Abstract
Nepal is committed to eliminating visceral leishmaniasis as a public health problem by 2030. The scattered distribution of VL cases across wide geo-ecological regions, including areas previously considered unsuitable for the survival of vectors and the transmission of the pathogen, poses a major threat to Nepal’s national VL elimination programme. Regular monitoring and accurate identification of sand fly species are essential for implementing tailored vector control interventions. Hence, this study aimed to update the distribution of sand fly species with a focus on Leishmania vectors and evaluate DNA barcoding as a complementary tool for their identification. Sand flies were collected from 43 districts with active VL cases across the country between 2017 and 2022. The mitochondrial COI gene was amplified for DNA barcoding analysis. The primary vector, Phlebotomus argentipes, was present in all except three districts. Potential vectors, Ph. (Adlerius) spp. and Ph. major, were found common in high-altitude regions. The species identification success rate of generated COI barcode sequences based on the “Best Close Match” was 97%, indicating high accuracy in delineating sand flies to the species level. The information on the distribution of phlebotomine sand flies and the potential use of DNA barcoding for their identification could be milestones for sand fly research and help to guide the vector control interventions in support of VL elimination in Nepal.
Similar content being viewed by others
Introduction
In the Indian subcontinent, sand flies belong to two genera, Phlebotomus and Sergentomyia, with more than 70 species identified1,2. Of these, only one species, Phlebotomus (Euphlebotomus) argentipes (Annandale and Brunetti, 1908) sensu lato, is incriminated as a main vector of the Leishmania donovani complex (Kinetoplastida: Trypanosomatidae) and is responsible for the spread of anthroponotic visceral leishmaniasis (VL), also known as “kala-azar” in Nepal, India and Bangladesh3,4,5,6. This same vector-parasite duo is implicated in transmitting cutaneous leishmaniasis (CL) in the Western Ghats of India7 and Sri Lanka8. Other phlebotomine species, Ph. (Larrousius) major (Annandale, 1910) s.l. and Ph. (Adlerius) longiductus (Parrot, 1928) were recently identified as suspected vectors of L. donovani complex, causing CL in northwestern India bordering the far western region of Nepal9. It is noteworthy that CL has been emerging from the same region of Nepal in recent years10,11 and an in-depth investigation into epidemiology, serology, and entomology to assess the local transmission is currently underway. Other vector species transmitting the Leishmania parasite in the Indian sub-continent are Ph. (Phlebotomus) papatasi (Scopoli, 1786), Ph. (Phlebotomus) salehi (Mesghali, 1965) and Ph. (Paraphlebotomus) sergenti (Parrot, 1917). The former two species transmit L. major, the causative agent of zoonotic cutaneous leishmaniasis (ZCL) and the latter transmits L. tropica causing ZCL in arid parts of Northwest India12,13,14. Given the diversity in vector and parasites in the region, we aimed to collect up-to-date information on the distribution of phlebotomine sand flies, with a focus on Leishmania vectors, across a wide range of climatic and ecological settings in Nepal, regardless of their endemicity status.
In the context of the Indian subcontinent, VL has been slated for elimination as a public health problem by lowering the disease incidence to less than one case in 10,000 at the district level in Nepal, and at the sub-district level in India and Bangladesh by 2026 (most likely to be extended to 2030)15,16. Until 2019, VL was endemic in 18 out of 77 districts in Nepal, where local transmission occurred with reports of autochthonous cases; with records of asymptomatic infection in humans, and with the presence of a competent vector population17. In the same year, 53 non-endemic districts were labeled as endemicity doubtful districts due to the presence of VL cases but without evidence of local transmission18,19,20. The trend in the geographical expansion of VL cases has been observed from eastern to western parts and from lower (< 600 m asl) to higher altitudes (> 2000 m asl), particularly in areas that otherwise were considered ecologically unfavorable for the survival of the known vector species, Ph. argentipes21. Currently, VL cases have not been reported from only five of the 77 districts22,23. Further, the co-existence of VL along with CL at higher altitudes (> 1000 m asl) in hilly and mountainous areas poses an additional threat to the national VL elimination program, as the vector and parasite species present in the local human population remain unexplored11,24. Hence, integrated surveillance (disease, parasite and vectors) to monitor the circulating vectors and parasites in broader areas encompassing various ecological regions is deemed essential for planning and implementing tailored interventions with disease and vector control measures.
Focusing on vector surveillance, which integrates the process of collection, identification and reporting of sand fly species of public health importance, is key to the prospective entomological research aiming at controlling leishmaniasis25. Conventional approaches for the species identification of the phlebotomine sand flies are labor-intensive and time-consuming. They are based on minute morphological and anatomical characteristics that require skilled taxonomists26. Additionally, species complexes and phenotypic plasticity complicate morphological identifications. To overcome these difficulties, integrative taxonomic approaches, including morphology as well as the use of molecular tools such as DNA barcoding, are promising for species identification27,28. To this end, mitochondrial DNA genes display interesting features, especially the cytochrome c oxidase subunit I (COI) gene, which is extensively used in delineating sand fly species complexes worldwide29 as well as in Southeast Asia30,31,32,33,34,35,36.
In this study, we updated the geo-ecological distribution of phlebotomine sand fly species, including the known vector of the L. donovani, potential vectors and other non-vectors in Nepal, aimed to provide significant baseline information to the national VL elimination program. We also assessed the use of the DNA barcoding method as a complementary tool for sand fly species identification and evaluated the genetic variation within and among the Phlebotomus species collected across Nepal.
Results
Distribution and diversity of sand flies in surveyed districts
Based on ecological region classification, the 43 districts included in this study comprised 14 in lowlands, 22 in hills and 7 in mountainous regions (Fig. 1). Altitudes of the collection sites ranged from 70 to 308 m in lowlands, 364–1680 m in hills and 1182–2960 m in mountainous districts.
