Abstract
The growing global demand for sustainable and nutritious food sources has spurred interest in alternative proteins, including those from insects. Cricket (CK) protein, in particular, has emerged as a promising candidate due to its high nutritional value and relatively environmentally friendly production. This study investigates the potential of CK protein and its derived peptides as functional food ingredients. We focus on the effects of CK protein consumption and the specific impacts of peptides generated through enzymatic hydrolysis, a process that breaks down proteins into smaller molecules and bioactive peptides. Our results suggest that CK protein, which is hydrolyzed by pepsin enzymes in the body, may reduce lipid accumulation without significantly affecting bone formation. However, enzymatic hydrolysis using pepsin or Alcalase enzymes can yield peptides with distinct biological effects. Specifically, pepsin-derived peptides promote osteogenesis, while Alcalase-derived peptides enhance adipogenesis. Moreover, we found that Alcalase-peptides significantly induced brown fat formation in vitro. These results highlight the potential of cricket protein-derived peptides as functional food ingredients to modulate cellular differentiation and metabolism.
Similar content being viewed by others
Introduction
There has been a growing interest in exploring alternative protein sources due to increasing global demand and environmental concerns. Edible insects offer a sustainable and nutritious protein option. Cricket protein stands out among other edible insect proteins due to its superior nutritional profile, ease of large-scale and sustainable farming, higher consumer acceptance especially for western markets. Cricket protein demonstrates high protein quality, a balanced essential amino acid profile, and rich micronutrient content. It consistently achieves elevated DIAAS and PDCAAS scores, reflecting efficient digestibility, and in some cases surpasses the protein quality of yellow mealworm (Tenebrio molitor) and matches or exceeds that of migratory locust (Locusta migratoria)1,2. Furthermore, crickets are notably abundant in vitamin B12, iron, and zinc compared with mealworms and grasshoppers3, underscoring their superior role as a sustainable and nutrient-dense protein source.
Moreover, peptides derived from processing edible crickets have emerged as a promising dietary supplement due to their high protein content and potential health benefits4,5,6 . While research on the biological effects of cricket peptides is still in its early stages, emerging studies have highlighted their potential health benefits including antioxidant7,8,9, anti-inflammatory7,8,10,11,12, and antimicrobial properties12,13 antihypertensive9 , and anticancer properties14. These properties suggest that cricket peptides may have the potential to modulate cellular processes, including cell proliferation, differentiation, and apoptosis. However, the underlying mechanisms by which cricket peptides regulate cellular activity are not well characterized. Therefore, we conducted this study to elucidate the functions of cricket peptides in human cells.
Human Mesenchymal Stem Cells (hMSCs) are multipotent stromal cells capable of differentiating into osteoblasts, adipocytes, and chondrocytes15. Due to their roles in tissue regeneration16,17, immune modulation16,18,19, and antioxidant activity20,21, hMSCs serve as a physiologically relevant in vitro model for investigating the biological effects of bioactive compounds, including peptides and protein hydrolysates. Previous studies have shown that protein hydrolysates from sources such as fish collagen and milk can modulate hMSC behavior by enhancing proliferation, migration, and osteogenic differentiation, suggesting that bioactive peptides can modulate stem cell behavior22,23. Similarly, Ganguly et al.11 reported that cricket protein isolate (CPI) promotes osteogenic differentiation in a concentration-dependent manner in human bone marrow-derived mesenchymal stem cells. These findings suggest that dietary proteins and their hydrolysates may exert functional effects on stem cell fate and tissue remodeling. Although animal studies have reported that cricket protein can modulate fat mass, muscle growth, and inflammation via amino acids-mediated activation of the mTORC1 pathway, a central regulator of lipid metabolism and protein synthesis24. However, evidence from human cell models remains limited.
This study aims to address this gap by assessing the effects of crude cricket (CK) protein and its enzymatically derived peptides on osteo-adipogenic fate determination in hMSCs. Three forms of cricket-derived macromolecules were prepared: (i) crude CK protein isolate, (ii) peptides hydrolyzed with pepsin (PS), simulating human gastrointestinal digestion, and (iii) peptides hydrolyzed with Alcalase (A), a commercially available protease. These variants were used to investigate their effects on hMSC behavior, with particular focus on antioxidant activity and the balance between osteogenic and adipogenic differentiation. By exploring the impact of cricket protein hydrolysates on hMSC differentiation and cellular function, this study provides mechanistic insights into the potential of insect-derived bioactive compounds for applications in regenerative medicine, wound healing, medical nutrition therapy, and functional foods targeting metabolic health.
Materials and methods
Cricket (CK) protein preparation
Cricket (CK) powder was obtained from JR Unique Foods, Ltd. (Thailand), and crude CK protein was extracted according to the protocol described by Summart et al.25. CK powder was mixed with deionized water at a 1:10 (w/v) ratio. Its pH was adjusted to 12 with 2.5 M NaOH. The mixture was then stirred for 1 h at room temperature prior to centrifugation at 8,000 g for 10 min and 4 °C after which the supernatant was collected. The pH of the supernatant was adjusted to 4 using 2.5 M HCl, followed by stirring for 30 min at room temperature. The mixture was then centrifuged at 9,000 g for 20 min at 4 °C. The precipitate was collected and freeze-dried. It was then kept at -20 °C. Prior to supplementation of the hMSCs, an MTT assay was performed to determine its cytotoxicity (Supplementary Fig. 1). Subtoxic doses were chosen for further experiments.
