Abstract
Accurate localization and segmentation of the liver based on CT images are crucial for the early diagnosis and staging of liver cancer. However, during CT scanning, the liver’s position may vary due to periodic respiratory motion, and its boundaries are often unclear due to its close proximity to surrounding organs, which increases the difficulty of segmentation. To address these challenges, this paper proposes an end-to-end liver image segmentation network–EDNe, based on a residual structure. The model introduces an automated feature fusion module (ECAdd) and a residual structure, enhancing the network’s ability to extract multi-scale features from liver CT images. Additionally, a Deep Feature Enhancement (DFE) attention module is incorporated during the decoding phase to improve the network’s ability to capture fine-grained details, thereby ensuring an effective improvement in segmentation accuracy. EDNet was validated on the LiTS2017 and 3D-IRCADb-01 datasets, achieving Dice scores of 0.9651 and 0.9683, and IoU scores of 0.9330 and 0.9385 on the LiTS2017 and 3D-IRCADb-01 datasets, respectively. Experimental results show that EDNet not only exhibits significant advantages in segmentation performance but also demonstrates high robustness across different datasets, providing a reliable and effective solution for liver CT image segmentation tasks.
Similar content being viewed by others
Introduction
Liver cancer is the sixth most common malignant tumor worldwide. According to the latest global cancer data released by the International Agency for Research on Cancer (IARC) of the World Health Organization in 2022, there were approximately 9.74 million cancer-related deaths globally, including 5.43 million males and 4.31 million females. Among these, liver cancer accounted for 760,000 deaths, ranking third in cancer-related mortality. Accurate liver segmentation plays a crucial role in precisely identifying the liver and its lesions, thereby improving the accuracy of lesion detection1. Moreover, in preoperative planning and personalized treatment, clinical applications such as liver tumor resection, liver transplantation, and radiofrequency ablation rely on accurate preoperative measurements of liver and lesion volumes. Precise liver segmentation not only enhances surgical safety but also assists physicians in developing personalized surgical strategies for different patients. Therefore, accurate liver segmentation holds significant importance in the precise diagnosis and treatment of liver diseases2.
To improve the efficiency and accuracy of segmentation, researchers have proposed various automatic or semi-automatic medical image segmentation methods, such as edge detection, template matching, statistical shape models, and active contour models3. While these methods have achieved some success in specific scenarios, liver CT images typically contain more complex features compared to conventional RGB images, presenting ongoing challenges in the field of medical image segmentation. To enhance the quality of liver CT images before segmentation, researchers often apply multiple preprocessing strategies. Among them, Contrast Limited Adaptive Histogram Equalization (CLAHE)4 is an effective approach. CLAHE performs histogram equalization within small local regions (tiles) of the image while limiting contrast amplification to reduce noise, thereby improving local contrast while preserving image details. For common issues in liver CT images such as low contrast and indistinct boundaries, CLAHE effectively enhances the visibility of liver contours and lesion regions, providing clearer input data for downstream segmentation models. This method has been widely adopted to improve the robustness and accuracy of deep learning models, especially in complex scenarios where lesions are small or grayscale differences are subtle. Integrating CLAHE into the segmentation pipeline as a preprocessing step not only improves edge recognition but also enhances detail extraction, making it an important component in achieving high-precision liver and liver tumor segmentation.
With the development of deep learning technologies, medical image segmentation has been widely studied, using state-of-the-art methods to train convolutional neural networks (CNNs) in a supervised manner. Research achievements in liver segmentation have also increased. In 2015, U-Net, an encoder-decoder structure, used skip connections to enhance the restoration of high-resolution features, enabling precise pixel-level segmentation5. Many researchers have further improved and optimized U-Net, making it a mainstream method in complex medical image segmentation tasks. U-Net and its variants have been widely applied to liver, lung, brain, heart, prostate, and other medical image tasks.