Map of Nepal with sampling locations of sand flies and their ecological regions. VL endemicity status at the district level based on 2016–2019 data17. The map was produced with QGIS (version 3.36.3) with open access shapefile downloaded from https://opendatanepal.com/dataset/new-political-and-administrative-boundaries-shapefile-of-nepal#.
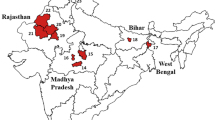
A total of 8,132 sand flies were collected from all the surveyed districts. The known vector Ph. argentipes was recorded from all except three districts (Fig. 2a). These three districts were located in the mountainous region. Phlebotomus argentipes represented 45.18% of the total collection, followed by Ph. major s.l. (10.85%) and Ph. (Adlerius) spp. (9.49%). Other species were Ph. papatasi and Sergentomyia spp. from lowlands and hills (Fig. 2b, Supplementary Excel file S1). Ecological regions imposed a significant effect on sand fly abundance. Abundance of all sand fly species and Ph. argentipes per district were lower in hills (IRR = 0.49, CI at 95% = 0.24–0.94 and IRR = 0.22, CI at 95% = 0.10–0.45, respectively) and mountains (IRR = 0.47, CI at 95% = 0.20–1.26 and IRR = 0.06, CI at 95% = 0.02–0.18, respectively) as compared to lowlands. Results also indicate the higher abundance of Ph. (Adlerius) spp. and Ph. major s.l. in the mountains (IRR = 4.76, CI at 95% = 1.45–20.94 and IRR = 1.88, CI at 95% = 0.80–5.09, respectively) as compared to hills. There were negligible collections of these two species from the lowlands (Fig. 2b).
Diversity and distribution of sand flies in Nepal; (a) in 43 surveyed districts and (b) in three ecological regions of the country based on the cross-sectional entomological survey conducted from 2017 to 2022. The map was produced with QGIS (version 3.36.3) with open access shapefile downloaded from https://opendatanepal.com/dataset/new-political-and-administrative-boundaries-shapefile-of-nepal#.
DNA-based species identifications
PCR and Sanger sequencing were successful for all 316 sand flies except one. We succeeded in the identification of two genera (Phlebotomus and Sergentomyia), seven subgenera (Euphlebotomus, Phlebotomus, Larroussius, Adlerius, Parrotomyia, Neophlebotomus, Sergentomyia) and six species (Ph. argentipes, Ph. papatasi, Se. babu, Se. iyengari, Se. punjabensis, Se. bailyi) based on morphology and the query results of the generated COI sequences against the available sequences in the BOLD and GenBank reference databases. However, 11 specimens of Ph. major s.l., a morphologically confirmed species, and six specimens of subgenus Adlerius were poorly matched (91.24% − 92.49%) with the available reference sequences in the BOLD/GenBank online databases. Species-level identification of one Phlebotomus and four Sergentomyia specimens remained inconclusive both by morphology and sequence analysis. Overall, 96.5% (304/315) of the sand fly specimens were successfully identified based on morphological characteristics, while 93% (293/315) had more than 96% pairwise identity match with the reference sequences available in the open-access databases (Table 1).
Among the generated sequences, the majority (84.4%; 266/315) were extracted from female sand flies, of which 207 were from the Ph. argentipes, the primary vector species of L. donovani (Table 1).
Genetic diversity estimates
In 315 successfully generated sequences of phlebotomine sand flies (both Phlebotomus and Sergentomyia), 101 haplotypes were described, with a haplotype diversity of 0.933 ± 0.008, a nucleotide diversity (Pi) of 0.078 ± 0.006, an average number of nucleotide differences of 49.99, and parsimony informative sites of 228 (with two variants – 137, three variants – 74 and four variants – 17). Haplotype and nucleotide diversities of Ph. argentipes, Ph. papatasi, Ph. (Adlerius) sp., Ph. major s.l., Se. babu, Se. iyengari, Se. punjabensis, Se. bailyi and Se. (Un3) sp. were 0.88 and 0.004, 0.98 and 0.007, 0.73 and 0.008, 0.93 and 0.007, 0.82 and 0.010, 1.00 and 0.024, 1.00 and 0.014, 1.00 and 0.015, and 1.00 and 0.003, respectively. The average number of nucleotide differences (k) for genus Phlebotomus was highest in Ph. (Adlerius) sp. (k = 5.13) followed by Ph. papatasi (k = 4.78), P. major s.l (k = 4.40) and Ph. argentipes (k = 2.78). The “k” value in the genus Sergentomyia was highest in Se. iyengari (k = 15.60) followed by Se. bailyi (k = 9.33), Se. punjabensis (k = 9.07), Se. babu (k = 6.44) and Se. (Un3) sp. (k = 2). The genetic diversity in individual phlebotomine species at the collection site (district) is shown in Supplementary Table S2.
Species identification efficiency and barcoding gap analysis
The species identification success rate of generated COI barcode sequences (n = 315) based on the “Best Close Match” was 97% (305/315). The overall mean genetic distance within our database of generated sequences was 8.89% ± 0.66% and the maximum pairwise K2P distance was 24.25% ± 2.18% (Supplementary Excel file S2). The mean intraspecific K2P distance within our database of generated sequences ranged from 0.31% ± 0.22% in Se. (Un3) sp. to 2.51% ± 0.42% in Se. iyengari (Table 2). All the species in this study showed a relatively large genetic variation (deep intraspecific divergence; mean K2P distance > 0.25), while interspecific divergence ranged from 12.23% ± 1.39% (Se. babu and Se. punjabensis) to 23.45% ± 2.06% (Ph. argentipes and Se. (Un1) sp.) (Supplementary Excel file S3).