CK protein hydrolysate preparation
Alcalase hydrolysis
Alcalase-based CK protein hydrolysates were prepared by hydrolyzing crude CK protein powder (12% w/v) with 5% Alcalase. The pH of the crude protein and Alcalase mixture was adjusted to 9.0. This mixture was incubated in a water bath at 50 °C for 3 h with continuous shaking at 120 rpm. Then, the reaction was stopped by heating the mixture in water at 100 °C for 10 min. It was then centrifuged at 10,000 g for 10 min at 4 °C. The supernatant was collected and freeze dried25.
Pepsin hydrolysis
Pepsin-based CK protein hydrolysates were prepared with slight modifications of the method described by Tejano et al.26. CK protein powder (12% w/v) was hydrolyzed with 4% pepsin. The pH of the crude protein and pepsin mixture was adjusted to 2. Then, the mixture was incubated in a water bath at 37 °C for 3 h with continuous shaking at 120 rpm. The reaction was stopped by heating the mixture in water at 100 °C for 10 min. Finally, the mixture was centrifuged at 10,000 g for 10 min at 4 °C. The supernatant was collected and freeze-dried.
Antioxidant activity assay
Radical scavenging activity by DPPH assay
The radical scavenging activity of CK protein hydrolysate was determined using a 1,1-diphenyl-2-picrylhydrazyl (DPPH•) assay, as previously described27. Briefly, 100 µL of a 0.2 mM DPPH solution was added to a mixture of 20 µL of each protein hydrolysate sample with 80 µL of 0.1 M Tris–HCl buffer (pH 7.4) in a 96-well microplate. The mixtures were shaken vigorously and incubated in the dark at room temperature for 30 min. Absorbance was measured at 514 nm. Trolox at a concentration of 0.1 mg/mL served for standard curve preparation.
Radical scavenging activity by ABTS assay
The radical scavenging activity of cricket protein hydrolysate was determined using a 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) (ABTS• +) method, according to a previously described procedure27. Briefly, 7 mM ABTS reagent was incubated with 2.45 mM potassium persulfate in a 1:1 ratio for 12–18 h in the dark at room temperature to generate ABTS• + radicals. Two-hundred µl of an ABTS working solution and 10 µL of CK protein hydrolysate were added in a 96-well plate and incubated at room temperature for 5 min. Absorbance was measured at 734 nm. Trolox was used as a standard.
Ferric-reducing antioxidant power assay (FRAP)
A FRAP assay was used according to the previously described procedure28. Briefly, 10 µL of peptide fractions were dissolved in DI water and mixed with 180 µL of a fresh FRAP solution. Reaction mixtures were incubated at 37 °C for 5 min. After that, their OD was measured at 593 nm and the antioxidant activities of CK protein hydrolysates were compared with a Trolox standard curve.
Isolation and culture of human mesenchymal stem cells
After receiving written informed consent from their mothers, postnatal tissues of the healthy newborns were collected. Chorionic tissue was isolated and washed in phosphate buffer saline (PBS), cut into small pieces prior to treatment with 0.25% Trypsin–EDTA (GIBCO™; Invitrogen Corporation, Carlsbad, CA, USA) for 30 min at 37 °C. The digested cells were harvested and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) before being plated into tissue culture vessels (Corning Incorporated, Corning, NY, USA). Cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2 in air. The culture medium was changed every other day. Cells were subjected to immunophenotypic characterization as previously described by Lorthongpanich et al. (2024)29. This was done to determine the presence of surface molecules specific to hMSCs, including CD73, CD90, and CD105 and the absence of hematopoietic surface markers CD34 and CD45 using flow cytometry (FACSCantoTM or FACSCaliburTM analyzer; BD Biosciences, San Jose, CA, USA). All antibodies used in this assay were 1:100 dilutions in PBS.
Osteogenic differentiation and mineralization assay
The hMSCs were cultured in DMEM-high glucose supplemented with 10% (v/v) FBS until their density reached 90% confluence. At this stage, the DMEM was replaced with the NH OsteoDiff® Medium (Miltenyi Biotec, Bergisch Gladbach, Germany) to induce osteogenic differentiation of human hMSCs, according to the manufacturer’s instructions. An NH OsteoDiff® medium was supplemented with various concentrations of crude CK proteins and hydrolyzed CK peptides, in concentrations ranging from 0 to 5 mg. Cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2 in air. The medium was replaced every 3 days. Calcium deposition was determined using the Alizarin Red S staining assay as previously described. Briefly, the differentiated hMSCs were fixed with 4% (w/v) Paraformaldehyde (PFA), washed twice with deionized water, and stained with 40 mM Alizarin Red S (Sigma-Aldrich, USA) for 20 min at room temperature (RT). Levels of calcium deposition were quantified using the amount of Alizarin Red S retained by the cells. Quantification of Alizarin Red S was done as previously described by Lorthongpanich et al.29.
Adipogenic differentiation
The hMSCs at a density of 5 × 104 cells were seeded into each well of the6-well plates (Corning) and cultured for 21 days in an adipogenic differentiation medium. This comprised DMEM high-glucose (Gibco) supplemented with 1 µM dexamethasone (Sigma-Aldrich), 0.5 µM isobutylmethylxanthine (Sigma-Aldrich), 5 µg/ml insulin, and 50 µM indomethacin (Sigma-Aldrich). The medium was supplemented with various concentrations of CK peptides (2.5 and 5 mg/ml). Differentiation efficiency was determined by qRT-PCR for the expression of adipogenic-specific genes23 and Nile red staining.
Nile Red staining
Nile Red staining and fluorescence measurements of adipogenesis were carried out using a 50 mM DMSO stock solution maintained at -20 °C and shielded from light. Cells were treated with CK peptides at final concentrations of 2.5 and 5 mg/ml. After treatment, the cells were rinsed with PBS, fixed with 4% paraformaldehyde for 10 min, and washed twice with PBS. Dye was then applied directly to the cells at a final concentration of 10 µM in PBS, incubated for 10 min at room temperature, and washed twice with PBS. The fluorescent signal was measured using a microplate reader (VICTOR Nivo Multimode plate readers, PerkinElmer) with excitation (485 nm) and emission (572 nm) spectra. The data was then averaged and standardized to a control.
