Abstract
Hyperspectral imaging has shown significant applicability in the medical field, particularly for its ability to represent spectral information that can differentiate specific biomolecular characteristics in tissue samples. However, the complexity of analyzing HSI data, due to its high dimensionality and the large volume of information, presents significant challenges. At the same time, deep learning, particularly convolutional neural networks and recurrent neural networks, has become an essential tool in medical diagnostics, providing detailed analysis across various contexts. These techniques enable the analysis of complex information often unattainable through traditional methods. This paper introduces a novel approach that integrates micro-FTIR spectroscopy with three different deep learning architectures, namely RNN, FCNN, and 1D-CNN, to compare their performance in region-based classification of thyroid tissues, including goiter, cancerous, and healthy types. The proposed deep learning methods were developed on a dataset of 60 patients and evaluated using grouped 10-fold cross-validation. The 1D-CNN achieved the highest scores in classifying the spectral data provided by micro-FTIR, enabling more precise and accurate region-based tissue classification. The 1D-CNN achieved an accuracy of 97.60%, while RNN and FCNN achieved 96.88% and 93.66%, respectively. These results highlight the effectiveness of this approach in enhancing the precision of thyroid pathology analysis.
Similar content being viewed by others
Introduction
Machine learning, particularly deep learning techniques, has become an important tool in daily medical procedures. These advancements support a wide variety of applications, from large language models (LLMs) that guide personalized healthcare recommendations1, to medical imaging diagnostics, such as detecting brain cancer in MRI through image analysis2.
Among the most widely used deep learning architectures, convolutional neural networks (CNNs) have gained significant attention due to their performance in tasks such as image classification, object detection, and segmentation. CNNs have been particularly valuable in medical imaging, where they have been applied to radiological scans3, pathology slides4, and other medical images to assist in accurate and efficient diagnosis.
CNNs are comprised of convolutional layers that perform convolutions across the input image, extracting filters that represent patterns such as edges and textures. These patterns capture different levels of complexity, highlighting important characteristics of the data. Such mechanisms have been adapted for other types of input data, including one-dimensional data, such as signals and vector data, as well as three-dimensional data, such as volumes and videos5.
One-dimensional CNNs (1D-CNNs) operates similarly to its 2D version but is designed to process one-dimensional data. In a 1D-CNN, convolutional layers slide filters along the input sequence to detect patterns across the data points. These filters capture essential features, such as peaks, trends, or repetitive patterns, which are then used to build a feature map5,6. This mechanism is particularly effective for spectral analysis, where the network can identify subtle variations and characteristics within the spectrum.
Long short-term memory recurrent neural networks (LSTM-RNNs) are another deep learning architecture widely used in medical applications, especially in the analysis of time-series data, such as ECG and EEG signal processing7. The LSTM-RNN works by incorporating memory cells that are capable of maintaining information in memory for long periods of time. This capability allows the network to capture long-term dependencies and patterns in sequential data, making it particularly effective for tasks where context from earlier in the sequence is crucial for current decisions8.
Fully-connected neural networks (FCNN), on the other hand, are primarily composed of dense layers that connect sequentially between preceding and subsequent layers of neurons. Unlike CNNs and LSTM-RNNs, FCNNs do not benefit from a specific type of data such as feature maps in CNNs or temporal changes in RNNs. Instead, they treat each input feature independently, which simplifies the overall pipeline. However, this approach might require additional preprocessing to extract meaningful features, potentially limiting their effectiveness in capturing complex patterns9.
Spectral analysis, particularly in the context of hyperspectral imaging (HSI), presents significant challenges due to the large volume and complexity of the data. A single HSI typically contains thousands of spectral voxels, each representing a small spatial region with its own unique spectral signature10. Besides, these spectra are often composed of over 500 intensity values across different frequency ranges, leading to a high-dimensional dataset. This large volume of data significantly increases the demand for computational resources, making the analysis both time-consuming and computationally expensive.
Traditional computer vision techniques typically rely on digital images of sample slides stained with hematoxylin and eosin (H&E) to represent image content as feature vectors, which are then used to develop machine learning models for classification. While these methods can perform well on small, homogeneous samples, they often struggle with larger samples that contain multiple pathologies11,12. In contrast, HSI employs a spectrometer to capture images with a large number of spectral channels, measuring how biological tissues interact with light across a range of frequencies, including those beyond human vision10. This technology provides detailed absorbance spectra that reveal the molecular and histochemical composition of tissues.
