Abstract
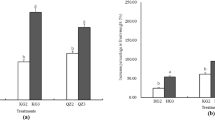
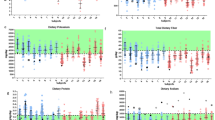
One of NASA’s goals is growing edible crops in spaceflight to supplement the astronauts’ diet with fresh, safe-to-eat vegetables. Growing plants in spaceflight presents challenges to achieve optimal plant growth and productivity. It is important to understand the optimal conditions for crops grown in spaceflight. The Advanced Plant Habitat (APH), an enclosed, environmentally controlled plant growth facility on the International Space Station (ISS) has been used to grow plants and monitors the environment. A technology demonstration mission on the ISS used the APH to grow chile peppers, Capsicum annuum, cv. NuMex Española Improved to maturity. Over 25 peppers were successfully harvested as a mix of red and green peppers at 109 and 137 days after initiation. Half of the fruit were consumed by the crew, and half were frozen at − 80 °C and returned with the science carrier (SC) to Kennedy Space Center, FL to determine microbial load and food safety. Bacterial and fungal counts, ranged from below detection limit to 111 CFU/gram fresh weight found on a single pepper only. Foodborne pathogens were not detected. Investigation of the communities using the V4 region of the 16S rRNA gene revealed taxonomic variations between the SC quadrants as well as between hardware components and plant tissues, indicating possible vertical or horizontal transfer of some bacteria. An investigation into the bacterial communities indicated 13 genera and 1 unidentified microbe as possible core microbiome components. In addition, bacteria such as Burkholderia and Sphingomonas previously identified from ISS water samples were present in the communities. This technology demonstration growing a long duration fruiting crop on the ISS provided verification of the pre-determined environmental conditions, contributed to the validation of the APHs’ capability to support fruiting crops in space, and provided the crew a safe-to-eat addition to their pre-packaged diet. This technology demonstration may serve as a baseline for future space crop production systems or long duration spaceflight missions.
Similar content being viewed by others
Data availability
The datasets generated and analyzed during this study have been deposited and are available in the NASA GeneLab repository [https://osdr.nasa.gov/bio/repo/data/studies/OSD-772], **DOI**: [https://doi.org/10.26030/mjvk-v435]. This data set can be identified as OSD-772. The direct link is https://doi.org/10.26030/mjvk-v435. An alternative method for data access is to contact the corresponding authors, Drs. Anirudha Dixit or Christina LM Khodadad@nasa.gov.
References
Zhou, W. et al. Performance of the Advanced Astroculture Plant Growth Unit During ISS-6A/7A Mission. In 2002-01–2280 https://doi.org/10.4271/2002-01-2280 (2002).
Link, B. M., Durst, S. J., Zhou, W. & Stankovic, B. Seed-to-seed growth of Arabidopsis thaliana on the International Space Station. Adv. Space Res. 31, 2237–2243 (2003).
Zhou, W. Advanced Astroculture Plant Growth Unit: Capabilities and Performances. In 2005-01-2840. https://doi.org/10.4271/2005-01-2840 (2005).
Evans, C. A. et al. International Space Station Science Research Accomplishments During the Assembly Years: An Analysis of Results from. International Space Station NASA/TP-2009-213146-Rev A (2009).
Stutte, G. W., Monje, O., Goins, G. D. & Tripathy, B. C. Microgravity effects on thylakoid, single leaf, and whole canopy photosynthesis of dwarf wheat. Planta 223, 46–56 (2005).
Morrow, R. C. & Crabb, T. M. Biomass production system (BPS) plant growth unit. Adv. Space Res. 26, 289–298 (2000).
Sychev, V. N., Levinskikh, M. A., Gostimsky, S. A., Bingham, G. E. & Podolsky, I. G. Spaceflight effects on consecutive generations of peas grown onboard the Russian segment of the International Space Station. Acta Astronaut. 60, 426–432 (2007).
Anderson, M. S. et al. Key gaps for enabling plant growth in future missions. In: AIAA SPACE and Astronautics Forum and Exposition (American Institute of Aeronautics and Astronautics, Orlando, FL). https://doi.org/10.2514/6.2017-5142 (2017).
Bingham, G. E., Topham, T. S., Mulholland, J. M. & Podolsky, I. G. Lada: The ISS Plant Substrate Microgravity Testbed. In 2002-01-2388.https://doi.org/10.4271/2002-01-2388 (2002).