In recent years, researchers have made several improvements to U-Net in liver CT image segmentation tasks to better address the challenges of liver CT image processing. Li et al.6 proposed a method combining 2D DenseUnet and 3D structure, where 2D DenseUnet extracts slice-level features, and 3D structure is used to hierarchically aggregate volumetric information, achieving precise segmentation of the liver and its lesions. Additionally, Jiang et al.7 adopted a cascaded structure for liver tumor segmentation, combining soft and hard attention mechanisms and further enhancing the model’s adaptability to complex regions using long-short skip connections. A joint Dice loss function was designed to reduce false-positive cases and improve segmentation accuracy. Lei et al.8 proposed a deformable encoder-decoder network (DefED-Net) for liver and liver tumor segmentation, using deformable convolutions to enhance the network’s feature representation ability. Through modifications to the U-Net structure, these studies effectively improved the accuracy and robustness of liver CT image segmentation, providing stronger methodological support for automatic liver and liver tumor segmentation.
Despite the significant progress made by U-Net and its variants in liver segmentation, models solely relying on CNNs still have limitations, such as insufficient recognition of small targets and missing details in the generated results. To address these issues, researchers have begun exploring deep learning methods that better recover fine-grained details. Among them, Generative Adversarial Networks (GANs) have emerged as an important direction in medical image segmentation. By adversarial training between a generator and a discriminator, GANs can effectively restore fine details in images, improving segmentation accuracy9. GANs have been widely applied to multi-organ tasks in medical image segmentation. For example, Taha et al.10 proposed Kid-Net for the segmentation of renal vasculature (arteries, veins, and collecting system), assisting doctors in making accurate decisions before surgery. Sekuboyina et al.11 proposed Btrfly Net, an improved GAN-based method focusing on spinal structure segmentation. Han et al.12 developed Spine-GAN for multi-task bone marrow segmentation. In liver CT image segmentation, Wei et al.13 combined GAN with a mask-based CNN architecture, enhancing pixel-level classification ability and optimizing image aspect ratio using k-means clustering, significantly improving liver segmentation performance. GANs have also been used in auxiliary training processes. For instance, Biswas et al.14 generated diverse liver data using GANs to augment the training dataset, thereby indirectly enhancing the model’s generalization ability. These studies show that GANs not only enable efficient segmentation but also optimize model training through strategies like data augmentation, demonstrating strong adaptability in medical scenarios.
With the further development of deep learning, large-scale pre-trained models and Transformer structures have begun to show powerful capabilities in medical image segmentation. In 2021, Cao et al.15 proposed the Swin-UNet network, a Transformer-based segmentation architecture that uses a hierarchical Swin Transformer with shifted windows as an encoder to extract contextual features, achieving excellent performance in multi-organ segmentation tasks. Ugur Demir et al.16 combined Transformer and GAN, using GAN to classify the confidence of generated liver segmentation masks, offering more reliable segmentation results. Jiajie et al.17 proposed a novel hybrid network that integrates U-Net and Transformer architectures. In the encoding structure, a dual-path method was used, with CNN and Transformer networks separately extracting features, achieving high precision in liver segmentation tasks.
At the same time, researchers have found that as the depth of neural networks increases, networks can extract higher-level features, learning semantic information from low to high levels. However, deeper networks tend to lose shallow local features, which can affect network performance. To address this, researchers have proposed multi-scale feature fusion strategies, using residual structures and skip connections to integrate shallow local features with deep global semantic features, minimizing information loss. Rahman et al.18 proposed a network model called ResU-Net, combining ResNet and U-Net architectures, which effectively integrates features from different scales and achieves accurate liver CT image segmentation. Wang et al.19 improved the U-Net model by using EfficientNet-B4 as the encoder and a U-Net-like decoder to enrich semantic information. To better integrate multi-scale features, the convolutional units in U-Net were replaced with residual units, and attention gates were added at each skip connection to focus on relevant liver regions. Liu et al.20 proposed Multi-scale Feature Extraction and Enhancement U-Net (mfeeU-Net), which uses Res2Net blocks as the basis of the encoder to effectively extract multi-scale features from liver images. Additionally, SE blocks were incorporated in the encoder to amplify information from specific channels, solving the problem of blurred liver boundaries. Kushnur and Talbar21 proposed Feature Fusion and Recalibration U-Net (HFRU-Net), which enhances skip connections through local feature reconstruction and feature fusion mechanisms. The fused features are adaptively recalibrated through a channel dependency learning process. The bottleneck layer adds a Spatial Pyramid Pooling (ASPP) module, enhancing the representation of detailed contextual information in high-level features. Wang et al.22 developed a U-Net variant called SAR-UNet for liver segmentation. Its main architecture is similar to the encoder-decoder structure of U-Net, but it uses compression and excitation (SE) blocks, spatial pyramid pooling (SPP), and residual learning.