MTT assay
hMSCs at a density of 5 × 103 cells were plated into each well of a 96-well plate and incubated overnight at 37 °C in a 5% CO2 atmosphere. These hMSCs were treated with different concentrations of cricket peptides ranging from 0–5 mg/ml. After 72 h of incubation, MTT solution (0.5 mg/ml final concentration) was added into each well and incubated at 37 °C for 3 h. After removing the medium from the plate, 100 µl of DMSO was added to each well to dissolve the purple formazan crystals. A Bio Tek Quant spectrophotometer was used to measure the cells at 570 nm and a reference wavelength of 630 nm. The vitality of the cells was determined as a percentage ratio and compared to the control cells.
Brown adipose tissue (BAT) differentiation
hMSCs were seeded at a density of 5 × 104 cells per well in a 12-well plate, allowing for growth to approximately 70% confluence. The culture medium was then replaced with BAT induction medium that included DMEM, 10% FBS, 10% horse serum (Gibco), 1% penicillin–streptomycin, 100 nM dexamethasone, 0.45 mM isobutyl methyl xanthine (Sigma Aldrich), 3.0 µg/mL insulin (Sigma-Aldrich), and 1.0 μM rosiglitazone (BRL49653). The BAT induction medium was supplemented with A-peptides at 1.25 and 2.5 mg/ml. The medium was changed every 3 days with appropriate supplementation.
Scratch wound migration assay
hMSCs at an 8 × 104 cells/well concentration were seeded into 24-well plates and cultured overnight. At 80–90% cell confluence, the cells were wounded by scratching the surface of the culture well with a sterile 1000 ml pipette tip. Cells were washed with 1xPBS to remove detached cells and other cellular debris before adding fresh culture medium supplemented with various concentrations of cricket peptides (0–5 mg) and incubated at 37 °C under a 5% CO2 atmosphere for 48 h. Inverted microscopy was used to capture images of the closing wounds at 0, 24, and 48 h.
Western blot analysis
hMSCs, treated at appropriate times, were lysed in RIPA buffer supplemented with protease inhibitors. Total proteins were loaded and separated on a 7.5% SDS-PAGE gel. Following electrophoresis, the proteins were transferred to a PVDF membrane (Merck Millipore), blocked in 5% (w/v) skim milk, and incubated overnight at 4 °C with primary antibodies. The antibodies were as follows: Collagen 1 and OPN (both from Cell Signaling Technology) diluted to 1:1,000 and anti-β-actin peroxidase (ACTB; Sigma-Aldrich) diluted to 1:10,000. The membrane was washed with Tris-buffered saline/Tween 3 times before incubation with secondary antibodies for 2 h at room temperature. Signals were visualized using a Lumina crescendo western horseradish peroxidase substrate (Millipore). The antibodies used in this study are listed in Supplementary Table S1.
Real-time quantitative reverse transcription polymerase chain reaction (Real-Time qRT-PCR)
RNA was isolated from the cells and subjected to reverse-transcription using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). qRT-PCR was conducted according to the manufacturer’s instructions. Real-time qRT-PCR was performed using a Real-Time PCR Master Mix (Applied Biosystems) with a CFX384 Real-Time PCR System (Bio-Rad Laboratories, Hercules, CA, USA). The primers used in this study are listed in Supplementary Table S2.
Statistical analysis
All experiments were conducted in triplicate. Results are presented as mean values ± standard deviations. One-way ANOVA and Duncan’s multiple-range tests were applied for mean comparison using SPSS 22.0 (IBM Corp., NY). One-way ANOVA and Duncan’s multiple-range tests were applied for mean comparisons of chemical determinations using SPSS 22.0. The Mann–Whitney U test was used to compare non-parametric variations between groups. A p-value < 0.05 was considered statistically significant. The data were analyzed using GraphPad Prism software version 3.0 for Windows (GraphPad Software, La Jolla, CA, USA).
Results
Distinctive effect of cricket peptides on stem cell proliferation
To evaluate the effects of CK, P and A-peptides on cell proliferation, a wound healing assay was performed. The degree of wound closure was observed at 0, 24, and 48 h. As shown in Fig. 1A, A-peptides significantly inhibited cell migration at both 24 and 48 h, especially at a 5 mg/ml concentration, which reduced migration by more than 50% compared to the control. In contrast, cells treated with CK and PS exhibited migration patterns that were not significantly different from the control group. Quantitative analysis of wound closure supports these findings. Cells treated with 5 mg/ml of A-peptides had significantly lower wound closure than all other conditions (P < 0.05). No significant differences were observed between CK, PS, and the control group at any time (Fig. 1B). These results suggest that A-peptides have a strong inhibitory effect on cell migration, whereas CK and PS do not significantly alter the migratory capacity of cells.
A-peptides and PS-peptides in the scratch wound migration assay. Imagery of wound closure at 0, 24, and 48 h (A). The percentage of wound closure for A-peptides and PS-peptides at 0, 2.5, and 5 mg/ml after 24 and 48 h (B). *P < 0.05, ns = not significantly different. The scale bar represents 500 µm.