Hummerick, M., Garland, J., Bingham, G., Sychev, V. & Podolsky, I. Microbiological analysis of lada vegetable production units (VPU) to define critical control points and procedures to ensure the safety of space grown vegetables. In: 40th International Conference on Environmental Systems. (American Institute of Aeronautics and Astronautics, Barcelona, Spain). https://doi.org/10.2514/6.2010-6255 (2010).
Hummerick, M. et al. A hazard analysis critical control point plan applied to the Lada vegetable production units (VPU) to ensure the safety of space grown vegetables. In: 41st International Conference on Environmental Systems (American Institute of Aeronautics and Astronautics, Portland, Oregon). https://doi.org/10.2514/6.2011-5277 (2011).
Massa, G. D. et al. VEG-01: Veggie hardware validation testing on the International Space Station. Open Agric. 2, 33–41 (2017).
Massa, G. D., Wheeler, R. M., Morrow, R. C. & Levine, H. G. Growth chambers on the International Space Station for large plants. Acta Hortic. https://doi.org/10.17660/ActaHortic.2016.1134.29 (2016).
Monje, O. et al. Hardware validation of the advanced plant habitat on ISS: Canopy photosynthesis in reduced gravity. Front. Plant Sci. 11, 673 (2020).
Morrow, R. C. et al. A New Plant Habitat Facility for the ISS. 46th International Conference on Environmental Systems, ICES-2016-320 (2016).
Matta, F. B. & Nakayama, R. M. Española improved Chile pepper. horts 19, 454 (1984).
Ichijo, T. et al. Bacterial bioburden and community structure of potable water used in the International Space Station. Sci. Rep. 12, 16282 (2022).
La Duc, M. T., Venkateswaran, K., Sumner, R. & Pierson, D. Characterization and Monitoring of Microbes in the International Space Station Drinking Water. In 2003-01-2404. https://doi.org/10.4271/2003-01-2404 (2003).
Be, N. A. et al. Whole metagenome profiles of particulates collected from the International Space Station. Microbiome 5, 81 (2017).
Venkateswaran, K. et al. Microbial Characteristics of ISS Environmental Surfaces, 47th International Conference on Environmental Systems, ICES-2017-131 (2017).
Venkateswaran, K. et al. International Space Station environmental microbiome—microbial inventories of ISS filter debris. Appl. Microbiol. Biotechnol. 98, 6453–6466 (2014).
Sielaff, A. C. et al. Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces. Microbiome 7, 50 (2019).
Checinska, A. et al. Microbiomes of the dust particles collected from the International Space Station and spacecraft assembly facilities. Microbiome 3, 50 (2015).
Kliss, M., Heyenga, G., Hoehn, A. & Stodieck, L. Toward the Development of a “Salad Machine”. In 2000-01-2476. https://doi.org/10.4271/2000-01-2476 (2000).
Kliss, M. & MacElroy, R. D. Salad Machine: A Vegetable Production Unit for Long Duration Space Missions. In 901280 https://doi.org/10.4271/901280 (1990).
Massa, G. D., Newsham, G., Hummerick, M. E., Morrow, R. C. & Wheeler, R. M. Plant pillow preparation for the veggie plant growth system on the International Space Station. Gravit. Space Res. 5, 24–34 (2017).
Wheeler, R. M., Mackowiak, C. L. & Sager, J. C. Soybean stem growth under high-pressure sodium with supplemental blue lighting. Agron. J. 83, 903–906 (1991).
Barillot, C. D. C., Sarde, C.-O., Bert, V., Tarnaud, E. & Cochet, N. A standardized method for the sampling of rhizosphere and rhizoplan soil bacteria associated to a herbaceous root system. Ann. Microbiol. 63, 471–476 (2013).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Andersen, K. S., Kirkegaard, R. H., Karst, S. M. & Albertsen, M. ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. Preprint at https://doi.org/10.1101/299537 (2018).
Shannon, C. E. A Mathematical Theory of Communication. Bell Syst. Tech. J. XXVII, 3 (1948).
Bray, J. R. & Curtis, J. T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349 (1957).
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46 (2001).
Oliveros, J. C. Venny. An Interactive Tool for Comparing Lists with Venn Diagrams. https://bioinfo.gp.cnb.csic.es/tools/venny/index.html (accessed 9/2024).
Hummerick, M. E. et al. Spatial characterization of microbial communities on multi-species leafy greens grown simultaneously in the vegetable production systems on the international space station. Life 11, 1060 (2021).