The common goal of these methods is to ensure that key features of the liver boundaries are not missed during segmentation and to improve sensitivity to small lesions. In terms of deep features, these methods enhance the model’s understanding of complex anatomical structures by modeling global semantics and extracting information on the liver’s overall shape and spatial relationships, thus avoiding segmentation biases caused by insufficient local features. This improves segmentation accuracy and provides strong data support for clinical decision-making, having a profound impact on the field of medical image analysis. Based on the above, the contributions of this paper are as follows:
-
An innovative end-to-end segmentation network, named EDNet, is designed in this work. The network combines a residual structure, an automated feature fusion module (ECAdd), and a deep feature enhancement attention module (DFE) to improve the accuracy and robustness of liver CT image segmentation.
-
In the multi-scale feature fusion phase, we adopted a weighted fusion approach and designed an automated feature fusion module (ECAdd). The core idea of this module is to allow the network to automatically select and weight the most relevant features based on the current task’s needs. This approach not only enhances the expressiveness of the features but also reduces the impact of redundant features on the decoder, thereby improving both the efficiency and accuracy of the network. Compared to traditional fusion methods, the ECAdd module intelligently adjusts the feature weights, ensuring that the network can focus on more representative information at different stages.
-
To further enhance the network’s ability to handle image details, particularly in the precise identification of liver boundaries, we designed a Deep Feature Enhancement (DFE) attention module. This module strengthens the network’s attention to spatial information, enabling it to more sensitively capture subtle differences in liver boundaries. The DFE module significantly improves the segmentation capability for liver details, especially in areas with blurred boundaries or high noise, demonstrating exceptional performance in these challenging regions.
Methods
The overall network architecture
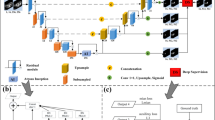
As shown in Fig. 1, this paper proposes the EDNet network model, an end-to-end segmentation model based on a residual structure. The numbers at the top of each feature block represent the number of channels, while the numbers at the bottom indicate the spatial dimensions \(H \times W\) of the feature maps. Since the input to the network is three-dimensional data, the depth \(D\) is not explicitly specified, as it was not uniformly cropped during the preprocessing stage.
Each residual block consists of two convolutional layers with a kernel size of 3 and an automated feature fusion module (ECAdd). The residual block is designed to effectively integrate multi-level feature information, thereby enhancing the network’s representational capacity. Within each residual block, the input feature map first undergoes two convolutional operations to extract local features and deeper semantic information. Then, the ECAdd module fuses the shallow local features with the deep semantic features to generate the residual block’s output, enabling the retention of multi-level information within the same feature map. As shown in Eqs. (1) and (2) , \(X'\) represents the features extracted by the second convolutional layer.
The feature maps output by the residual blocks are further downsampled. In this paper, convolutional operations are employed for downsampling instead of traditional pooling operations. Downsampling via convolution not only reduces spatial resolution but also preserves more semantic information, which helps retain richer contextual information.
To further enhance the network’s ability to extract and represent depth features in 3D images, a Depth Feature Enhancement (DFE) attention module is introduced in the last two layers of the encoder and all layers of the decoder. This module takes the outputs of two residual blocks as input. The specific processing procedure is shown in the Eq. (3) , where \(X_i^{Res}\) represents the feature vector output by the residual block.
