or presence of increasing doses of TNF (Fig. 6C). These observations suggest that LIF can antagonize pro-inflammatory signals mediated by NFKBRE activation.
LIF antagonizes pro-inflammatory signals in microglia. (A) Sim-A9 cells expressing the luciferase gene under the control of the NF-kBRE were incubated with or without either (B) Tnf or (C) Lif and with different concentrations of Lif (B) or Tnf (C) for 24 h. Values represent average ± SEM, compared by one-way ANOVA followed by Holm-Sidak test for multiple comparisons. *= adj. p-value≤ 0.05, **= adj. p-val≤ 0.01, ***= adj. p-val≤ 0.001, ****= adj p-val ≤0.0001. (n = 3 independent experiments, 15 biological replicates) (NF-kBRE – nuclear factor kappa-light-chain-enhancer of activated B cells response element, TNF – tumor necrosis factor).
Next, we tested whether this mechanism affects other microglia-related genes by treating SIM A9 cells with or without Tnf in the presence or absence of Lif for 24 h and using RT-qPCR to measure changes in gene expression. We generated primers for Tnf and Il-6 (two inflammatory cytokines), Cdk1 (a marker of proliferation), and P2ry12 (a microglia homeostatic marker). Tnf treatment increased the expression of both Tnf and Il-6, which was antagonized by the presence or Lif (Fig. 7A, B). In contrast, adding Lif increased the expression of P2ry12, which was antagonized by the presence of Tnf (Fig. 7C). Finally, although the addition of neither Tnf or Lif changed the relative expression of Cdk1, treatment with both Tnf and Lif simultaneously significantly decreased Cdk1 expression (Fig. 7D). These results demonstrate that Lif levels can modulate the expression of microglial genes involved in multiple functions. This effect can significantly impact the overall functional state of microglia during neuroinflammation.
Tnf and Lif regulate microglia pro-inflammatory, homeostatic, and proliferative genes in vitro. We stimulated Sim A9 microglia cells with Tnf, Lif, or Tnf in the presence of Lif for 24 h. RT-qPCR for inflammatory genes (A,B), homeostasis (C) and proliferation (D) were used to quantify gene expression changes. Values represent average ± StDev. Values were compared with a one-way ANOVA followed by Holm-Sidak test for multiple comparisons. *= adj p-val≤ 0.05, **= adj p-val≤ 0.01, ***= adj. p-val≤ 0.001 (n = 3 independent experiments).
Discussion
Our study demonstrated that delivery of an anti-inflammatory viral gene can limit retinal damage induced by chronic RPE oxidative damage and inflammation in a mouse model of GA. A single intravitreal injection of our AAV vector delivering a secretable and cell-penetrating M013 tagged with a V5 moiety decreased retinal inflammation in the EIU mouse model, similar to non-tagged secretable and cell-penetrating M013, as previously described10. When injected into the RPESod2cKO mouse model of GA, this vector preserved the amplitudes of the a-, b-, and c-waves from three to nine months of age compared to the control injections. More importantly, the average amplitudes at nine months in TatM013v5-treated mice were similar to or higher than those observed in sGFP-treated animals at three months. In addition, the TatM013v5 treatment blocked the retinal activation of microglia and Müller glia, in contrast to control injected animals. The significant decrease in the expression of M1-associated mononuclear phagocytic genes within these retinal samples further validates the anti-inflammatory effects of TatM013v5. Longer photoreceptor inner and outer segments accompanied these changes in sGFP-TatM013v5-treated retinas compared to those in sGFP-treated retinas. Also, the expression of TatM1013v5 in the retina of this mouse model increased IL9, IL-10, and LIF levels compared to control injected retinas. Using an in vitro model to study microglia activation, we demonstrated that Lif treatment can antagonize Tnf-mediated pro-inflammatory gene expression. Our results indicate that retinal M013-mediated inhibition of RelA/p65 decreases retinal inflammatory pathways and stimulates neuroprotective pathways through an uncharacterized mechanism that involves microglia gene expression changes in the retina.