Effect of crude CK protein on the differentiation capacity of hMSCs to osteo- and adipogenic lineage differentiation
It is well-established that hMSCs must maintain a delicate balance for their differentiation, especially for differentiation into fat and bone30. As bone induction factors, such as RUNX2, can inhibit adipogenic gene peroxisome proliferator-activated receptor γ (PPARγ) expression, and vice versa31, the balance of these lineages should be studied when treating hMSCs with different molecules. In this study, we investigated the effect of CK protein on hMSC differentiation towards osteogenic and adipogenic lineages. The hMSCs were subjected to osteogenic differentiation as previously described by Lorthongpanich et al.32,33. CK protein was supplemented into the osteogenic differentiation media at concentrations of 0, 2.5, and 5 mg/ml (Fig. 2). Differentiated cells were collected to study the effect of CK protein on osteogenic cell commitment using qPCR.
Effect of CK protein on osteogenic differentiation of hMSCs. Transcriptional expression of osteogenic specific genes, including RUNX2, ALP and COL1 were analyzed upon treatment with CK protein at 0, 2.5, and 5 mg/ml (A). The concentration of Alizarin Red as spectrophotometrically measured (B), and representative images of osteogenic cells at Day 21 of differentiation for each treatment (C). Protein expression of COL1, OPN, and ACTIN were determined by western blot (D) and relative quantification of target protein/ACTIN band intensity (E). *P < 0.05, ns = not significantly different. The scale bar represents 100 µm. Full-length gels associate with Fig. 2D are presented in Supplementary Fig. 2.
Transcript analysis revealed no significant differences in the expression of osteogenic genes, RUNX2, and COL1 genes, while slight differences were observed in ALP gene expression compared to the non-supplemented control (Fig. 2A). Calcium deposition capacity was evaluated on Day 14 of differentiation using Alizarin Red S staining. The results showed no significant difference between the control and CK protein-treated cells (Fig. 2B and C). Consistently high expression of COL1, an early marker of osteoblast differentiation, was observed across all treatments, although a slight reduction in OPN protein levels, a late-stage osteogenic marker, was observed in the CK treatments (Fig. 2D and E).
For adipogenic study, we found that supplementing CK protein (2.5–5 mg/ml) into the adipogenic differentiation media slightly increased the expression of adipogenic specific genes PPARG, AdipoQ and aP2 genes (Fig. 3A-C). However, lipid droplet accumulation of hMSCs was not significantly different as determined by Oil Red O staining (Fig. 3D-E). These results demonstrated that crude CK protein do not significantly affect the osteogenic and adipogenic cell formation of hMSCs. However, to reflect the real consumption of CK protein, it must be considered that the protein is hydrolyzed by pepsin enzymes (PS) upon ingestion. The effect of PS-peptides was evaluated in further experiments.
Effect of CK protein on adipogenic differentiation of hMSCs. Adipogenic specific genes, PPARG (A), aP2 (B), and AdipoQ (C) expression in CK protein treated cells. Representative images (D) and absorbance analysis (E) of Oil Red O staining on the adipogenic differentiated cells (Day 21 of differentiation) after treatment with 0–5 mg/ml of CK. *P < 0.05, ns = not significantly different. The scale bar represents 100 µm.
Generation of pepsin (PS) and Alcalase (A) hydrolysate peptides upon pepsin enzyme treatment and its antioxidant activity
The physicochemical properties of A-peptide and PS-peptide obtained from enzymatic hydrolysis were analyzed. The degree of hydrolysis (%DH) was 18.69% and 7.77% for A-peptide and PS-peptide, respectively. Protein concentrations were approximately 51 mg/mL and 47 mg/mL, respectively. Solubility analysis revealed that the crude cricket protein (CK) exhibited the lowest solubility at pH 4 and the highest at pH 10. In contrast, A-peptide and PS-peptide showed solubility ranges of 41–60% and 53–64%, respectively, within the pH range of 2–10. The molecular weights of both peptide types were in the range of 130–465 Da, indicating that they are predominantly short-chain peptides.
The antioxidant activities of cricket protein hydrolysate treatments with pepsin (PS) and Alcalase (A) enzyme were evaluated for free radical scavenging activity using DPPH and ABTS assays. Ferric-reducing activity was determined using a FRAP assay. As shown in Fig. 4, these cricket protein hydrolysates presented DPPH (Fig. 4A) and ABTS (Fig. 4B) radical scavenging activities and had ferric-reducing activity (Fig. 4C). PS and A-hydrolyzed peptides showed the same DPPH radical scavenging activities (P > 0.05). The highest ABTS radical scavenging activity was found in A-hydrolyzed peptide (P < 0.05). Additionally, A-hydrolyzed peptides also displayed a significantly higher ferric-reducing capability than PS based on the FRAP assay (Fig. 4C). These results imply that the cricket protein hydrolysates have antioxidant properties, indicating their potential for protective roles against oxidative damage from free radicals.
Pepsin-derived peptides significantly reduced adipogenesis in human MSCs
To determine the biological activity of CK protein after hydrolysis with a pepsin enzyme, the resulting peptides, hereafter referred to as PS, were supplemented into osteogenic and adipogenic differentiation media. Then, they were used for culturing and differentiating human MSCs into osteocytes and adipocytes, respectively. After 21 days of osteogenic cell differentiation, cells were collected and evaluated for the expression of osteogenic genes and calcium mineralization capacity. Transcriptional analysis of osteogenic-specific genes showed no significant differences between the non-treated and treated groups (Fig. 5A). The Alizarin Red S staining assay was used to assess calcium deposits in cultured cells. Results showed no significant calcium deposition in the cells treated with PS-peptides, as shown in Fig. 5B. Quantification of Alizarin Red S extracted from each sample confirmed no significant differences in the calcium mineralization capacity of these cells (Fig. 5C). The effect of PS on adipogenesis of human MSCs was also evaluated. After 21 days of adipogenic differentiation with various concentrations of PS-peptides and with no supplementation, adipogenic gene expression was determined. These results showed significant inhibition of adipogenic-specific genes, including PPARG, Adipoq, and aP2 (Fig. 5D). A reduction in lipid droplet formation upon treatment with PS-peptides (Figs. 5E and F) correspond to the gene expression results. These results indicate that CK protein, when hydrolyzed with a pepsin enzyme, can yield peptides that effectively reduce lipid production. This is an intriguing finding, as it could potentially lead to novel approaches for managing lipid levels and promoting overall health using insect-based proteins.