Excluding sequences of Sergentomyia but including Old World vector phlebotomine sand flies sequences mined from online repositories to the database (n = 1,400), the overall average divergence was 14.90% ± 1.05% and the maximum pairwise K2P distance was 24.95% ± 2.36% (Supplementary Excel file S4). Mean intraspecific genetic divergence was the lowest in Ph. transcaucasicus (0.08% ± 0.07%) and the highest in Ph. major (5.90% ± 0.67%) (Table 2). The lowest value of pairwise mean genetic divergence was assessed to identify the nearest neighbor among the species: Ph. perfiliewi and Ph. transcaucasicus were the closest species with a mean interspecific K2P distance of 2.26% ± 0.36%; the nearest species to Ph. argentipes was the unidentified Phlebotomus sp. with a mean interspecific K2P distance of 16.51% ± 1.71% (Table 2, Supplementary Excel file S5). The pairwise inter- and intraspecific divergence is presented as a boxplot (Fig. 3).
Boxplot displaying inter- and intraspecific genetic distances of 1,400 COI sequences (BOLD, GenBank and generated), including 27 vector species of Old World phlebotomine sand flies (Phlebotomus). The calculation was based on the K2P nucleotide substitution model, using the package Spider and the software R. Thick horizontal lines inside the boxes represent the median and vertical dashed lines show the range. The boxes themselves represent the upper and lower quartiles. Outliers are displayed as open circles.
Neighbor-Joining tree
Neighbor-Joining tree shows the distinct branching in the two genera and 12 species of sand flies collected in Nepal (Fig. 4). The clustered nodes of the sequences from individuals of the same species were supported by high bootstrap values (99% – 100%). Based on the NJ tree, the unspecified Phlebotomus species clustered closely with Ph. argentipes, supported by an 89% bootstrap value.
Additionally, most haplotypes from particular species or species complexes clustered together on the NJ tree constructed based on the database of all Phlebotomus vector species reported from Old World countries. Exceptions are sequences of Ph. major s.l. from Nepal, Jordan and Turkey, forming three supported clusters. Sequences of Ph. bergeroti and Ph. papatasi are divided into two highly supported clusters (Fig. 5, Supplementary PDF file S1).
Haplotype and nucleotide diversities, and distribution of Ph. argentipes
Based on the 231 generated sequences of Ph. argentipes from six districts in Nepal, we found 36 haplotypes, with a haplotype diversity of 0.876 ± 0.013, a nucleotide diversity of 0.0043 ± 0.0002, and an average number of nucleotide differences of 2.78 (Supplementary Table S2). The dataset included 34 variable sites (11 singletons and 23 parsimony informative sites) (Supplementary Fig. S1). Among these haplotypes, H_2 was the most frequent (n = 62), followed by H_6 (n = 38) and H_4 (n = 30) (Supplementary Table S3). Similarly, the district-wise distribution of these haplotypes is shown in Supplementary Table S4.
Median-joining analysis
Of 422 sequences of Ph. argentipes (231 generated and 191 mined from BOLD and GenBank), 82 haplotypes were identified and included in the median-joining analysis. Haplotype H_2 was the most frequent one (n = 179), occurring in the Morang, Sunsari and Saptari districts of Nepal, Bihar, West Bengal, Kerala and Pondicherry regions in India and Delft island in Sri Lanka. Haplotype H_6 was the second most frequent (n = 38) and occurred in three districts of Nepal. Haplotype H_4 (n = 32) was present in five districts in Nepal, and Bihar and Kerala states in India. Less frequent haplotypes in terminal nodes represented more recently derived ones (Fig. 6). Locations in Nepal, India and Sri Lanka where COI sequences of Ph. argentipes were generated, or available from previous work (BOLD, GenBank) are shown in Supplementary Fig. S2.
Median-joining network of Ph. argentipes (n = 422) from Nepal, India, Sri Lanka and Israel (i.e., for which sequences were available from online repositories) showing genetic relationships among COI haplotypes. The sizes of circles are proportional to haplotype frequency and vertical lines are proportional to the number of nucleotide substitutions separating the connected haplotypes. Circles were colored according to the geographical origin of the barcoded specimens (i.e., district and country, as displayed in the figure legend).
Discussion
The current study documents the distribution of the primary competent vector of L. donovani, Ph. argentipes, in most of the surveyed districts with reported VL cases in Nepal. Other potential vectors, Ph. (Adlerius) spp. and Ph. major s.l., were abundant in the Himalayan foothills (hills and mountainous regions). The DNA barcoding method successfully allocated sand flies to seven morphologically validated species, while five taxa were identified up to the genus level. Our results provide strong evidence supporting the DNA barcoding as a complementary method for identification of major vector species of L. donovani, Ph. argentipes. This is critically important for vector surveillance and control efforts aligned with sustaining the VL elimination in Nepal.
COI sequences of Ph. argentipes, Ph. papatasi, Se. babu, Se. iyengari, Se. punjabensis and Se. bailyi had high pairwise identity (97–100%) with available COI sequences of respective species in DNA online repositories. Phlebotomus major s.l. collected from Nepal had poor pairwise identity match with available sequences, but were morphologically confirmed. Such variation might result from genetic differentiation linked, for example, to isolation by distance, leading to the formation of distinct geographical clusters37,38. The other five taxa have poor consensus with the available sequences, showing the limitation of the online databases, which are resourced with the reference sequences of only 20% of the described sand fly species worldwide39. Also, the morphological identification of these remained unsuccessful, possibly due to insufficiently mounted specimens or poorly established keys, or complicated anatomical characteristics that were difficult to interpret.
In the Neighbor-Joining tree, haplotypes of Ph. argentipes as well as other phlebotomine species were grouped with high bootstrap values32,36,40. Congeneric as well as conspecific clustering in the NJ tree supported the morphological identifications. Likewise, conspecific sequences clustered at the species level with high bootstrap values on the NJ tree, including all phlebotomine vector species from Old World countries. Some supported intra-specific branching was also found to be geographically structured by region or country of collection. For example, Ph. kandelakii is divided into two supported clusters (98%), related to Turkey, and India and Azerbaijan, respectively. Three clusters were also observed in the widely distributed Ph. major s.l. species sampled in Nepal, Jordan and Turkey (root bootstrap value 79%)41,42,43,44. In addition, Ph. perfiliewi and Ph. transcaucasicus clustered together, supporting their species complex status45,46. A similar situation was observed between Ph. martini and Ph. celiae47. Additional investigations might be required to determine if some sequences mined from online repositories were not initially misidentified (Fig. 5, Supplementary PDF file S1).