Khodadad, C. L. M. et al. Microbiological and nutritional analysis of lettuce crops grown on the International Space Station. Front. Plant Sci. 11, 199 (2020).
Singh, N. K., Wood, J. M., Karouia, F. & Venkateswaran, K. Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces. Microbiome 6, 204 (2018).
Nelson, E. B. The seed microbiome: Origins, interactions, and impacts. Plant Soil 422, 7–34 (2018).
Wilpiszeski, R. L. et al. Soil aggregate microbial communities: Towards understanding microbiome interactions at biologically relevant scales. Appl. Environ. Microbiol. 85, e00324-e419 (2019).
Ahkami, A. H., Allen White, R., Handakumbura, P. P. & Jansson, C. Rhizosphere engineering: Enhancing sustainable plant ecosystem productivity. Rhizosphere 3, 233–243 (2017).
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Van Themaat, E. V. L. & Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838 (2013).
Gaiero, J. R. et al. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 100, 1738–1750 (2013).
Berendsen, R. L., Pieterse, C. M. J. & Bakker, P. A. H. M. The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012).
Lindow, S. E. & Brandl, M. T. Microbiology of the Phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883 (2003).
Upper, C. D. & Hirano, S. S. Aerial dispersal of bacteria. In: Assessing Ecological Risks of Biotechnology. pp. 75–93 (Elsevier). https://doi.org/10.1016/B978-0-409-90199-3.50011-5 (1991).
Compant, S., Samad, A., Faist, H. & Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37 (2019).
Kinkel, L. L., Wilson, M. & Lindow, S. E. Plant species and plant incubation conditions influence variability in epiphytic bacterial population size. Microb. Ecol. 39, 1–11 (2000).
O’Brien, R. D. Effect of plant species and environmental conditions on epiphytic population sizes of Pseudomonas syringae and other bacteria. Phytopathology 79, 619–627 (1989).
Lebeis, S. L. Greater than the sum of their parts: Characterizing plant microbiomes at the community-level. Curr. Opin. Plant Biol. 24, 82–86 (2015).
Wallenstein, M. D. Managing and manipulating the rhizosphere microbiome for plant health: A systems approach. Rhizosphere 3, 230–232 (2017).
Schreiter, S. et al. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5, 144 (2014).
Acharya, S. M. et al. Fine scale sampling reveals early differentiation of rhizosphere microbiome from bulk soil in young Brachypodium plant roots. ISME Commun. 3, 54 (2023).
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T. & Singh, B. K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621 (2020).
Tabrett, A. & Horton, M. W. The influence of host genetics on the microbiome. F1000Res 9, 84 (2020).
Dawson, W., Hör, J., Egert, M., Van Kleunen, M. & Pester, M. A small number of low-abundance bacteria dominate plant species-specific responses during rhizosphere colonization. Front. Microbiol. 8, 975 (2017).
Turner, T. R., James, E. K. & Poole, P. S. The plant microbiome. Genome Biol 14, 209 (2013).
Choi, K., Khan, R. & Lee, S.-W. Dissection of plant microbiota and plant-microbiome interactions. J. Microbiol. 59, 281–291 (2021).
Correa-Galeote, D., Bedmar, E. J. & Arone, G. J. Maize endophytic bacterial diversity as affected by soil cultivation history. Front. Microbiol. 9, 484 (2018).
Castro, V. A., Thrasher, A. N., Healy, M., Ott, C. M. & Pierson, D. L. Microbial characterization during the early habitation of the International Space Station. Microb. Ecol. 47, 119–126 (2004).
Bruce, R. J., Ott, C. M., Skuratov, V. M. & Pierson, D. L. Microbial Surveillance of Potable Water Sources of the International Space Station. In 2005-01-2886. https://doi.org/10.4271/2005-01-2886 (2005).
Pierson et al. Microbial monitoring of the ISS. (2013).
Van Houdt, R., Mijnendonckx, K. & Leys, N. Microbial contamination monitoring and control during human space missions. Planet. Space Sci. 60, 115–120 (2012).
O’Rourke, A., Lee, M. D., Nierman, W. C., Everroad, R. C. & Dupont, C. L. Genomic and phenotypic characterization of Burkholderia isolates from the potable water system of the International Space Station. PLoS ONE 15, e0227152 (2020).
Matsumoto, H. et al. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 7, 60–72 (2021).