The DFE module is introduced to improve the model’s perception and understanding of depth information in 3D images. Specifically, when dealing with 3D image data, the DFE module effectively captures the spatial structure characteristics of objects by integrating multi-scale depth information. This enables the network to enhance both detail preservation and semantic representation, providing richer feature representations for the subsequent decoding process.
Automated feature fusion module
As shown in Fig. 2, the ECAdd module consists of two EC modules, each processing feature maps of different sizes. The detailed processing procedure of the EC module is illustrated in Fig. 3.
The EC module is designed based on the principles of the efficient channel attention mechanism, aiming to optimize the feature fusion process in the residual structure. As shown in Fig. 3, the EC module adaptively assigns weights to each channel based on its importance.
As presented in Eqs. (4) and (5), each input feature map \(X_i\) undergoes adaptive average pooling to ensure that the sizes of the two feature maps are consistent with the smallest feature map size within the residual block.
\(w_i\) is the channel attention weight vector, generated through a convolution operation. In Eq. (5), the kernel size k is determined by the number of channels \(C\) in the input feature map. There exists a mapping relationship between k and \(C\), expressed as \(C = \emptyset (\textrm{k})\), as shown in Eqs. (6) and (7).
Equation (7) is derived from taking the logarithm of Eq. (6).
To prevent the convolution kernel from becoming excessively large, the parameters in this experiment are set as \(r = 2\) and \(b = 1\).
As shown in Eqs. (8) and (9), \(w_i'\) represents the attention weight obtained after the sigmoid activation, while \(X_i^{EC}\) denotes the weighted feature map processed by the EC module. Input of different sizes is processed by the corresponding EC module, making the sizes of the two feature maps consistent before performing element-wise fusion, as described in the Eq. (10).
The output of the ECAdd module does not undergo dimensionality reduction. Instead, by employing weighted fusion, it ensures the maximum retention of important information and adaptively selects features relevant to the target task, thereby further enhancing the representational capacity of the network.
Deep feature enhancement attention module
As shown in Fig. 4, the DFE module receives input from two different residual blocks. The processing method before fusion differs between the encoder and decoder. The lighter regions in the figure illustrate the DFE module during the encoding process, where the larger feature maps are processed through adaptive average pooling to match the size of the smaller feature maps. The specific process in Eq. (11)
As shown in Eq. (12), during the decoding process, the smaller feature maps are upsampled to larger dimensions using trilinear interpolation.
This separate design ensures that feature information is not lost significantly during the decoding process.
Taking the encoding process as an example, the output of the first residual block, after processing, is aligned with \(X_2^{Res}\) in both size and number of channels, with dimensions \((H_{X_2^{Res}}, W_{X_2^{Res}}, D_{X_2^{Res}})\) and \(C\) channels. In Eq. (13), the interpolated feature maps \(X_1'\) and \(X_2'\) are concatenated along the channel dimension to obtain \(F_{cat}\). The dimensions of \(F_{cat}\) are \((H_{X_2^{Res}}, W_{X_2^{Res}}, D_{X_2^{Res}})\), with \(2C\) channels.
Subsequently, a 3D convolution with a kernel size of 1 is applied to reduce the channel dimension from \(2C\) to \(C\), generating the fused feature map \(F\), as shown in Eq. (14).
The concatenation operation preserves the differences in multi-scale features, while the convolutional layers adaptively learn the spatial-channel dependencies between features, thus preventing feature confusion caused by direct addition.
The fused feature map \(F\) undergoes two convolution operations with a kernel size of 3 to expand the receptive field and capture more complex semantic features. Using Eq. (15), the final attention feature vector is computed.