Despite its complexity, HSI holds significant potential to assist in pathology analysis, particularly in the study of thyroid tissue. Various approaches have been explored in the literature to leverage this potential. For instance, Halicek et al.13 modeled HSI data into 2D patches and used them to train a 2D-CNN based on the Inception-v4 architecture, achieving an AUC of 90%. Similarly, Tran et al.14 employed a reflectance-based spectroscopy method to generate HSI from the thyroid gland, and then used portions of the frequency data to train a VGG-19 architecture, another form of 2D-CNN, achieving a higher AUC of 96.6%. On the other hand, Wang et al.15 developed a 3D-CNN to detect patterns across the HSI volume, utilizing a custom version of the VGG-16 architecture with 3-dimensional convolutional layers. This method, also based on reflectance spectroscopy, achieved an accuracy of 84.63%. Tran et al.16 developed a method based on the TimeSformer network, a Vision Transformer-based architecture, to classify tumor and normal cells using transmittance spectroscopy, achieving an accuracy of 90.87%. And recently, Xiaoqing et al.17 proposed a deep learning approach to differentiate Hashimoto’s thyroiditis and papillary thyroid carcinoma using HSI. Their proposed model achieved an accuracy of 88.36% using low-wavelength spectral features (400–500 cm−1).
Although advancements have been made in HSI analysis for thyroid pathology, a significant gap remains in the exploration of various spectroscopic techniques. Additionally, while previous studies have predominantly focused on 2D modeling of HSI data or employed 3D classification using the volume, the potential of one-dimensional modeling still requires further investigation. And finally, most literature treats thyroid tissue classification as a binary problem, detecting patterns for cancerous and normal tissue, while goiter, another thyroid pathology present in the tissue, still demands much more extensive research.
This paper addresses the gap in the current literature by exploring the application of micro-FTIR spectroscopy for thyroid pathology analysis, focusing on a comparative analysis of three deep learning techniques: two previously developed methods18,19 and a newly developed one-dimensional convolutional neural network (1D-CNN). Specifically, this study develops and evaluates the 1D-CNN developed for spectral data, capable of classifying each voxel of hyperspectral images by leveraging the detailed spectral information provided by micro-FTIR. The goal is to demonstrate the effectiveness of the 1D-CNN in improving the accuracy and precision of thyroid tissue classification from a multiclass perspective, including goiter tissue, and to compare its performance against those of the established RNN and FCNN approaches, traditionally used in 2D and 3D modeling techniques.
The main contribution of this paper is the application and comparative analysis of micro-FTIR spectroscopy combined with three deep learning models: a newly developed 1D-CNN, and two previously developed architectures, for voxel-based classification of thyroid tissue. Micro-FTIR provides highly detailed spectral information at a microscopic level20, which has not been fully leveraged in previous studies. By utilizing these models, particularly the 1D-CNN, this approach allows for the classification of each voxel within the hyperspectral image, effectively utilizing the fine spectral details captured by micro-FTIR. Additionally, this method addresses the problem in a multiclass format, enabling the classification of various thyroid tissue types, including goiter, cancer, and healthy tissue, rather than limiting the analysis to a binary classification. The comparative analysis aims to demonstrate the relative effectiveness of these models in enhancing the accuracy and precision of thyroid tissue classification.
Materials and methods
Dataset description
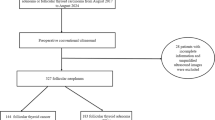
We used a dataset of 60 tissue microarrays (TMAs), each containing a single core per case, with 20 cases representing each diagnosis (cancer, healthy and goiter). These TMAs were acquired from MicroArray Inc., Rockville, USA, and comprises both male and female patients, with ages ranging from 17 to 80 years, ensuring diversity across age and sex. The cancer group includes cases of both papillary and follicular thyroid carcinomas, while the goiter group consists of a variety of benign conditions, such as nodular goiter, diffuse toxic goiter, Hashimoto’s thyroiditis, subacute thyroiditis, and fibrous thyroiditis. The tissue samples were initially pre-labeled by pathologists from the company and subsequently verified and confirmed by two experienced pathologists specializing in thyroid tissue: one from the Department of Pathology at the Ribeirão Preto School of Medicine, University of São Paulo, and the other from the Pathology Service at the Clinical Hospital of Ribeirão Preto.