Nucleotide diversity of Ph. argentipes population was very low in Nepal (Pi = 0.004), in contrast to the high genetic variation reported from Sri Lanka (Pi = 0.427), however, the haplotype diversity was found to be equivalent in both countries (Hd = 0.88)32. The median-joining network supported a demographic expansion of Ph. argentipes populations, with a star-like topology of haplotypes identified in Nepal and other countries. The most frequent haplotype, H_2, was reported from southern India, Sri Lanka and eastern Nepal. Some unique haplotypes are reported from hilly districts situated in the central and western part of Nepal (Palpa and Surkhet). Sri Lanka Island did not share the maximum of its prevalent haplotypes (except three collected from Delft Island; H_40, H_49 and H_50) with India and Nepal, representing a genetically distinct/isolated Ph. argentipes population48. Haplotype diversities of other important phlebotomine sand flies recorded from Nepal, i.e., Ph. (Adlerius) sp., Ph. major s.l. and Ph. papatasi were also very high (0.73 to 0.98), reflecting the representative populations from varying ecological regions49.
Based on the intra and interspecific K2P distance and NJ tree, species of interest were successfully delimited to their predicted group, leaving a few species unidentified. However, in molecular identification techniques, use of a single mtDNA marker is not ideal; therefore, analyzing multiple markers is recommended for the speciation of closely related species39.
In Nepal, phlebotomine fauna is least explored in terms of their diversity and distribution owing to the complexity of morphological identification. About 14 species of phlebotomine sand flies (eight species of Phlebotomus and six species of Sergentomyia) were reported from Nepal till 200014,50 and no further investigation on the sand fly diversity and distribution has ever been conducted since then. Over the last three decades, most investigations on sand flies were focused on the Ph. argentipes sand fly due to its significance in L. donovani transmission. In recent years, visceral as well as cutaneous forms of leishmaniasis have spread to wide geo-ecological regions (hills and mountains), even in areas that were once considered unsuitable for the transmission of the disease. Using morphology and DNA-based techniques (though on a small scale), we confirmed the occurrence of Ph. argentipes in most of the surveyed districts. There are significant implications of correctly identifying vector species for tailored vector control interventions. This study validated the use of DNA barcoding for the successful identification of vector sand flies, especially the females. The large number of sequences obtained from female Ph. argentipes was a part of the validation process of morphologically identified specimens subjected for Leishmania infection and blood meal analysis, as described elsewhere51. Thus, the method proves to be useful for epidemiological and vector surveillance activities, especially in situations where taxonomic expertise is unavailable.
The presence of potential vectors from Adlerius group (though species-level identification, both morphologically and molecularly, is still pending) and Ph. major s.l. from areas above 1,000 m asl with rocky terrain and warm humid conditions confirms the records of previous findings from Nepal and its neighboring countries14,21,44,52. These competent vectors in high altitude areas likely play a role in the Leishmania transmission, as it has been recorded from similar geo-ecological regions in the bordering state (Uttarakhand) in India9,53.
For other poorly identified specimens, most of them were single, and hence, it is suggested to collect more sand flies from the same areas where these were collected during previous survey and perform integrated taxonomic approach for the species level identification. The current study demonstrated the phlebotomine sand fly diversity and distribution based on the investigation primarily targeted for the collection of the known vector. A nationwide general survey of sand flies across diverse habitats and representative geo-ecological regions along defined altitudinal gradients is needed to obtain a more precise picture of phlebotomine sand fly diversity and distribution.
Importantly, the cost of the DNA barcoding method remains a limiting factor for its large-scale application. It is recommended to select 5–10% of the sand fly samples collected during vector surveillance activities for molecular identification to accurately determine the sand fly species.
Conclusion
The study demonstrated the presence of the incriminated vector in most of the areas with active VL cases, accompanied by other potential vectors at high altitudes. This finding advocates the necessity of systematic entomological surveillance to sustain disease elimination. The potential of the DNA barcoding method to identify major vector species was highly successful and can be utilized in epidemiological investigations and surveillance, for example, in situations where sand fly taxonomy experts are not available. The generated sequences during this study contribute to the enrichment of the public reference DNA barcode databases. The current investigation suggests advanced studies on sand fly biodiversity in Nepal, both at the morphological and molecular levels.
Methods
Sand fly collection and morphological identification
Sand flies were collected from 43 districts (15 endemic and 28 endemicity doubtful) with reported cases of VL, as part of epidemiological and entomological assessments coordinated through the National VL elimination program during 2017 and 2022. The entomological collections were part of a number of activities, including cross-sectional transmission assessment, longitudinal surveillance for seasonality studies and insecticide resistance monitoring in Ph. argentipes. The geo-ecological settings in the surveyed districts varied from lowlands – “Terai” – with a tropical savannah climate to high hills and mountains experiencing a temperate climate with dry winters and hot or warm summers54. Lowlands in Nepal (67–300 m asl) are primarily a fertile Gangetic plain area, rich in agricultural lands, dense vegetation and water bodies. Hilly districts encompass a wide range of geography (> 300 m – 2,500 m asl), including undulating terrain, deep valleys with scattered agricultural terraced lands and dense subtropical and coniferous forests. Mountainous districts are situated in the inner and high Himalayan region with altitudes ranging from roughly 2,500 m to more than 8,000 m asl and is characterized by a steep, rugged landscape with high peaks, deep valleys with scattered dwarf shrubs and alpine meadows55 (Fig. 1, Supplementary Table S1). In each surveyed district, we selected two or more villages, depending on the objectives of the individual studies. Sand flies were collected from households with or without cohabiting cattle, using the Center for Disease Control and Prevention (CDC) light traps installed inside the dwellings. In addition, we manually searched and aspirated resting sand flies from inside corners of rooms, cracks and crevices of walls of houses and cattle sheds, cattle tying poles and around the cattle feeding troughs. All sand flies were preserved in 80% ethanol and transferred to the entomology laboratory at B.P. Koirala Institute of Health Sciences (BPKIHS - Dharan, Nepal) for further laboratory processing. These sand flies were identified based on the morphological and anatomical characteristics of male and female genitalia, pharyngeal teeth, cibarium and antennal segments with the help of the regional sand fly species identification keys14,56 a stereoscope and a light microscope.