Experiments and analysis
Dataset and preprocessing
The proposed network model’s segmentation performance is validated on the LiTS2017 and 3D-IRCADb-01 datasets in this study. The input images are shown in Fig. 5, and each CT image is paired with its corresponding ground truth segmentation for performance evaluation. The ground truth segmentation results are manually annotated by expert doctors and provided in the form of binary masks. As shown in Fig. 6, these masks indicate the liver region and the tumor region within the liver.
As shown in Fig. 7, since the focus of this study is liver segmentation, the first step is to merge the liver and liver tumor labels from the ground truth into a single label. A bilateral threshold is set to (-200, 200), and pixel values outside this threshold are truncated. Then, the image width and height are cropped to 256×256 to match the mask size. It is important to note that the 3D-IRCADb-01 dataset provides images in DICOM format, which were converted to NII format using the Numpy library prior to preprocessing for easier and uniform processing.
As shown in Figs. 8 and 9, this study selects four liver images, labeled as a, b, c, and d. Among them, images a and d have larger liver regions with relatively regular boundaries, whereas images b and c exhibit more complex liver boundaries. According to the study in reference4, after normalization, contrast-limited adaptive histogram equalization (CLAHE) was further applied to enhance image contrast. CLAHE is an improved version of adaptive histogram equalization (AHE) that effectively suppresses the noise amplification problem potentially caused by AHE. From the processing results, it can be observed that CLAHE significantly enhances contrast in both images with regular boundaries and those with complex boundaries, providing clearer input features for subsequent segmentation tasks.
Evaluation indicators
We chose the Dice coefficient, Jaccard coefficient, Volume Overlap Error (VOE), and Hausdorff Distance (HD) as evaluation metrics. The Dice coefficient, as a commonly used similarity measure, quantifies the overlap between the algorithm’s segmentation results and the ground truth labels, as shown in Eq. (16).
Jaccard coefficient, also known as intersection over union (IoU), is another key metric used to measure the region consistency between the algorithm’s segmentation result and the ground truth label, as shown in Eq. (17).
VOE measures the discrepancy between the segmented volume and the ground truth, evaluating how consistently the model preserves the shape and size of the target structures. A lower VOE value indicates higher agreement and more accurate volume delineation, as shown in Eq. (18).
The Hausdorff distance is commonly used to assess the boundary consistency between the model segmentation results and manual annotations, as shown in Eq. (19).
By comprehensively analyzing these metrics, we can compre hensively and deeply evaluate the performance of the algorithm. In the analysis of experimental results, we will focus on the numerical changes of these indicators in order to more accurately evaluate the advantages and disadvantages of the proposed algorithm and provide a scientific and reliable evaluation basis for our research work.
Experimental setup
In this study, an NVIDIA RTX 3090 GPU was employed for both training and inference in segmentation tasks. The batch size was set to 8, and the learning rate was initialized at 1e-4. The Adam optimizer was utilized to update the model parameters. The model was trained for a maximum of 200 epochs, with early stopping applied based on validation performance, using a patience of 15 epochs to prevent overfitting and reduce unnecessary computation.
Comparison experiments
In the liver segmentation task, this study employed the LITS2017 and 3D-IRCADb-01 datasets and conducted a comparative analysis with several state-of-the-art methods. Evaluation was based on three metrics: Intersection over Union (IoU), Dice coefficient, and Volumetric Overlap Error (VOE). The results demonstrate that the proposed EDNet consistently outperforms the competing methods across both datasets, thus affirming its effectiveness and superiority in liver segmentation tasks.
As shown in Table1, on the LITS2017 dataset, mfeeU-Net performs exceptionally well, achieving an IoU of 0.9100, a Dice coefficient of 0.9530, and a VOE of 0.0900. Its excellent segmentation performance is attributed to the accurate localization of the liver region and fine detail capture. AttentionV-Net performs moderately on the same dataset. Despite the incorporation of an attention mechanism, it still exhibits limitations in global feature and contextual information integration, which restricts its segmentation performance. On the 3D-IRCADb-01 dataset, F-UNet achieves an IoU of 0.8877, a Dice coefficient of 0.9405, and a VOE of 0.1123. This method performs well in feature extraction but fails to adequately fuse deep features, resulting in a gap in segmentation accuracy compared to advanced methods. TransUNet also performs excellently on the LITS2017 dataset, with an IoU of 0.9093 and a Dice coefficient of 0.9525, benefiting from the advantages of the Transformer in global feature modeling, which significantly improves segmentation accuracy. AttentionV-Net performs even better on the 3D-IRCADb-01 dataset, with an IoU of 0.9013 and a Dice coefficient of 0.9481. Compared to the LITS2017 dataset, AttentionV-Net performs better on this dataset.