AMD is a multifactorial disease that is associated with complex genetic and environmental factors. Variations in the CFH and CFB genes, mononuclear phagocytic cells in the retina, and complement deposition indicate that AMD is associated with chronic retinal inflammation and complement activation. These observations have sparked interest in the development of novel anti-inflammatory therapies. For example, a clinical study by Jaffe et al. demonstrated that monthly injection of a C5 inhibitor in AMD patients significantly slowed disease progression using differences in fundus autofluorescence in vivo38. Unfortunately, this group also found an increase in the incidence of macular neovascularization in patients treated with a C5a inhibitor.
Similarly, a clinical trial using a C3 inhibitor in AMD patients showed a significant reduction in the GA area. However, it also shows an increased incidence of macular neovascularization39. These observations support the idea that biologicals targeting the inflammatory pathways in the retina could be potential therapies. Further research is needed to better understand how complement inhibition in the retina leads to neovascularization. Interestingly, neovascularization was not observed in our mouse model of geographic atrophy. In addition, we observed no increase in the expression of pro-neovascular (M2) genes following TatM1013v5 treatment; though we did not determine whether prolonged treatment leads to choroidal neovascularization.
The early increase in LIF, IL-10, and IL-9 expression suggests the activation of Stat3 signaling. Ligand engagement of these cytokines is known to activate Stat340,41,42. However, the mechanism by which RelA/p65 activation inhibition leads to cytokine increases remains elusive. Simultaneous deletion of RelA and Stat3 in mice hepatocytes was demonstrated to abrogate the acute phase response to pathogenic bacteria43. This RelA and Stat3 co-deletion in hepatocytes was also capable of augmenting the severity of colitis in a mouse model44. Interestingly, Mangum et al. showed that macrophages from diabetic patients express high levels of Stat3 and of NF-kB-induced pro-inflammatory cytokines (e.g., IL-1βand IL-6), thus suggesting a potential mechanistic interaction between NF-kB and Stat3 signaling45. In addition, gene delivery of a drug-tunable form of LIF significantly slowed retinal degeneration in the rd10 mouse model of inherited retinal degeneration46. Thus, further understanding of how anti-inflammatory and neuroprotective pathways in the retina interact to maintain homeostasis is required. As a first step, our future studies will address the dynamic mechanism between RelA and Stat3 and other potential factors in the retina.
Conclusions
Retinal inflammation has been recognized as a part of the pathobiology of AMD. We demonstrated that retinal inhibition of p65/RelA and the NLRP3 inflammasome significantly delays the loss of retinal function in a mouse model of AMD geographic atrophy. Our observations suggest retinal inflammation is a vital disease modifier in dry AMD. Furthermore, locally inhibiting inflammatory pathways, such as NF-kB, holds great promise as targets to slow vision loss in dry AMD and can significantly impact the quality of life of patients afflicted with this disease. Future studies will elucidate the cellular mechanisms underlying how the inhibition of RelA/p65 and the delay of retinal degeneration.
Data availability
The data analyzed in the current study is available from the corresponding author upon reasonable request.
References
Jonas, J. B., Cheung, C. M. G. & Panda-Jonas, S. Updates on the epidemiology of age-related macular degeneration. Asia Pac. J. Ophthalmol. 6, 493–497 (2017).
Crabb, J. W. The proteomics of Drusen. Cold Spring Harb Perspect. Med. 4, a017194 (2014).
Winkler, T. W. et al. Genome-wide association meta-analysis for early age-related macular degeneration highlights novel loci and insights for advanced disease. BMC Med. Genomics. 13, 120 (2020).
Lorés-Motta, L. et al. Common haplotypes at the CFH locus and low-frequency variants in CFHR2 and CFHR5 associate with systemic FHR concentrations and age-related macular degeneration. Am. J. Hum. Genet. 108, 1367–1384 (2021).
Mao, H. et al. Mitochondrial oxidative stress in the retinal pigment epithelium leads to localized retinal degeneration. Invest. Ophthalmol. Vis. Sci. 55, 4613–4627 (2014).
Young, B. M. et al. Expression of a CARD slows the retinal degeneration of a geographic atrophy mouse model. Mol. Ther. Methods Clin. Dev. 14, 113–125 (2019).
Rahman, M. M. & McFadden, G. Myxoma virus lacking the pyrin-like protein M013 is sensed in human myeloid cells by both NLRP3 and multiple Toll-like receptors, which independently activate the inflammasome and NF-κB innate response pathways. J. Virol. 85, 12505–12517 (2011).