These tissues were later used to measure absorbance spectra for each small portion corresponding to a voxel. These measurements used a PerkinElmer Spotlight 400 micro-FTIR spectrometer, equipped with a microscope to assess the absorbance intensity of thyroid tissues across the infrared spectrum range of 778–1800 cm−1. Most of the images captured were approximately 300 × 300 pixels in size, with slight variations in dimensions. Through spectral mapping, data were collected at two wavenumber intervals, resulting in 512 intensity measurements per voxel. The system provided a spectral resolution of 4 cm−1 and a spatial resolution of 6.25 μm. To ensure precision, hyperspectral imaging (HSI) data were acquired under strictly controlled conditions, with relative humidity maintained below 5% using dry air.
Preprocessing
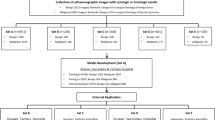
To enhance data quality and remove irrelevant information, we developed a five-step preprocessing pipeline (Fig. 1). The goal of this pipeline was to segment the region of interest (ROI), such as cellular structures, while eliminating unwanted components (e.g., other thyroid structures like the colloid). First, each spectral band was normalized to a range of 0 to 1 using Min-Max scaling. The resulting spectra were then multiplied by 255 and converted to integers to generate pixel values, allowing each spectral band to be visualized as a grayscale image.
Next, we selected a representative spectral band (Fig. 1–Step 1) that best delineated the internal structures of the tissue, particularly distinguishing cells from other elements such as the colloid. We chose the 1624 cm−1 band, which lies within the spectral region known as Amide I (approximately 1590–1690 cm−1), associated with the C = O stretching vibrations of proteins. This absorption is strongly expressed in cellular structures due to the high protein content of thyroid follicular cells. In contrast, the colloid is predominantly composed of glycoproteins, which contain fewer peptide bonds and exhibit weaker absorption in this region21. When converting each spectral band into a grayscale image, we observed that the 1624 cm−1 band provided the greatest visual contrast between the cells (high absorption) and the colloid (low absorption).
Afterward, we transformed the grayscale image at 1624 cm−1 into the frequency domain (Fig. 1–Step 2) using the fast Fourier transform (FFT). This transformation enabled the analysis of the image based on its spatial frequency components. Subsequently, we applied a high-pass filter to emphasize high-frequency components and suppress low-frequency ones (Fig. 1–Step 3). This step was important to enhance the visibility of fine tissue details and cellular boundaries.
Following the high-pass filtering, we applied a threshold with a value of 127, isolating the background and the region of interest (Fig. 1–Step 4). This process primarily highlighted the cell nuclei, allowing us to focus on significant features within the tissue. However, to capture a more comprehensive spectrum representative of the entire cell structure, we employed morphological operations to dilate the detected regions (Fig. 1–Step 5), using a 3 × 3 dilation kernel over one iteration. This dilation enhanced the visibility of the cell structures, enabling us to obtain a more accurate representation of the cellular architecture within the tissue.
We selected the voxels corresponding only to the white regions in the segmentation mask, as these areas were deemed more representative of the cellular structures within the tissue. This focused approach ensured that our analysis was based on the most relevant data. The selected voxels were then utilized for pattern recognition using the 1D-CNN. In total, we obtained 104,107 voxels for this analysis, providing a robust dataset that enhances the accuracy and effectiveness of our pattern recognition efforts.
Finally, we augmented the dataset by adding Gaussian noise to the existing samples. This addition of Gaussian noise is beneficial for increasing the robustness of the model, as it helps to simulate real-world variations and improve the model’s ability to generalize to unseen data. By incorporating this noise, we effectively doubled our training dataset, enhancing its variability and contributing to the overall model’s generalization. It is important to note that we applied augmentation exclusively to the training samples, ensuring that the testing dataset remained unaltered and retained only the original preprocessing.
Classification methodology
Our comparative study analyzes three distinct deep learning architectures applied to the classification of thyroid tissue using hyperspectral imaging: the FCNN (Fig. 2a) and LSTM-RNN (Fig. 2b), which have been previously developed and tested for thyroid classification, alongside a newly developed architecture, the 1D-CNN (Fig. 2c). These architectures were developed through empirical experimentation. For the FCNN, we conducted a grid search to optimize key hyperparameters such as activation functions, loss functions, optimizers, and the number of neurons per layer. These optimized configurations provided a strong baseline, which we then used as a reference point to guide the fine tuning of the LSTM-RNN and the newly proposed 1D-CNN architectures.