This article references literature26,27,28,29 and conducted statistical tests. As shown in Table 2, the statistical test results indicate that the p-values between EDNet and other comparison methods are all less than 0.05, demonstrating that EDNet’s segmentation performance is significantly better than other methods in terms of statistical significance. EDNet exhibits greater stability and accuracy, further validating the effectiveness of its module design and its potential for practical application.
As shown in Figs. 10 and 11, the green lines represent the mask regions, while the red lines indicate the segmentation results.The proposed EDNet demonstrates outstanding performance on both the LITS2017 and 3D-IRCADb-01 datasets. Although there is some discrepancy between the segmentation results and the ground truth masks, EDNet outperforms other liver segmentation methods in terms of detail handling, closely aligning with the mask data. This indicates that the proposed method offers significant advantages in feature extraction, global information capture, and multi-scale feature fusion, effectively handling complex 3D data while maintaining consistent high performance across different datasets.
As shown in Fig. 12, in terms of Dice coefficient, all models started with a relatively low value, around 0.5, but improved rapidly during training. EDNet and Biformer exhibited a faster initial increase, demonstrating stronger learning capability. Ultimately, EDNet led the other models with a Dice coefficient close to 0.96.
Overall, EDNet demonstrates the best performance in both loss optimization and Dice coefficient improvement. In the loss curve, its optimization efficiency and stability outperform other methods; in the Dice coefficient curve, its segmentation accuracy remains exceptional in both the early and later stages of training. These results highlight the effectiveness and robustness of EDNet in model training and semantic segmentation tasks, indicating its high potential for practical applications.
Ablation experiment
According to the results in Table 3, first, after removing the ECAdd module, the model’s IoU drops to 0.9144, Dice coefficient decreases to 0.9552, and VOE increases to 0.0856, indicating that without the ECAdd module, the model is unable to effectively weight and fuse important features, leading to a decline in segmentation accuracy. After removing the residual structure, the IoU further decreases to 0.9071, Dice coefficient drops to 0.9512, and VOE rises to 0.0929. The residual structure plays a crucial role in effectively fusing features at different scales, helping the model extract multi-level semantic information. Removing it weakens the feature fusion capability, resulting in a further decline in segmentation performance. After removing the DFE module, the model’s IoU drops to 0.8772, Dice coefficient decreases to 0.9346, and VOE increases to 0.1228. The DFE module effectively fuses features extracted from two residual blocks at different scales, preserving fine-grained patterns at the liver boundaries. Without it, the model’s ability to handle fine details at the liver boundaries is weakened, leading to a significant performance drop. The ablation experiments demonstrate that the introduction of the residual structure, ECAdd module, and DFE module significantly enhances segmentation accuracy, while the final performance of EDNet reflects the synergistic benefits of these modules working together.