Garg, R. R. et al. Myxoma virus M013 protein antagonizes NF-κB and inflammasome pathways via distinct structural motifs. J. Biol. Chem. 294, 8480–8489 (2019).
Rahman, M. M., Mohamed, M. R., Kim, M., Smallwood, S. & McFadden, G. Co-regulation of NF-kappaB and inflammasome-mediated inflammatory responses by myxoma virus pyrin domain-containing protein M013. PLoS Pathog. 5, e1000635 (2009).
Ildefonso, C. J. et al. Gene delivery of a viral anti-inflammatory protein to combat ocular inflammation. Hum. Gene Ther. 26, 59–68 (2015).
Ridley, R. B. et al. AAV mediated delivery of myxoma virus M013 gene protects the retina against autoimmune uveitis. J. Clin. Med. ;8 (2019).
Marneros, A. G. Role of inflammasome activation in neovascular age-related macular degeneration. FEBS J. 290, 28–36 (2023).
Ambati, M. et al. Identification of fluoxetine as a direct NLRP3 inhibitor to treat atrophic macular degeneration. Proc. Natl. Acad. Sci. USA. 118 (2021).
Allingham, M. J., Loksztejn, A., Cousins, S. W. & Mettu, P. S. Immunological aspects of age-related macular degeneration. Adv. Exp. Med. Biol. 1256, 143–189 (2021).
Weaver, C., Cyr, B., de Rivero Vaccari, J. C. & de Rivero Vaccari, J. P. Inflammasome proteins as inflammatory biomarkers of age-related macular degeneration. Transl Vis. Sci. Technol. 9, 27 (2020).
Celkova, L., Doyle, S. L. & Campbell, M. NLRP3 inflammasome and pathobiology in AMD. J. Clin. Med. 4, 172–192 (2015).
Doyle, S. L. et al. NLRP3 has a protective role in age-related macular degeneration through the induction of IL-18 by Drusen components. Nat. Med. 18, 791–798 (2012).
Biswal, M. R. et al. Timing of antioxidant gene therapy: implications for treating dry AMD. Invest. Ophthalmol. Vis. Sci. 58, 1237–1245 (2017).
Biswal, M. R. et al. Daily zeaxanthin supplementation prevents atrophy of the retinal pigment epithelium (RPE) in a mouse model of mitochondrial oxidative stress. PLoS ONE. 13, e0203816 (2018).
Zolotukhin, S. et al. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods 28, 158–167 (2002).
Ridley, R. B., Walsh, E. M. & Ildefonso, C. J. Molecular design and production of AAV viral vectors for gene therapy. Methods Mol. Biol. 2225, 77–92 (2021).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods 25, 402–408 (2001).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Nagamoto-Combs, K., Kulas, J., Combs, C.K. A novel cell line from spontaneously immortalized murine microglia. J. Neurosci. Methods. 233, 187–198 (2014).
Brown, E. E., DeWeerd, A. J., Ildefonso, C. J., Lewin, A. S. & Ash, J. D. Mitochondrial oxidative stress in the retinal pigment epithelium (RPE) led to metabolic dysfunction in both the RPE and retinal photoreceptors. Redox Biol. 24, 101201 (2019).
Biswal, M. R. et al. Conditional induction of oxidative stress in RPE: A mouse model of progressive retinal degeneration. Adv. Exp. Med. Biol. 854, 31–37 (2016).
Long, K. O. & Aguirre, G. D. The cone matrix sheath in the normal and diseased retina: cytochemical and biochemical studies of peanut agglutinin-binding proteins in cone and rod-cone degeneration. Exp. Eye Res. 52, 699–713 (1991).
Blanks, J. C. & Johnson, L. V. Specific binding of peanut lectin to a class of retinal photoreceptor cells. A species comparison. Invest. Ophthalmol. Vis. Sci. 25, 546–557 (1984).
Yadav, S., Dwivedi, A. & Tripathi, A. Biology of macrophage fate decision: implication in inflammatory disorders. Cell. Biol. Int. 46, 1539–1556 (2022).
Jiang, K. et al. STAT3 promotes survival of mutant photoreceptors in inherited photoreceptor degeneration models. Proc. Natl. Acad. Sci. USA. 111, E5716–E5723 (2014).