The FCNN is designed with eight layers: one input layer, six hidden dense layers, and one output layer. The input layer contains 100 neurons, corresponding to the 100 frequencies selected using the SelectKBest algorithm. This method was the only one that used dimensionality reduction; the LSTM and the 1D-CNN did not use it due to their nature of handling long sequences of data. Each of the FCNN’s six hidden layers features 86 neurons, all employing the rectified linear unit (ReLU) activation function. The output layer comprises three neurons, each representing one of the study’s classes: cancerous, goiter, and healthy tissues, with a softmax activation function.
The LSTM-RNN model is structured with two layers: the first containing 256 LSTM units and the second 128 units. Each LSTM layer is followed by a dropout layer that deactivates 20% of the neurons. The model processes the full spectrum of hyperspectral data, which consists of 512 intensity values per frequency. The output layer contains three neurons to address the three classification labels.
Finally, the newly proposed 1D-CNN architecture is set up as follows. It consists of six layers in total. The input layer receives 512 intensity values, followed by two convolutional layers that extract 16 and 32 filters, respectively, both utilizing a filter size of three. These values were empirically defined based on experimental results, aiming to achieve a balance between architectural complexity and model performance. After each convolution operation, a one-dimensional spatial dropout is applied, randomly deactivating 40% of the neurons in that layer, along with a one-dimensional max pooling operation with a size of two. This sequence aims to reduce the dimensionality of the data while preserving essential characteristics, thereby reducing the risk of overfitting.
The combination of these operations yields a total of 4032 one-dimensional feature maps from the convolutional layers. These features are subsequently flattened to prepare for training with a multilayer perceptron (MLP). The MLP consists of three dense layers: the first layer has 4032 neurons, the second has 100 neurons, and the third has three neurons. The hidden layer employs the ReLU activation function, while the output layer uses the softmax function to perform classification by assigning probabilities to each input, indicating its likelihood of belonging to one of three predefined classes: cancer, healthy tissue, or goiter.
Additional parameters include the Adam optimizer, chosen for its efficiency in adapting the learning rate, making it particularly suitable for large datasets with numerous parameters. L2 regularization is implemented to minimize the risk of overfitting by adding a penalty term that encourages lower weight values within the network. The batch size is set to be automatic, allowing the system to determine the most efficient value to optimize computational resource usage. Finally, the network is trained over 5000 epochs to ensure robust learning and model performance.
Experiments and results
The experiments were conducted using the grouped k-fold cross-validation protocol, which ensures that the data is divided into k parts. In each iteration, one part is used for testing, while the remaining \(k - 1\) parts are used for training. For our experiments, we set \(k = 10\) to thoroughly evaluate various train-test subset combinations and avoid scenarios where specific combinations might overperform. Additionally, patient IDs were used to separate the groups for the grouped k-fold, ensuring that no data from the same patient was split between the testing and training subsets.
During each iteration of the cross-validation, five evaluation metrics were computed: accuracy, precision, sensitivity, specificity, and AUC (area under the receiver operating characteristic curve). Each metric provides a comprehensive overview of the model’s performance. For example, sensitivity measures the model’s ability to correctly identify true positive cases, indicating how well it detects actual instances of the target class. Specificity, on the other hand, measures the ability to correctly identify true negative cases, demonstrating how effectively the model can avoid false positives. The AUC represents the overall performance of the model by showing how well it distinguishes between positive and negative classes across different threshold values.
We conducted the experiments on a Linux server equipped with two Intel Xeon Silver 4310 processors (48 threads in total), 192 GB of DDR4 RAM, two NVIDIA RTX A4000 GPUs with 16 GB of VRAM each, and a 2 TB NVMe SSD. The implementation was developed in Python 3.9 using TensorFlow 2.10, OpenCV 4.5.5, and Scikit-Learn 1.1.
Results
The results shown on Table 1, obtained from the 1D-CNN architecture for classifying thyroid tissue, demonstrate its high performance across various evaluation metrics. The model achieved an average accuracy of 97.60%, highlighting its robustness and consistency in correctly predicting tissue conditions in most test cases. This high accuracy is supported by a relatively low standard deviation of ± 2.57, indicating that the model performed consistently across all 10 folds without significant fluctuations in results.