The impact of preprocessing on the experiment
The data in Tables 4 and 5 indicate that the combined preprocessing approach of normalization and CLAHE plays a vital role in enhancing the model’s segmentation performance. Normalization effectively reduces distribution differences across images by mapping pixel values to a fixed range, thereby improving the standardization of input data. CLAHE enhances local contrast, making blurred regions and fine details more distinct, which further strengthens the model’s ability to capture object boundaries and intricate features. The integration of these two methods significantly boosts segmentation performance, achieving higher IoU and Dice coefficients, as well as notably reducing VOE and HD values across both datasets. These findings demonstrate that the optimized preprocessing pipeline not only improves the model’s segmentation accuracy but also enhances its adaptability to complex boundary regions, which is crucial for overall performance improvement in medical image segmentation tasks
Discussion
This paper presents a multi-scale feature fusion network, EDNet, for liver CT image segmentation. By combining residual structures, the Depth Feature Enhancement (DFE) module, and the automated feature fusion (ECAdd) module, EDNet significantly improves segmentation performance. The Dice coefficients of EDNet on the LiTS2017 and 3D-IRCADb-01 datasets reach 0.9651 and 0.9683, respectively, with IoU values of 0.9330 and 0.9385, outperforming existing state-of-the-art methods. The core advantage of EDNet lies in the introduction of residual structures and convolutional downsampling, which effectively alleviates the issue of feature loss. Additionally, the DFE module enhances the capture of details in blurry boundary regions through multi-scale pooling and depth feature enhancement. Furthermore, the ECAdd module utilizes channel attention mechanisms to adaptively fuse multi-scale features, improving the model’s efficiency in extracting key information. The combined CLAHE preprocessing strategy further optimizes image contrast, enhancing segmentation accuracy. Comprehensive ablation and comparative experiments also validate the effectiveness of each module.
However, the introduction of multiple modules (such as DFE, ECAdd, and residual structures) may increase the network’s architectural complexity. The study does not provide a detailed analysis of model size, inference speed, or memory consumption, which are critical factors for assessing the feasibility of clinical deployment.
Future work could focus on further optimizing model efficiency, expanding validation on multi-center datasets, and exploring hybrid architectures with Transformers to further enhance performance.
Data availability
The datasets analyzed during the current study are available in the Codalab repository, https://competitions.codalab.org/competitions/17094 and Ircad repository, https://www.ircad.fr/research/data-sets/liver-segmentation-3d-ircadb-01/
References
Tang, F. et al. Cmunext: an efficient medical image segmentation network based on large kernel and skip fusion. In 2024 IEEE International Symposium on Biomedical Imaging (ISBI) 1–5 (IEEE, 2024).
Lu, Z., She, C., Wang, W. & Huang, Q. Lm-net: A light-weight and multi-scale network for medical image segmentation. Comput. Biol. Med. 168, 107717 (2024).
Gong, Y., Qiu, H., Liu, X., Yang, Y. & Zhu, M. Research and application of deep learning in medical image reconstruction and enhancement. Front. Comput. Intell. Syst. 7, 72–76 (2024).
Ansari, M. Y., Yang, Y., Meher, P. K. & Dakua, S. P. Dense-psp-unet: a neural network for fast inference liver ultrasound segmentation. Comput. Biol. Med. 153, 106478 (2023).
Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation. In Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 2015, proceedings, part III 18 234–241 (Springer, 2015).
Li, X. et al. H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes. IEEE Trans. Med. Imaging 37, 2663–2674 (2018).
Jiang, H., Shi, T., Bai, Z. & Huang, L. Ahcnet: an application of attention mechanism and hybrid connection for liver tumor segmentation in ct volumes. Ieee Access 7, 24898–24909 (2019).
Lei, T. et al. Defed-net: deformable encoder-decoder network for liver and liver tumor segmentation. IEEE Trans. Radiat. Plasma Med. Sci. 6, 68–78 (2021).
Goodfellow, I. et al. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 27, 856 (2014).
Taha, A., Lo, P., Li, J. & Zhao, T. Kid-net: convolution networks for kidney vessels segmentation from ct-volumes. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part IV 11 463–471 (Springer, 2018).
Sekuboyina, A. et al. Btrfly net: Vertebrae labelling with energy-based adversarial learning of local spine prior. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part IV 11 649–657 (Springer, 2018).
Han, Z., Wei, B., Mercado, A., Leung, S. & Li, S. Spine-gan: semantic segmentation of multiple spinal structures. Med. Image Anal. 50, 23–35 (2018).