Effect of pepsin-derived peptides (PS-peptides) on osteo-adipogenic differentiation of hMSCs. Expression of RUNX2, ALP, and COL1 genes on Day 21 of hMSCs after being subjected to osteogenic differentiation supplemented with 0–5 mg/ml of PS-peptides (A). Representative images of Alizarin Red S stained cells on Day 21 of differentiation (B), and Alizarin Red S concentration after extraction from each culture condition (C). Transcriptional analysis of adipogenic specific genes, PPARG, Adipoq, and aP2 on hMSCs-derived adipogenic cells treated with 0–5 mg/ml of PS-peptides (D). Oil Red O staining on the hMSCs-derived adipogenic cells (E), and absorbance analysis of Oil Red O in each sample as spectrophotometrically determined (F). *P < 0.05, ns = not significantly different. The scale bar represents 100 µm.
Alcalase-derived peptides increase lipid production
Alcalase, an enzyme that cleaves internal peptide bonds in proteins, is widely used in various industries, especially in the manufacture of foods and beverages. CK proteins were hydrolyzed with Alcalase (A) to generate A-peptides. To assess the impacts of A-peptides on human mesenchymal stem cell (hMSC) differentiation, hMSCs were cultured in osteogenic or adipogenic differentiation media supplemented with various concentrations of A-peptides (0–5 mg/ml).
Results indicated that a 5 mg/ml concentration of A-peptides was cytotoxic to MSCs when added continuously over 21 days. Therefore, only 0 and 2.5 mg/ml concentrations were used for further experiments. Treatment with 2.5 mg/ml of A-peptides had no significant effect on osteogenic lineage commitment (Fig. 6A) but rather impacted maturation as determined by COL1 protein expression (Fig. 6B) and calcium mineralization of the induced osteogenic cells (Fig. 6C-D). Conversely, when A-peptides were added to adipogenic differentiation media, a large increase in lipid droplet formation was observed as early as Day 1 (Fig. 6E). The number of lipid droplets continued to increase before declining on Day 14 (Fig. 6E-F) while a control group showed significantly slower lipid formation and fewer lipid droplets in the culture. This phenotypic change correlated with increased expression of adipogenic specific genes PPARG and AdipoQ (Fig. 6G–H). These findings demonstrate that enzymatic hydrolysis of CK proteins can generate bioactive peptides with distinct biological effects. In this study, A-peptides were shown to potently stimulate adipogenesis while exerting a subtler influence on osteogenesis.
Effect of Alcalase-derived peptides (A-peptides) on adipo-osteogenic differentiation of hMSCs. Osteogenic specific gene RUNX2, ALP, and COL1 expression on Day 21 of hMSCs after being subjected to osteogenic differentiation supplemented with 0- 2.5 mg/ml of A-peptides (A). Western blot shows COL1 protein expression in hMSCs –derived osteogenic cells supplemented with 0–2.5 mg/ml of A-peptides (B). Alizarin Red staining (C) and its concentration of the differentiated cells as spectrophotometrically measured (D). Representative images of lipid droplets stained with Oil Red O (E), and measurement of Oil Red O (F) on hMSCs-derived adipogenic cells supplemented with 2.5 mg/ml of A-peptides from Days 1 to 21 of differentiation. Expression of PPARG (G) and AdipoQ (H) in differentiated cells treated with A-peptides. *P < 0.05, **P < 0.01, ns = not significantly different. The scale bar represents 100 µm. Full-length gels associate with (B) are presented in Supplementary Fig. 3.
Alcalase-derived peptides stimulate lipid formation in a cytokine-free culture environment
To determine whether A-peptides could induce lipid formation in human MSCs in a cytokine-free environment, complete MSC culture media (DMEM + 10% FBS) supplemented with 2.5 mg/ml of A-peptides was used for culturing human MSCs. Results showed that even with no supplementation using adipogenic inducing cytokines, A-peptides supplementation alone was sufficient to induce adipogenesis, as evidenced by the increased expression of adipogenic genes (Fig. 7A–B) and the abundant lipid droplets observed as early as Day 1 of culture, determined by Oil Red O staining (Fig. 7C–D). Lipid accumulation increased throughout the culture period and declined on Day 14 due to adipogenic cell death. This result suggests that A-peptides may contain important components that can induce adipogenic lineage differentiation. However, further study is required to characterize the adipogenic induction factors of A-peptides.
A-peptides stimulate lipid droplet production independent of external cytokine supplementation. The hMSCs were cultured in a DMEM + 10% Fetal Calf Serum (FCS) supplemented with 0 and 2.5 mg/ml of A-peptides for 21 days. The expression of adipogenic genes PPARG (A) and AdipoQ (B) is shown. Absorbance analysis of Oil Red O at each time is presented (C). Representative images of lipid droplets stained with Oil Red O after the hMSCs were cultured in DMEM + 10% FCS supplemented with 0 or 2.5 mg/ml of A-peptides (D). *P < 0.05, ns = not significantly different. The scale bar represents 100 µm.