Precision and sensitivity, both achieved around 97.69% and 97.60%, respectively, reflecting the model’s ability to identify true positive cases without a substantial increase in false positives or false negatives. The mean specificity of 97.60% underlines the model’s strong capability to accurately recognize negative cases, preventing incorrect classifications of healthy tissue as diseased. The AUC metric, with an average of 99.61% and a standard deviation of ± 0.60, confirms the model’s overall performance in distinguishing between different tissue classes.
The best results were achieved in folds 1 and 4, where the accuracy, precision, and sensitivity each exceeded 99%. On the other hand, fold 8 displayed the lowest performance but still performed well, reaching an accuracy and sensitivity above 90%.
Discussion
Deep learning has shown very promising results when applied to voxel-based classification of hyperspectral images. All three architectures achieved an overall accuracy and sensitivity above 90%, demonstrating their potential to assist pathologists in diagnosing and detecting cancer patterns in histological samples
Comparing the 1D-CNN to the other architectures previously developed (Table 2), the 1D-CNN outperformed both the FCNN and LSTM-RNN in terms of classification metrics for a voxel-based thyroid tissue analysis. The 1D-CNN achieved the highest accuracy, precision, sensitivity, and AUC among the models evaluated. While the LSTM-RNN also showed strong performance with an accuracy of 96.88% and AUC of 98.87%, the 1D-CNN’s results indicate a more refined ability to classify HSI data. The FCNN, although effective, exhibited the lowest performance metrics, with an accuracy of 93.66% and an AUC of 98.62%.
Besides, the 1D-CNN offers several advantages compared to other architectures. It is more computationally efficient than the 2D and 3D version commonly used in the literature, achieving excellent classification results with significantly lower resource requirements. Compared to previously developed architectures, the 1D-CNN completes its training in significantly less time than the LSTM-RNN, requiring only 2191 sec per fold compared to the RNN’s 16,500 sec. However, it is slower than the FCNN, which completes each fold in just 500 sec. Additionally, the 1D-CNN uses considerably less RAM, requiring only 16 GB compared to the 24 GB and 77 GB needed for the FCNN and RNN models, respectively.
Extending the comparison to traditional computer vision techniques for analyzing digital images of thyroid histological samples, the HSI analysis also achieved better results. In a related study12, we noted the challenges in analyzing histological images due to various factors, such as samples containing regions of both pathological and healthy tissue and the morphological similarity between healthy and goiter cells. On the other hand, HSI analysis proved to be a very effective solution in addressing these problems and was able to represent the information in a manner that allowed deep learning models to sufficiently discern patterns to accurately differentiate between the three pathological states.
We also performed an external validation experiment using nine samples, comprising three cases for each diagnosis category. These samples were entirely excluded from the training and testing datasets. The classification was conducted using the FCNN, LSTM-RNN, and 1D-CNN models. Figure 3 presents the results organized by columns, where the first three represent healthy tissue, the middle three depict goiter, and the last three show cancerous tissue. Each row corresponds to a different step: the first row displays the H&E-stained adjacent slides for visual reference, followed by the ROI masks used for classification. The subsequent three rows show the voxel-wise classification maps generated by each model: FCNN, LSTM-RNN, and 1D-CNN, respectively. The predicted classes are color-coded as follows: green for healthy tissue, blue for goiter, and red for cancer.
Voxel-wise classification results for nine external validation samples not used during training or testing. The columns are grouped by diagnostic category: the first three columns show healthy tissue samples, the next three represent goiter, and the final three correspond to cancerous tissue. The first row displays the H&E-stained adjacent histological slides for visual reference, and the second row shows the binary masks of the regions of interest (ROI) used in the classification. The remaining rows present the classification maps generated by the three deep learning models: FCNN, LSTM-RNN, and 1D-CNN. In the classification maps, each voxel is colored according to its predicted class: green for healthy tissue, blue for goiter, and red for cancer.
The results of this experiment were consistent with the quantitative outcomes observed in earlier evaluations, with each model exhibiting similar loss values and accuracy. The 1D-CNN delivered the best visual performance, attributed to its strong generalization ability and high accuracy, precision, and AUC scores, which enabled more reliable classification. However, the LSTM-RNN misclassified some healthy voxels as goiter and presented wrong cancer targets as well, while the FCNN incorrectly labeled certain goiter regions as normal and some cancerous regions as goiter.