Frank, A. C., Saldierna Guzmán, J. P. & Shay, J. E. Transmission of bacterial endophytes. Microorganisms 5, 70 (2017).
Wang, J. et al. Comparative genomics of degradative novosphingobium strains with special reference to microcystin-degrading Novosphingobium sp. THN1. Front. Microbiol. 9, 2238 (2018).
Harmsen, M., Yang, L., Pamp, S. J. & Tolker-Nielsen, T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 59, 253–268 (2010).
Pericolini, E. et al. Real-time monitoring of Pseudomonas aeruginosa biofilm formation on endotracheal tubes in vitro. BMC Microbiol. 18, 84 (2018).
Crabbé, A. et al. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol. 12, 1545–1564 (2010).
Kim, W. et al. Spaceflight promotes biofilm formation by Pseudomonas aeruginosa. PLoS ONE 8, e62437 (2013).
Preston, G. M. Plant perceptions of plant growth-promoting Pseudomonas. Phil. Trans. R. Soc. Lond. B 359, 907–918 (2004).
Salido, R. A. et al. The International Space Station has a unique and extreme microbial and chemical environment driven by use patterns. Cell 188, 2022-2041.e23 (2025).
Sah, S., Krishnani, S. & Singh, R. Pseudomonas mediated nutritional and growth promotional activities for sustainable food security. Curr. Res. Microb. Sci. 2, 100084 (2021).
Kim, H. W. & Rhee, M. S. Space food and bacterial infections: Realities of the risk and role of science. Trends Food Sci. Technol. 106, 275–287 (2020).
Liu, H. et al. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytol. 229, 2873–2885 (2021).
Lee, S. A. et al. Chitinophaga agri sp. nov., a bacterium isolated from soil of reclaimed land. Arch. Microbiol. 203, 809–815 (2021).
Lu, Z. et al. A polysaccharide utilization locus from Chitinophaga pinensis simultaneously targets chitin and β-glucans found in fungal cell walls. mSphere. 8, e00244-e323 (2023).
Kim, D. Y. et al. Biocontrol potential of Chitinophaga flava HK235 producing antifungal-related peptide chitinocin. Front. Microbiol. 14, 1170673 (2023).
Zhang, X. et al. Dynamics of rice microbiomes reveal core vertically transmitted seed endophytes. Microbiome 10, 216 (2022).
Dixit, A. R. et al. Persistence of Escherichia coli in the microbiomes of red Romaine lettuce (Lactuca sativa cv. ‘Outredgeous’) and mizuna mustard (Brassica rapa var. japonica)-does seed sanitization matter?. BMC Microbiol. 21, 289 (2021).
Deng, X. et al. Soil microbiome manipulation triggers direct and possible indirect suppression against Ralstonia solanacearum and Fusarium oxysporum. npj. Biofilms Microbiomes 7, 33 (2021).
Acknowledgements
The authors would like to thank the Redwire Space staff for their science operations at Kennedy Space Center, the Sierra Space staff for facility operations and NASA astronauts Megan McArthur, Mark Van de Hei, Shane Kimbrough, Kayla Barron, and ESA astronaut Thomas Pesquet for nurturing the pepper plants during the growth period. The authors are grateful for the fertilizer counsel and support of Ed Rosenthal and Florikan ESA.
Funding
Funding was provided by NASA’s Biological and Physical Sciences which funded the ground and development work. The International Space Station Program funded PH-04 flight study.
Author information
Authors and Affiliations
Contributions
CLK- experimental design, sample processing, data collection, data analysis, manuscript development; ARD- experimental design, sample processing, data collection, data analysis; manuscript development; MEH- experimental design, sample processing, data collection, data analysis; LES- experimental design, horticulture and crop management, project development; CJS- sample processing, data collection, manuscript development; JJT-project development, data collection, horticulture and crop management; OM- data collection, experimental design, project development; JTR-project management, project development, data collection, experimental design; JLG- sample processing, data collection; ABC- sample processing, data collection; GDM- project development, data collection, experimental design; ND- project management, project development; LP-experimental design, horticulture and crop management, data collection; MWR- project development, project management, experimental design, funding support; RMW- project development, experimental design, funding support. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Khodadad, C.L.M., Dixit, A.R., Hummerick, M.E. et al. Evaluating microbial community profiles of Chile peppers grown on the International Space Station provides implications for fruiting crops. Sci Rep (2026). https://doi.org/10.1038/s41598-025-20440-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-20440-9