Wei, X., Chen, X., Lai, C., Zhu, Y. & Yang, H. Du y. automatic liver segmentation in ct images with enhanced gan and mask region-based cnn architectures. hindawi[internet] (2021).
Biswas, A., Maity, S. P., Banik, R., Bhattacharya, P. & Debbarma, J. Gan-driven liver tumor segmentation: enhancing accuracy in biomedical imaging. SN Comput. Sci. 5, 652 (2024).
Cao, H. et al. Swin-unet: Unet-like pure transformer for medical image segmentation. In European conference on computer vision 205–218 (Springer, 2022).
Demir, U. et al. Transformer based generative adversarial network for liver segmentation. In International Conference on Image Analysis and Processing 340–347 (Springer, 2022).
Ou, J. et al. Restransunet: an effective network combined with transformer and u-net for liver segmentation in ct scans. Comput. Biol. Med. 177, 108625 (2024).
Rahman, H. et al. A deep learning approach for liver and tumor segmentation in ct images using resunet. Bioengineering 9, 368 (2022).
Wang, J., Zhang, X., Lv, P., Zhou, L. & Wang, H. Ear-u-net: Efficientnet and attention-based residual u-net for automatic liver segmentation in ct. arxiv 2021. arXiv preprint arXiv:2110.01014 (2021).
Liu, J. et al. mfeeu-net: a multi-scale feature extraction and enhancement u-net for automatic liver segmentation from ct images. Math. Biosci. Eng. MBE 20, 7784–7801 (2023).
Kushnure, D. T. & Talbar, S. N. Hfru-net: high-level feature fusion and recalibration unet for automatic liver and tumor segmentation in ct images. Comput. Methods Programs Biomed. 213, 106501 (2022).
Wang, J., Lv, P., Wang, H. & Shi, C. Sar-u-net: Squeeze-and-excitation block and atrous spatial pyramid pooling based residual u-net for automatic liver segmentation in computed tomography. Comput. Methods Programs Biomed. 208, 106268 (2021).
Liu, X., Yin, R. & Yin, J. Attention v-net: a modified v-net architecture for left atrial segmentation. Appl. Sci. 12, 3764 (2022).
Chen, J. et al. Transunet: transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021).
Han, Y. & Ye, J. C. Framing u-net via deep convolutional framelets: application to sparse-view ct. IEEE Trans. Med. Imaging 37, 1418–1429 (2018).
Agarwal, R. et al. Deep quasi-recurrent self-attention with dual encoder-decoder in biomedical ct image segmentation. IEEE J. Biomed. Health Inf. (2024).
Agarwal, R., Ghosal, P., Sadhu, A. K., Murmu, N. & Nandi, D. Multi-scale dual-channel feature embedding decoder for biomedical image segmentation. Comput. Methods Programs Biomed. 257, 108464 (2024).
Mandal, B. Optimization of quadratic curve fitting from data points using real coded genetic algorithm. In Emerging Technologies in Data Mining and Information Security: Proceedings of IEMIS 2020, Volume 1 419–428 (Springer, 2021).
Agarwal, R., Ghosal, P., Murmu, N. & Nandi, D. Spiking neural network in computer vision: Techniques, tools and trends. In International Conference on Advanced Computational and Communication Paradigms 201–209 (Springer, 2023).
Acknowledgements
This research is funded by the Doctoral Research Start-up Project, “Research on Intelligent Analysis Method of Massive Turbulence Data” (40615037), and the Horizontal Project, “Digital Intelligent Warehouse Visualization Management Platform Based on Image Processing” (29023307).
Author information
Authors and Affiliations
Contributions
D.Z. conceived the experiments, D.Z. and T.Y.M. conducted the experiments, Y.F.W. and S.B.H. analysed the results,L.T.Z and J.Y.S.guided the deployment experiment. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, D., Ma, T., Zhang, L. et al. Liver CT image segmentation network based on multi-scale feature fusion. Sci Rep 15, 33115 (2025). https://doi.org/10.1038/s41598-025-18049-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-18049-z