Although requiring specialized equipment such as spectrometers to capture absorbance spectra and create hyperspectral images, HSI offers significant advantages in the analysis of histological samples. HSI provides enhanced accuracy, substantially reducing the rates of false positives and false negatives, making it a powerful tool for detailed and precise pathological examinations. On the other hand, traditional computer vision systems, while not requiring additional equipment beyond what is typically available in most laboratories, such as cameras and microscopes, are easier to implement in most hospitals and clinics, and can perform satisfactorily well although these systems face challenges in handling overlapping pathological and healthy tissues and in distinguishing between morphologically similar tissue states.
While the 1D-CNN outperforms other architectures in terms of speed and memory usage, it may not capture complex spatial relationships as effectively as 2D or 3D CNNs, which can analyze multi-dimensional data structures. Additionally, the entire pipeline was specifically developed for thyroid tissue, so applying it to other tissue types, such as breast or prostate, may require modifications due to anatomical differences. This presents an opportunity for future studies to develop a more generalizable pipeline capable of handling various tissue types effectively.
Another limitation of this study is the relatively small dataset, composed of 60 patients. This constraint is common in biomedical hyperspectral imaging due to the complexity and cost of both sample acquisition and spectral data collection. To address this limitation, we adopted strategies mentioned in the method section, such as Gaussian noise-based data augmentation, dropout layers, and shallow network architectures, which help reduce the risk of overfitting and improve generalization. While the current results are promising, we expect that larger datasets in future studies will further enhance model robustness and performance.
The 1D-CNN model has promising potential for clinical applications, particularly in assisting pathologists with the diagnosis of thyroid conditions. By integrating this model with a hyperspectral imaging system connected to a spectrometer, pathologists can gain a powerful tool for more accurately and efficiently detecting tumoral regions. Such a system would enhance the diagnostic process by providing real-time, reliable classifications, allowing pathologists to identify abnormal tissue with greater precision. This integration could significantly speed up workflow, reduce human error, and lead to more timely and effective patient care, supporting pathologists in making efficient and faster decisions.
Conclusion
Hyperspectral imaging classification is a significantly challenging task due to its large volume of data and its high dimensionality. Deep learning models, however, provide various approaches to address these complexities. Based on previously developed architectures, we found that the custom 1D-CNN is the most accurate and efficient model for this purpose. It has demonstrated strong performance in classifying thyroid tissue samples into goiter, normal, and cancer categories, showcasing its capability to handle HSI data effectively and efficiently. These promising results highlight that deep learning approaches can overcome the inherent difficulties of HSI classification. For future work, we plan to develop an integrated tool that can be incorporated into spectrometers, facilitating real-time pathological analysis and enhancing diagnostic workflows with HSI technology. We also intend to explore practical aspects of clinical deployment, including real-time processing, workflow integration, and compliance with regulatory standards to support translational applicability.
Data availability
The datasets used and/or analysed during the current study is available from the corresponding author on reasonable request.
References
Yu, P., Xu, H., Hu, X. & Deng, C. Leveraging generative AI and large language models: A comprehensive roadmap for healthcare integration. Healthcare 11, 2776 (2023) ((MDPI)).
Aamir, M. et al. A deep learning approach for brain tumor classification using MRI images. Comput. Electr. Eng. 101, 108105 (2022).
Iqbal, A., Usman, M. & Ahmed, Z. Tuberculosis chest x-ray detection using CNN-based hybrid segmentation and classification approach. Biomed. Signal Process. Control. 84, 104667 (2023).
Baranwal, N., Doravari, P. & Kachhoria, R. Classification of histopathology images of lung cancer using convolutional neural network (CNN). In Disruptive Developments in Biomedical Applications 75–89 (CRC Press, London, 2022).
Kiranyaz, S. et al. 1D convolutional neural networks and applications: A survey. Mech. Syst. Signal Process. 151, 107398 (2021).
Kiranyaz, S., Ince, T., Abdeljaber, O., Avci, O. & Gabbouj, M. 1-D convolutional neural networks for signal processing applications. In ICASSP 2019 - 2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 8360–8364 (2019). https://doi.org/10.1109/ICASSP.2019.8682194.