Descriptive analysis of sand fly distribution and diversity
Sand flies collected during the cross-sectional survey in each of the 43 districts were analyzed for distribution and diversity. Diversity of the sand fly species was proportionately represented in pie charts engraved in the map of Nepal. We fitted generalized linear models (GLM) with a negative binomial distribution in the entomological data to assess the association of the sand fly abundance in function of the explanatory variables, like ecological regions. The model was fitted because the sand fly abundance data were over-dispersed and showed a non-normal distribution, i.e., variance was larger than mean value. The calculation was done using the function ‘glm.nb’ from a R package “MASS”57. Results of the analysis are interpreted as an incidence rate ratio (IRR) and confidence interval (CI) at 95%.
DNA extraction and PCR amplification
DNA extraction and PCR amplification were performed in 2019. Sand flies were collected from only 12 districts from across the country during 2017–2019. Among these, we selected sand flies from eight districts (five endemic and three endemicity doubtful) that represented a wide ecological regions of the country (East, West, and also variation in topography) (Fig. 1) for molecular analysis. Genomic DNA was extracted from 316 sand fly specimens using the DNeasy® Blood and Tissue Kit (QIAGEN, Hilden, Germany), following the manufacturer’s protocol. A fragment of the mitochondrial cytochrome c oxidase subunit I (COI) gene (658 bp) was amplified with the universal primer pair: LCO1490–5′-GGTCAACAAATCATAAAGATATTGG-3′ and HCO2198–5′-TAAACTTCAGGGTGACCAAAAAATCA-3′58. Each amplification was performed in a volume of 25 µl containing 12.5 µl GoTaq Green master mix with 2 mM MgCl2 (Promega, USA), 0.4 µM of each of the primers (Biolegio, The Netherlands), 6.7 µl of PCR grade water (Himedia, India) and 5 µl of DNA template. The PCR profile was as follows: denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 92 °C for 30 s, annealing at 50 °C for 45 s, extension at 72 °C for 60 s, and a final extension at 72 °C for 10 min. The PCR products, positive and negative controls were loaded on a 2% agarose gel, stained with ethidium bromide, and examined under a gel documentation system (BIORAD and SYNGENE). Positive amplicons were outsourced to BaseClear (The Netherlands) and Macrogen (South Korea) for purification and sequencing.
Sequence editing and data analysis
All generated sequences were checked and edited to resolve ambiguities in BioEdit Sequence Alignment Editor 7.0.5.359. Each of these sequences was checked for stop codons, and primer residues were trimmed off. Each of these edited sequences was saved as fasta data files and queried using the BOLD identification engine (www.boldsystems.org; Species Level Barcode Records option) and BLAST for GenBank (https://blast.ncbi.nlm.nih.gov; program option optimized for megablast). Multiple sequence alignment (ClustalW) and computation of nucleotide composition were performed in MEGA v.760. The number of haplotypes, polymorphic sites and nucleotide diversities were analyzed in DnaSP v.6 software61. The “Best Close Match” approach of identifying species based on DNA barcoding distances was used to estimate relative frequency of identification success and was calculated in R using the function “bestCloseMatch” available in the package “spider”62. This approach considers sequences with the smallest genetic distance to query all conspecific and within 95% of all intraspecific distances63. Mean genetic distance estimates, pairwise sequence divergence and tree construction60,62, including all generated sequences from the present investigation (n = 315), were performed based on the nucleotide substitutions Kimura-2-parameter (K2P) model with 1,000 bootstrap replications. A neighbor-joining tree was constructed from the identified haplotypes (n = 101), including both genera, Phlebotomus and Sergentomyia.
For further analyses, available COI sequences of Phlebotomus species – known vectors of Leishmania from Old World countries12,64 were mined from the public databases of BOLD and GenBank. Sequences less than 600 bp in length, species with singleton sequences and ambiguous sequences were discarded from the list. Generated Phlebotomus sequences (n = 276) and sequences from BOLD and GenBank that passed exclusion criteria (n = 1,124) were subsequently aligned (ClustalW) and trimmed to retain the overlapping standard barcode region. The mean genetic distance (K2P) between non-conspecific and conspecific sequences was computed in MEGA and R with the package “spider” using the function “sppDistMatrix”62. The barcoding gap assessment was based on the mean intraspecific and the minimum interspecific distances (the nearest neighbor)65. A haplotype Neighbor-Joining tree was further constructed, including phlebotomine vector species (BOLD/GenBank: 545 haplotypes, generated dataset: 70 haplotypes) and one outgroup taxon (Aedes aegypti; LC489421) (K2P; 1,000 bootstrap replications)66,67.
Finally, a median-joining network was built for Ph. argentipes, including the sequences generated in this study (n = 231) and available sequences from all other countries (n = 191), using the median-joining algorithm in NETWORK v.10.2 (fluxus-engineering.com)68.
Data availability
All datasets generated and analyzed during the study are included in the manuscript as tables, figures and supplementary files. The generated COI sequences of phlebotomine sand flies from Nepal are submitted in GenBank under accession numbers PV588139 – PV588236 and PV940754 – PV940756.