Rhee, K. D. et al. CNTF-mediated protection of photoreceptors requires initial activation of the cytokine receptor gp130 in Müller glial cells. Proc. Natl. Acad. Sci. USA. 110, E4520–E4529 (2013).
Ueki, Y., Wang, J., Chollangi, S. & Ash, J. D. STAT3 activation in photoreceptors by leukemia inhibitory factor is associated with protection from light damage. J. Neurochem. 105, 784–796 (2008).
Zhang, C. et al. STAT3 activation protects retinal ganglion cell layer neurons in response to stress. Exp. Eye Res. 86, 991–997 (2008).
Joly, S., Lange, C., Thiersch, M., Samardzija, M. & Grimm, C. Leukemia inhibitory factor extends the lifespan of injured photoreceptors in vivo. J. Neurosci. 28, 13765–13774 (2008).
Dong, S. et al. Leukemia inhibitory factor protects against degeneration of cone photoreceptors caused by RPE65 deficiency. Curr. Med. Chem. 31, 4022–4033 (2024).
Agca, C. & Grimm, C. Leukemia inhibitory factor signaling in degenerating retinas. Adv. Exp. Med. Biol. 801, 389–394 (2014).
Jaffe, G. J. et al. C5 inhibitor avacincaptad pegol for geographic atrophy due to age-related macular degeneration: A randomized pivotal phase 2/3 trial. Ophthalmology 128, 576–586 (2021).
Liao, D. S. et al. Complement C3 inhibitor pegcetacoplan for geographic atrophy secondary to age-related macular degeneration: A randomized phase 2 trial. Ophthalmology 127, 186–195 (2020).
Chucair-Elliott, A. J. et al. Leukemia inhibitory factor coordinates the down-regulation of the visual cycle in the retina and retinal-pigmented epithelium. J. Biol. Chem. 287, 24092–24102 (2012).
Donninelli, G. et al. Interleukin-9 regulates macrophage activation in the progressive multiple sclerosis brain. J. Neuroinflammation. 17, 149 (2020).
Riley, J. K., Takeda, K., Akira, S. & Schreiber, R. D. Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J. Biol. Chem. 274, 16513–16521 (1999).
Quinton, L. J. et al. Hepatocyte-specific mutation of both NF-κB rela and STAT3 abrogates the acute phase response in mice. J. Clin. Invest. 122, 1758–1763 (2012).
Jyotsna, S. B. et al. A hepatocyte-specific transcriptional program driven by rela and Stat3 exacerbates experimental colitis in mice by modulating bile synthesis. eLife. 12 (2024).
Mangum, K. D. et al. The STAT3/SETDB2 axis dictates NF-κB-mediated inflammation in macrophages during wound repair. JCI Insight. ;9 (2024).
Santiago, C. P. et al. A drug-tunable gene therapy for broad-spectrum protection against retinal degeneration. Mol. Ther. 26, 2407–2417 (2018).
Acknowledgements
We want to acknowledge the Ocular Gene Therapy Core’s help in generating the AAV vectors used in this study.
Funding
This work was funded by grants from the Bright Focus Foundation (M2017126), the National Eye Institute (EY026268), and a core equipment grant from NEI (S10OD028476). This study was also supported by an unrestricted grant from the Research to Prevent Blindness to the Department of Ophthalmology. The Shaler Richardson Professorship supported ASL.
Author information
Authors and Affiliations
Contributions
B.M.T: conducted experiments and collected data; R.B.R: conducted experiments and collected data; J.L: conducted experiments, collected, and analyzed data; E.W: conducted experiments and collected data; M.T.M: conducted experiments and collected data; A.S.L: analyzed data and manuscript preparation; C.J.I: conceived experiments, conducted experiments, collected data, analyzed data, and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The University of Florida Institutional Animal Care and Use (IACUC) Board approved all the animal experiments conducted for this research (Protocol 201709854). All procedures adhered to ARVO guidelines for the use of animals in biomedical research. The study is reported in accordance with ARRIVE guidelines.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.


Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ridley, R.B., Tischner, B.M., Lee, J. et al. Modulation of retinal inflammation delays degeneration in a mouse model of geographic atrophy. Sci Rep 15, 30153 (2025). https://doi.org/10.1038/s41598-025-15891-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-15891-z