Laitala, J. et al. Robust ECG R-peak detection using lstm. In Proceedings of the 35th Annual ACM Symposium on Applied Computing 1104–1111 (2020).
Medsker, L. & Jain, L. C. Recurrent Neural Networks: Design and Applications (CRC Press, London, 1999).
Bengio, Y., Goodfellow, I. & Courville, A. Deep Learning Vol. 1 (MIT Press, Cambridge, MA, 2017).
Lu, G. & Fei, B. Medical hyperspectral imaging: A review. J. Biomed. Opt. 19, 010901–010901 (2014).
Assaad, S. et al. Thyroid cytopathology cancer diagnosis from smartphone images using machine learning. Mod. Pathol. 36, 100129. https://doi.org/10.1016/j.modpat.2023.100129 (2023).
Baffa, M. F. O. et al. Histopathological analysis of fine-needle aspiration biopsies of thyroid nodules using explainable convolutional neural networks. In Proceedings of IFMBE International Federation for Medical and Biological Engineering (2022).
Halicek, M., Dormer, J. D., Little, J. V., Chen, A. Y. & Fei, B. Tumor detection of the thyroid and salivary glands using hyperspectral imaging and deep learning. Biomed. Opt. Express 11, 1383–1400 (2020).
Tran, M. H., Ma, L., Litter, J. V., Chen, A. Y. & Fei, B. Thyroid carcinoma detection on whole histologic slides using hyperspectral imaging and deep learning. In Proceedings of SPIE–the International Society for Optical Engineering, Vol. 12039 (NIH Public Access, 2022).
Wang, J. et al. Classification of benign-malignant thyroid nodules based on hyperspectral technology. Sensors 24, 3197 (2024).
Tran, M. H. et al. Detection and margin assessment of thyroid carcinoma with microscopic hyperspectral imaging using transformer networks. J. Biomed. Opt. 29, 093505–093505 (2024).
Xiaoqing, Y. et al. Application of hyperspectral imaging and machine learning for differential diagnosis of Hashimoto’s thyroiditis and papillary thyroid carcinoma. J. Biophotonics https://doi.org/10.1002/jbio.202500123 (2025).
Baffa, M. F. O. et al. Advancing thyroid pathologies detection with recurrent neural networks and micro-ftir hyperspectral imaging. In 2023 IEEE 36th International Symposium on Computer-Based Medical Systems (CBMS) 611–615 (IEEE, 2023).
Baffa, M. F. O. et al. Deep neural networks can differentiate thyroid pathologies on infrared hyperspectral images. Comput. Methods Programs Biomed. 247, 108100 (2024).
Bandeira, C. C. S. et al. Micro-Fourier-transform infrared reflectance spectroscopy as tool for probing igg glycosylation in COVID-19 patients. Sci. Rep. 12, 4269 (2022).
Movasaghi, Z., Rehman, S. & Ur Rehman, D. I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 43, 134–179 (2008).
Baffa, M. d. F. O. & Felipe, J. C. Caracterizaçao do tecido tireoidiano por meio da análise de imagens hiperespectrais utilizando aprendizagem profunda. In Simpósio Brasileiro de Computação Aplicada à Saúde (SBCAS) 163–168 (SBC, 2025).
Acknowledgements
This work is supported in part by The São Paulo Research Foundation (FAPESP) under grant 2021/00633-0 and partially supported by CNPq (INCT-INTERAS 406761/2022-1). M.F.O.B. is supported in part by Coordination for the Improvement of Higher Education Personnel (CAPES) under grant 88887.498626/2020-00 and 88887.695355/2022-00.
Author information
Authors and Affiliations
Contributions
M.F.O.B. conceptualized the idea, implemented the methodology, conducted the experiments, analyzed the data, and wrote the manuscript. D.M.Z. and L.B. conceptualized the idea, obtained the histological samples, collected the spectra, and analyzed the results. T.M.P. conceptualized the idea, obtained the histological samples, collected the spectra, preprocessed the data, and analyzed the results. J.C.F. coordinated the research, conceptualized the idea, analyzed the experiments and results, and wrote the manuscript. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Baffa, M., Zezell, D.M., Bachmann, L. et al. A comparative analysis of deep learning architectures for thyroid tissue classification with hyperspectral imaging. Sci Rep 15, 31351 (2025). https://doi.org/10.1038/s41598-025-16060-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-16060-y