References
Shah, H. K., Fathima, P. A., Kumar, N. P., Kumar, A. & Saini, P. Faunal richness and checklist of sandflies (Diptera: Psychodidae) in India. Asian Pac. J. Trop. Med. 16, 193–203. https://doi.org/10.4103/1995-7645.377740 (2023).
Wijerathna, T. & Gunathilaka, N. Morphological identification keys for adults of sand flies (Diptera: Psychodidae) in Sri Lanka. Parasit. Vectors. 13, 450. https://doi.org/10.1186/s13071-020-04305-w (2020).
Tiwary, P. et al. Seasonal variation in the prevalence of sand flies infected with leishmania donovani. PLoS One. 8, e61370. https://doi.org/10.1371/journal.pone.0061370 (2013).
Tiwary, P., Kumar, D., Singh, R. P., Rai, M. & Sundar, S. Prevalence of sand flies and leishmania donovani infection in a natural population of female phlebotomus argentipes in Bihar state, India. Vector Borne Zoonotic Dis. 12, 467–472. https://doi.org/10.1089/vbz.2011.0808 (2012).
Bhattarai, N. R. et al. Natural infection of phlebotomus argentipes with leishmania and other trypanosomatids in a visceral leishmaniasis endemic region of Nepal. Trans. R Soc. Trop. Med. Hyg. 103, 1087–1092. https://doi.org/10.1016/j.trstmh.2009.03.008 (2009).
World Health Organization. Leishmaniasis, (2023). https://www.who.int/news-room/fact-sheets/detail/leishmaniasis
Srinivasan, R., Kumar, N. P. & Jambulingam, P. Detection of natural infection of leishmania donovani (Kinetoplastida: Trypanosomatidae) in phlebotomus argentipes (Diptera: Psychodidae) from a forest ecosystem in the Western ghats, india, endemic for cutaneous leishmaniasis. Acta Trop. 156, 95–99. https://doi.org/10.1016/j.actatropica.2016.01.010 (2016).
Wijerathna, T., Gunathilaka, N., Gunawardena, K., Fujii, Y. & Gunasekara, D. Detection of leishmania donovani DNA within Field-Caught phlebotomine sand flies (Diptera: Psychodidae) in three cutaneous leishmaniasis endemic foci of Kurunegala district, Sri Lanka. J. Trop. Med. 2021 (6650388). https://doi.org/10.1155/2021/6650388 (2021).
Lata, S., Kumar, G., Ojha, V. P. & Dhiman, R. C. Detection of leishmania donovani in Wild-Caught phlebotomine sand flies in endemic focus of leishmaniasis in Himachal pradesh, India. J. Med. Entomol. 59, 719–724. https://doi.org/10.1093/jme/tjab202 (2022).
Bastola, A. et al. A case of high altitude cutaneous leishmaniasis in a non-endemic region in Nepal. Parasitol. Int. 74, 101991. https://doi.org/10.1016/j.parint.2019.101991 (2020).
Pandey, K. et al. Emergence of cutaneous leishmaniasis in Nepal. Trop. Med. Health. 49 https://doi.org/10.1186/s41182-021-00359-3 (2021).
Ready, P. D. Biology of phlebotomine sand flies as vectors of disease agents. Annu. Rev. Entomol. 58, 227–250. https://doi.org/10.1146/annurev-ento-120811-153557 (2013).
Killick-Kendrick, R. The biology and control of phlebotomine sand flies. Clin. Dermatol. 17, 279–289. https://doi.org/10.1016/S0738-081X(99)00046-2 (1999).
Kalra, N. L. & Bang, Y. H. Manuals on Entomology in Visceral Leishmaniasis (World Health Organization, SEARO, 1988).
World Health Organization. Regional office for South-East, A. Regional Strategic Framework for Accelerating and Sustaining Elimination of kala-azar in the South-East Asia Region: 2022–2026. (New Delhi: World Health Organization. Regional office for South-East Asia), Licence: CC BY-NC-SA 3.0 IGO. (2022).
World Health Organization. Ending the neglect to attain the sustainable development goals: a rationale for continued investment in tackling neglected tropical diseases 2021–2030. Report No. 978 92 4 005293 2, 50. World Health Organization, (2020).
Epidemiology and Disease Control Division. National Guideline on Kala-azar Elimination Program (Updated). (Department of Health Services, Ministry of Health and Population, Government of Nepal, Teku, Kathmandu. 2019). (2019).
Ruiz-Postigo, J. A. et al. World Health Organization,. Global leishmaniasis surveillance, 2022: assessing trends over the past 10 years. Report No. 40, 471–488 (2023).
Department of Health Services. Kala-azar in Annual Report 2077/78. 153–160 (Ministry of Health and Population, Government of Nepal, 2021). (2020)/21.
Ruiz-Postigo, J. A. et al. Global leishmaniasis surveillance: 2019–2020, a baseline for the 2030 roadmap. Rep. No. 35, 401–419 (2021).
Ostyn, B. et al. Transmission of leishmania donovani in the hills of Eastern nepal, an outbreak investigation in Okhaldhunga and Bhojpur districts. PLoS Negl. Trop. Dis. 9, e0003966. https://doi.org/10.1371/journal.pntd.0003966 (2015).
Jain, S. et al. World Health Organization,. Global leishmaniasis surveillance updates 2023: 3 years of the NTD road map. Report No. 45, 653–676 (2024).
Department of Health Services. Annual Health Report 2079/80. Ministry of Health and Population (MoHP), Government of Nepal, Teku, Kathmandu, Nepal. Report No. 978-9-93716-176-3. (2024).
Shrestha, M. et al. Visceral leishmaniasis from a non-endemic Himalayan region of Nepal. Parasitol. Res. 117, 2323–2326. https://doi.org/10.1007/s00436-018-5887-6 (2018).