Brown fat specific genes increased upon treatment with A-peptides
An increase in brown fat is preferred over white fat. We therefore determined the potential of A-peptides to induce brown fat production by supplementing with A-peptides (in concentrations ranging from 0–2.5 mg/ml) into brown fat differentiation media. The A-peptides-treated groups showed significantly more lipid in a dose-dependent manner (Fig. 8A). CD137 was identified as a marker of beige adipocyte precursors and mature beige adipocytes in microarray screens of immortalized obesity-resistant Sv129 mice cell lines. Furthermore, significant upregulation of the brown fat genes for uncoupling protein 1 (UCP1) and CD137 was observed, suggesting a pattern of differentiation compatible with beige-brown fat (Fig. 8B). This result suggests that CK protein hydrolyzed with Alcalase enzyme could promote beige-brown fat formation in vitro.
Discussion
This study demonstrates the potential of CK proteins and its hydrolyzed peptides as a valuable dietary supplement. CK peptides possess potent antioxidant properties and significantly influence cellular processes, including cell proliferation and differentiation of human mesenchymal stem cells (hMSCs). Additionally, the study reveals that enzymatic hydrolysis of CK proteins can yield peptides with diverse biological effects on hMSCs. While CK protein hydrolysis by pepsin, mimicking gastric digestion, may reduce lipid accumulation, hydrolysis with Alcalase results in an inverted phenotype. Reactive oxygen species (ROS) generation represents a key mechanism for suppressing the stemness of MSCs and inducing cellular senescence. Consequently, mitigating oxidative stress or enhancing antioxidant defenses has become a favored approach to enhance the stemness of hMSCs34. Oxidative stress, resulting from elevated levels of reactive oxygen species (ROS), can damage DNA, RNA, and cellular proteins, potentially triggering apoptosis in stem cells35. Numerous studies have demonstrated that antioxidants not only reduce oxidative stress and enhance stem cell survival but also influence the potency and differentiation of human stem cells36. A tenfold reduction in ROS levels with increased glutathione (GSH) and glutathione reductase (GR) activities were observed in induced pluripotent stem cells (iPSCs) compared to fibroblasts37 . Choi et al.38 demonstrated that adding ascorbic acid 2-phosphate (AAP) at different concentrations can influence the fate of hMSCs from adult human bone marrow. AAP significantly increased osteogenic differentiation at a 50 mM concentration, while a significant induction of adipogenic differentiation with oil droplet formation was noted at concentrations of 250 mM and higher. In the current study, our CK protein hydrolysates from pepsin and Alcalase showed antioxidant properties with DPPH and ABTS radical scavenging activities and ferric-reducing activity. From our previous report, peptides from CK protein hydrolyzed with Alcalase enzyme also enhance the activation of antioxidant enzymes, including SOD, CAT, GSR, and GPx9.
The molecular structural attributes of bioactive peptides, including molecular size, amino acid hydrophobicity, and peptide sequences, largely influence their antioxidant capabilities to reduce DPPH, ABTS, and FRAP radicals by donating electrons and deprotonating reactive functional groups39. Previously, tetrapeptides containing hydrophobic and positively charged amino acids from cricket protein hydrolysate were reported to provide good potential for antioxidant activity9. Then, using different enzymes for protein hydrolysis will result in differences in peptide sequences due to the specific digestion sites of each enzyme. This may be related to the capability of these peptides to stimulate adipogenesis while exerting a subtler influence on osteogenesis. Then, using antioxidant peptides can improve the viability and self-renewal capacity of stem cells and affect their differentiation potential.
In addition to their antioxidant properties, short peptides also exhibit biological activities, including regulation of proliferation, differentiation, apoptosis, innate immunity, tumorigenesis, and wound healing40. Results on the regulation of chondrogenic differentiation via various types of short peptides have been reported41. The W9 (YCWSQYLCY) exhibited peptide-induced osteogenesis through osteoblasts of the MC3T3-E1 line. W9 peptide also induced differentiation of osteoblasts in MC3T3-E1 strain cells and in human mesenchymal stem cells (hMSCs)42. The impacts of short peptides on cell differentiation vary depending on their structure and concentration. This study revealed that A-peptides exhibited a stronger capability to stimulate stem cell differentiation through adipogenesis and osteogenesis than PS-peptides. This suggests that Alcalase-generated peptides are more effective in inducing stem cell differentiation than those produced through pepsin proteolysis. The tetrapeptides, AEDG and AEDP, promote differentiation of pluripotent cells into epidermal, mesenchymal, and neural tissues, while peptides AEDL and KEWD specifically drive differentiation of lung and pancreatic cells, respectively43. In the current study, Alcalase proteolysis resulted in CK protein hydrolysates primarily consisting of small peptides with sizes under 1 kDa. Tetrapeptides derived from A-peptides were previously identified for their biological activities9. This is consistent with previous findings, which suggest that tetrapeptides may contribute to promoting stem cell differentiation. However, identifying specific short peptide fragments within the A-peptides could further elucidate the mechanisms underlying these effects and potentially lead to development of novel strategies for managing lipid levels and promoting overall health through insect-based protein consumption.
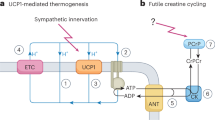
Brown adipose tissue (BAT) is a type of fat that can convert fat to heat. BAT plays a role in adaptive thermogenesis in response to cold environments and dietary intake44,45. Most studies have shown that exercise activates BAT formation and increases its density. A recent study by Vatner and colleagues showed that BAT can directly increase exercise performance46. Increasing brown fat improves exercise endurance and supports healthy aging. This may also help protect against illnesses such as obesity, diabetes, and cardiovascular disease45. Our study found that cricket peptides can also increase brown fat levels. Therefore, the hydrolysate A-peptides could potentially be used in a supplementary diet for athletes to increase their exercise endurance. However, the mechanism of A-peptide action on adipogenic differentiation, and increased brown fat should be further investigated.