Bates, P. A. et al. Recent advances in phlebotomine sand fly research related to leishmaniasis control. Parasites Vectors. 8, 131. https://doi.org/10.1186/s13071-015-0712-x (2015).
Lewis, D. J. The phlebotominae sandflies (Diptera: Psychodidae) of the Oriental region. Bull. Br. Museum (Natural History) Entomol. 37, 217–343 (1978).
Wang, W., Srivathsan, A., Foo, M., Yamane, S. & Meier, R. Sorting specimen-rich invertebrate samples with cost-effective NGS barcodes: validating a reverse workflow for specimen processing. Mol. Ecol. Resour. https://doi.org/10.1111/1755-0998.12751 (2018).
DAYRAT, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 85, 407–417. https://doi.org/10.1111/j.1095-8312.2005.00503.x (2005).
Rodrigues, B. L. & Galati, E. A. B. Molecular taxonomy of phlebotomine sand flies (Diptera, Psychodidae) with emphasis on DNA barcoding: A review. Acta Trop. 238, 106778. https://doi.org/10.1016/j.actatropica.2022.106778 (2023).
Chen, H. et al. Mitochondrial COI and Cytb gene as valid molecular identification marker of sandfly species (Diptera: Psychodidae) in China. Acta Trop. 238, 106798. https://doi.org/10.1016/j.actatropica.2022.106798 (2023).
Preativatanyou, K. et al. Species diversity of phlebotomine sand flies and sympatric occurrence of leishmania (Mundinia) martiniquensis, leishmania (Leishmania) donovani complex, and trypanosoma spp. In the visceral leishmaniasis focus of Southern Thailand. Acta Trop. 244, 106949. https://doi.org/10.1016/j.actatropica.2023.106949 (2023).
Pathirage, D. R. K., Weeraratne, T. C., Senanayake, S. C., Karunaratne, S. H. P. P. & Karunaweera, N. D. Genetic diversity and population structure of phlebotomus argentipes: vector of leishmania donovani in Sri Lanka. PLoS One. 16, e0256819. https://doi.org/10.1371/journal.pone.0256819 (2021).
Tharmatha, T., Gajapathy, K., Ramasamy, R. & Surendran, S. N. Morphological and molecular identification of cryptic species in the sergentomyia Bailyi (Sinton, 1931) complex in Sri Lanka. Bull. Entomol. Res. 107, 58–65. https://doi.org/10.1017/S0007485316000626 (2017).
Gajapathy, K. et al. DNA barcoding of Sri Lankan phlebotomine sand flies using cytochrome c oxidase subunit I reveals the presence of cryptic species. Acta Trop. 161, 1–7. https://doi.org/10.1016/j.actatropica.2016.05.001 (2016).
Gajapathy, K. et al. Molecular identification of potential leishmaniasis vector species within the phlebotomus (Euphlebotomus) argentipes species complex in Sri Lanka. Parasit. Vectors. 6, 302. https://doi.org/10.1186/1756-3305-6-302 (2013).
Kumar, N. P., Srinivasan, R. & Jambulingam, P. DNA barcoding for identification of sand flies (Diptera: Psychodidae) in India. Mol. Ecol. Resour. 12, 414–420. https://doi.org/10.1111/j.1755-0998.2012.03117.x (2012).
Absavaran, A. et al. Identification of sand flies of the subgenus Larroussius based on molecular and morphological characters in North Western Iran. Iran. J. Arthropod Borne Dis. 3, 22–35 (2009).
Adler, S. The sandflies of Cyprus (Diptera). Bull. Entomol. Res. 36, 497–511. https://doi.org/10.1017/S0007485300024111 (1946).
Depaquit, J. Molecular systematics applied to phlebotomine sandflies: review and perspectives. Infect. Genet. Evol. 28, 744–756. https://doi.org/10.1016/j.meegid.2014.10.027 (2014).
Piyasiri, S. B. et al. Comparative Analysis of Phlebotomus argentipes Vector of Leishmaniasis in India and Sri Lanka. Microorganisms 12, (2024). https://doi.org/10.3390/microorganisms12071459
Mukbel, R., Kanaan, D., Abu-Halaweh, M. & Amr, Z. Morphological, distributional, and molecular study of the phlebotomus sand flies of Jordan. J. Vector Ecol. 49 https://doi.org/10.52707/1081-1710-49.1.64 (2024).
Erisoz Kasap, O., Linton, Y. M., Karakus, M., Ozbel, Y. & Alten, B. Revision of the species composition and distribution of Turkish sand flies using DNA barcodes. Parasit. Vectors. 12, 410. https://doi.org/10.1186/s13071-019-3669-3 (2019).
Kasap, O. E., Votýpka, J. & Alten, B. The distribution of the phlebotomus major complex (Diptera: Psychodidae) in Turkey. Acta Trop. 127, 204–211. https://doi.org/10.1016/j.actatropica.2013.05.001 (2013).
Kamhawi, S., Abdel-Hafez, S. K. & Molyneux, D. H. A comprehensive account of species composition, distribution and ecology of phlebotomine sandflies in Jordan. Parasite 2, 163–172. https://doi.org/10.1051/parasite/1995022163 (1995).
González, M. A., Ruiz-Arrondo, I., Gutiérrez-López, R., Barceló, C. & Miranda, M. Á. First record of phlebotomus (Larroussius) perfiliewi (Diptera: Psychodidae), Vector of Leishmania infantum and Phleboviruses, in Spain. Diversity 15, 400 (2023).
Rassi, Y. et al. Phlebotomus perfiliewi transcaucasicus, a vector of leishmania infantum in Northwestern Iran. J. Med. Entomol. 46, 1094–1098. https://doi.org/10.1603/033.046.0516 (2009).
Gebre-Michael, T. & Lane, R. Distinguishing the females of phlebotomus (Synphlebotomus) martini and P. (S.) Celiae (Diptera: Phlebotominae), vectors of visceral leishmaniasis in southern Ethiopia. Bull. Entomol. Res. 83, 353–360. https://doi.org/10.1017/S0007485300029242 (1993).