Conclusion
This study demonstrates the biological efficacy of cricket (CK) protein hydrolysates, particularly those derived from Alcalase and Pepsin digestion, in promoting antioxidant activity and modulating the behavior of hMSCs. The peptides produced exhibit significant effects on cell proliferation and differentiation, with distinct responses depending on the enzymatic hydrolysis. Pepsin-derived peptides showed potential in reducing lipid accumulation, while Alcalase-derived peptides induced adipogenic differentiation and promoted brown adipose tissue (BAT) formation, suggesting potential applications in metabolic health support. These findings highlight the potential for the development of insect-derived peptides as sustainable bioactive ingredients in functional foods or nutraceuticals targeting regenerative health and metabolic support. Further studies are required to identify specific peptide sequences responsible for these effects and to elucidate the underlying molecular mechanisms.
Data availability
The data used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Hammer, L. et al. Mealworm larvae (Tenebrio molitor) and crickets (Acheta domesticus) show high total protein in vitro digestibility and can provide good-to-excellent protein quality as determined by in vitro DIAAS. Front Nutr 10, 1150581 (2023).
Nachtigall, L. et al. Proteins and amino acids from edible insects for the human diet-A narrative review. Nutrients 17, 1245 (2025).
Watanabe, F., Yabuta, Y., Bito, T. & Teng, F. Determination of vitamin B12 in four edible insect species by UHPLC. Food Chem 262, 60–64 (2018).
Montowska, M. et al. Nutritional value, protein and peptide composition of edible cricket powders. Food Chem 289, 130–138 (2019).
Nowakowski, A. C. et al. Potential health benefits of edible insects. Crit Rev Food Sci Nutr 62(13), 3499–3508 (2022).
Skotnicka, M. et al. Possibilities of the Development of Edible Insect-Based Foods in Europe. Foods 10(4), 766 (2021).
Quinteros, M. F. et al. Functional, Antioxidant, and Anti-Inflammatory Properties of Cricket Protein Concentrate (Gryllus assimilis). Biology (Basel) 11(5), 776 (2022).
Zielinska, E., Baraniak, B. & Karas, M. Antioxidant and Anti-Inflammatory Activities of Hydrolysates and Peptide Fractions Obtained by Enzymatic Hydrolysis of Selected Heat-Treated Edible Insects. Nutrients 9(9), 970 (2017).
Summart, R. et al. Characterization and molecular docking of tetrapeptides with cellular antioxidant and ACE inhibitory properties from cricket (Acheta domesticus) protein hydrolysate. Heliyon 10(15), e35156 (2024).
Fashakin, O. O. et al. Isolation and Identification of Antioxidant Peptides Derived from Cricket (Gryllus bimaculatus) Protein Fractions. Insects 14(8), 674 (2023).
Ganguly, K. et al. Naturally-derived protein extract from Gryllus bimaculatus improves antioxidant properties and promotes osteogenic differentiation of hBMSCs. PLoS ONE 16(6), e0249291 (2021).
Hall, F., Reddivari, L. & Liceaga, A. M. Identification and Characterization of Edible Cricket Peptides on Hypertensive and Glycemic In Vitro Inhibition and Their Anti-Inflammatory Activity on RAW 264.7 Macrophage Cells. Nutrients 12, 3588. https://doi.org/10.3390/nu12113588 (2020).
Rivero-Pino, F., Leon, M. J. & Montserrat-de la Paz, S. Potential applications of antimicrobial peptides from edible insects in the food supply chain: uses in agriculture, packaging, and human nutrition. Food Biosci. 62, 105396 (2024).
Summart, R., Imsoonthornruksa, S., Ketudat Cairns, M. & Udomsil, N. Exploring the anticancer potential of cricket derived peptides in human cancer cells; pro apoptotic effects via a caspase 3 pathway. J Funct Foods 127, 106760 (2025).
Pittenger, M. F. et al. Multilineage potential of adult human mesenchymal stem cells. Science 284(5411), 143–147 (1999).
Ayala-Cuellar, A. P. et al. Roles of Mesenchymal Stem Cells in Tissue Regeneration and Immunomodulation. Biomol Ther (Seoul) 27(1), 25–33 (2019).
Rehman, A. et al. Mesenchymal Stem Cells in Soft Tissue Regenerative Medicine: A Comprehensive Review. Medicina 59(8), 1449 (2023).
Li, P. et al. Immunomodulatory properties of mesenchymal stem cells/dental stem cells and their therapeutic applications. Cell Mol Immunol 20(6), 558–569 (2023).
Li, H., Dai, H. & Li, J. Immunomodulatory properties of mesenchymal stromal/stem cells: The link with metabolism. J Adv Res 45, 15–29 (2023).
Xia, C. et al. Emerging Antioxidant Paradigm of Mesenchymal Stem Cell-Derived Exosome Therapy. Front Endocrinol (Lausanne) 12, 727272 (2021).
Stavely, R. & Nurgali, K. The emerging antioxidant paradigm of mesenchymal stem cell therapy. Stem Cells Transl Med 9(9), 985–1006 (2020).
Chalamaiah, M., Yu, W. & Wu, J. Immunomodulatory and anticancer protein hydrolysates (peptides) from food proteins: A review. Food Chem 245, 205–222 (2018).
Zhang, L. et al. The anti-photoaging effect of antioxidant collagen peptides from silver carp (Hypophthalmichthys molitrix) skin is preferable to tea polyphenols and casein peptides. Food Funct 8(4), 1698–1707 (2017).
Lanng, S. K. et al. Influence of protein source (cricket, pea, whey) on amino acid bioavailability and activation of the mTORC1 signaling pathway after resistance exercise in healthy young males. Eur J Nutr 62(3), 1295–1308 (2023).
Summart, R. et al. Characterization and molecular docking of tetrapeptides with cellular antioxidant and ACE inhibitory properties from cricket (Acheta domesticus) protein hydrolysate. Heliyon 10(1), e35156 (2024).
Tejano, L. A., Peralta, J. P., Yap, E. E. S. & Chang, Y. W. Bioactivities of enzymatic protein hydrolysates derived from Chlorella sorokiniana. Food Sci Nutr 7, 2381–2390 (2019).
Rattanachitthawat, S. et al. Phenolic content and antioxidant activities in red unpolished Thai rice prevents oxidative stress in rats. Journal of Medicinal Plants Research 4, 796–801 (2010).
Liaqat, H. et al. Antioxidant Effect of Wheat Germ Extracts and Their Antilipidemic Effect in Palmitic Acid-Induced Steatosis in HepG2 and 3T3-L1 Cells. Foods 10(5), 1061 (2021).
Lorthongpanich, C. et al. Effect of the polyphenol flavonoids fisetin and quercetin on the adipogenic differentiation of human mesenchymal stromal cells. Biosci Rep 44(10), BSR20240623 (2024).
Chen, Q. et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts?. Cell Death Differ 23(7), 1128–1139 (2016).
Ge, C. et al. Reciprocal Control of Osteogenic and Adipogenic Differentiation by ERK/MAP Kinase Phosphorylation of Runx2 and PPARgamma Transcription Factors. J Cell Physiol 231(3), 587–596 (2016).
Lorthongpanich, C. et al. Fisetin Inhibits Osteogenic Differentiation of Mesenchymal Stem Cells via the Inhibition of YAP. Antioxidants (Basel) 10(6), 879 (2021).
Lorthongpanich, C. et al. YAP as a key regulator of adipo-osteogenic differentiation in human MSCs. Stem Cell Res Ther 10(1), 402 (2019).
Al-Azab, M. et al. Enhancers of mesenchymal stem cell stemness and therapeutic potency. Biomed Pharmacother 162, 114356 (2023).
Silva, J. P. & Coutinho, O. P. Free radicals in the regulation of damage and cell death - basic mechanisms and prevention. Drug Discov Ther 4(3), 144–167 (2010).
Shaban, S. et al. Effects of Antioxidant Supplements on the Survival and Differentiation of Stem Cells. Oxid Med Cell Longev 2017, 5032102 (2017).
Dannenmann, B. et al. Genome surveillance in pluripotent stem cells: Low apoptosis threshold and efficient antioxidant defense. Mol Cell Oncol 3(2), e1052183 (2016).
Choi, K. M. et al. Effect of ascorbic acid on bone marrow-derived mesenchymal stem cell proliferation and differentiation. J Biosci Bioeng 105(6), 586–594 (2008).
Wu, R. et al. Overview of Antioxidant Peptides Derived from Marine Resources: The Sources, Characteristic, Purification, and Evaluation Methods. Appl Biochem Biotechnol 176(7), 1815–1833 (2015).
Khavinson, V. K. et al. Peptide Regulation of Gene Expression: A Systematic Review. Molecules 26(22), 7053 (2021).
Renner, J. N. & Liu, J. C. Investigating the effect of peptide agonists on the chondrogenic differentiation of human mesenchymal stem cells using design of experiments. Biotechnol Prog 29(6), 1550–1557 (2013).
Otsuki, Y. et al. W9 peptide enhanced osteogenic differentiation of human adipose-derived stem cells. Biochem Biophys Res Commun 495(1), 904–910 (2018).
Lin’kova, N. S., Trofimov, A. V. & Dudkov, A. V. Peptides from the pituitary gland and cortex stimulate differentiation of polypotent embryonic tissue. Bull Exp Biol Med 151(4), 530–531 (2011).
van Marken Lichtenbelt, W. D. et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med 360(15), 1500–1508 (2009).
Fedorenko, A., Lishko, P. V. & Kirichok, Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 151(2), 400–413 (2012).
Vatner, D. E., Zhang, J. & Vatner, S. F. Brown adipose tissue enhances exercise performance and healthful longevity. Aging (Albany NY) 16(22), 13442–13451 (2024).
Acknowledgments
The authors thank the staff members of the Siriraj Center of Excellence for Stem Cell Research (SiSCR), Faculty of Medicine Siriraj Hospital, Mahidol University, Miss Ruechaphorn Buasamrit for administrative assistant.
Funding
This research was supported by Mahidol University (Basic Research Fund: fiscal year 2023) [FF-118/2566] to NU and Mahidol University (Strategic Research Fund): Fiscal year 2024 (R016720003) to CLo.
Author information
Authors and Affiliations
Contributions
N.U. Conceptualization, Formal analysis, Funding acquisition, Writing original draft; P.S. and S.M. Investigation and analysis; M.K.C supervise the project and editing manuscript; R.S. investigation and analysis; C.La. Methodology and Analysis, S.L. Supervise the project ; C.Lo. Conceptualization, Formal analysis, Investigation, Writing original draft and funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declarations
The hMSCs derivation in this study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand. (COA no. Si 112/2020 and the date of approval is February 7, 2020). Informed consent statement: Informed consent was obtained from all subjects involved in the study. The protocol of hMSC differentiation was approved by Mahidol University Central Institutional Review Board (MU-CIRB) with COA No. MU-CIRB 2023/087.0906.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Udomsil, N., Septham, P., Imsoonthornruksa, S. et al. Cricket protein peptides regulate adipogenic and osteogenic differentiation of human mesenchymal stem cells. Sci Rep 15, 35924 (2025). https://doi.org/10.1038/s41598-025-19713-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-19713-0