Pathirage, D. R. K., Karunaratne, S., Senanayake, S. C. & Karunaweera, N. D. Insecticide susceptibility of the sand fly leishmaniasis vector Phlebotomus argentipes in Sri Lanka. Parasit. Vectors. 13, 246. https://doi.org/10.1186/s13071-020-04117-y (2020).
Guernaoui, S., Hamarsheh, O., Garcia, D., Fontenille, D. & Sereno, D. Population genetics of phlebotomus Papatasi from endemic and nonendemic areas for zoonotic cutaneous leishmaniasis in morocco, as revealed by cytochrome oxidase gene subunit I sequencing. Microorganisms 8 https://doi.org/10.3390/microorganisms8071010 (2020).
Sharma, R. P. Studies on Sandfly Fauna in Bardiya and Their Ecological Relation Master thesis, Tribhuvan University, Kirtipur, Kathmandu, (2000).
Roy, L. et al. The ongoing risk of leishmania donovani transmission in Eastern nepal: an entomological investigation during the elimination era. Parasites Vectors. 16, 404. https://doi.org/10.1186/s13071-023-05986-9 (2023).
Lewis, D. J. The phlebotomine sand flies of West Pakistan (Diptera: Psychodidae). Bull. Br. Museum (Nat Hist.) Entomol. 19, 1–57 (1967).
Sharma, N. L. et al. The sandflies of the Satluj river valley, Himachal Pradesh (India): some possible vectors of the parasite causing human cutaneous and visceral leishmaniases in this endemic focus. J. Vector Borne Dis. 46, 136–140 (2009). http://www.mrcindia.org/journal/issues/462136.pdf
Karki, R., Talchabhadel, R., Aalto, J. & Baidya, S. New Climatic classification of Nepal. Theoret. Appl. Climatol. https://doi.org/10.1007/s00704-015-1549-0 (2015).
Carson, B., Shah, P. B. & Maharjan, P. L. Government of Nepal,. The Soil Landscapes of Nepal, Land Systems Report. 280 (1986).
Lewis, D. J. A taxonomic review of the genus Phlebotomus (Diptera: Psychodidae). Bull. Br. Museum Entomol. (Natural History). 45, 121–209 (1982).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. Fourth Edition (Springer, 2002).
HebertP.D., Cywinska, A., Ball, S. L. & deWaardJ.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. https://doi.org/10.1098/rspb.2002.2218 (2003).
Hall, T. A. & BioEdit A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Asids Symposium Series, 95–98 (1999).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetic analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. https://doi.org/10.1093/molbev/msx248 (2017).
BROWN, S. D. J. et al. Spider: an R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol. Ecol. Resour. 12, 562–565. https://doi.org/10.1111/j.1755-0998.2011.03108.x (2012).
Meier, R., Shiyang, K., Vaidya, G. & Ng, P. K. L. DNA barcoding and taxonomy in diptera: A Tale of high intraspecific variability and low identification success. Syst. Biol. 55, 715–728. https://doi.org/10.1080/10635150600969864 (2006).
Maroli, M., Feliciangeli, M. D., Bichaud, L., Charrel, R. N. & Gradoni, L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med. Vet. Entomol. 27, 123–147. https://doi.org/10.1111/j.1365-2915.2012.01034.x (2013).
Čandek, K. & Kuntner, M. DNA barcoding gap: reliable species identification over morphological and geographical scales. Mol. Ecol. Resour. 15, 268–277. https://doi.org/10.1111/1755-0998.12304 (2015).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. https://doi.org/10.1007/BF01731581 (1980).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Bandelt, H. J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036 (1999).
Acknowledgements
We would like to thank all the female community health volunteers from the respective study villages for guiding us through the village and facilitating communication. We would also like to thank the vector control officers from the respective district public health offices for helping us to select the study villages. In addition, we acknowledge the entomology technicians for their assistance in sand fly collections.
Funding
This research was supported by the doctoral fellowship (grant no. 716228/40/70, to LR) at the Institute of Tropical Medicine, Antwerp, Belgium. Entomological surveys were partly funded by Health Directorate of Koshi and Karnali Provinces, Vector Borne Disease Research and Training Centre, Hetauda, WHO Country Office and ASCEND programme in support to National VL elimination programme in Nepal.
Author information
Authors and Affiliations
Contributions
L.R.: Conceptualization, Methodology, Validation, Data curation, Formal analysis, Writing –Original draft, Writing-Review and Editing, Visualization, Funding acquisition; S.U.: Methodology, Writing –Original draft Funding acquisition; K.R.: Conceptualization, Methodology, Validation, Formal analysis; K.C.: Conceptualization, Methodology, Validation, Writing-Original draft; M.L.D.: Conceptualization, Methodology, Validation, Writing-Original draft; N.S.: Conceptualization, Methodology, Validation, Supervision, Writing-Original draft, W.V.B.: Conceptualization, Methodology, Validation, Formal analysis, Writing –Original draft, Writing-Review and Editing, Supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Ethics statement
Ethical clearance for the study was obtained from Nepal Health Research Council (ERB Protocol No. 228/2021 P) and the Institutional Review Committee of B.P. Koirala Institute of Health Sciences, Dharan, Nepal (Registration No. 475/073/074-IRC). Informed written consent was obtained from the household owners where sand flies were collected. We hereby confirm that the research does not involve any human or animal subjects. All methods of sand fly capture and molecular analysis were performed in accordance with the standard guidelines and regulations of the concerned Institute and the Government of Nepal.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Roy, L., Uranw, S., Rai, K. et al. Mapping the distribution of phlebotomine sand fly species with emphasis on Leishmania vectors in Nepal and exploring the potential of DNA barcoding for their identification. Sci Rep 15, 36356 (2025). https://doi.org/10.1038/s41598-025-20276-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-20276-3