Abstract
One of the common challenges in medical artificial intelligence (AI) applications using echocardiography is the lack of image data harmonization. This study aims to improve the prediction accuracy of left ventricular ejection fraction (LVEF) AI models by incorporating data augmentation (DA) techniques to address the variability in image data across different vendor machines. A database comprising 15,770 echocardiographic videos from 3154 patients across five different centers was utilized, with the data acquired using various vendor machines. We prepared datasets specific to GE healthcare (GE), philips (PH), and canon medical systems (CA) vendors, including 1911, 804, and 427 cohorts, respectively. A three-dimensional convolutional neural network (3D-CNN) was trained to predict LVEF, using videos consisting of 20 images per heartbeat as input, with training performed exclusively on data from GE machines (GE-based model). DA techniques, including gamma correction, scaling, median filtering, unsharp masking, translation, rotation, noise correction, and image conversion using a cycle generative adversarial network, were applied. A regression analysis using five different chamber views was performed on the GE test data. The accuracy of LVEF prediction using the GE-based model with DA—specifically gamma correction, scaling, and translation corrections—was excellent. The mean absolute error (root mean square error) in test cohorts was 4.33 (5.58) for GE, 4.42 (5.57) for PH, and 4.89 (6.57) for CA, which were comparable to the results of a model developed using data from all vendors, demonstrating the effectiveness of DA in harmonizing images. This study demonstrates that echocardiographic videos are transferable across vendors and that DA is highly effective in improving LVEF predictions from data acquired using different vendor machines.
Similar content being viewed by others
Introduction
Echocardiography is one of the most convenient and rapid tests for diagnosing cardiovascular diseases. It offers the significant advantage of not involving radiation exposure, which is a common concern in radiological diagnostics, and it can be performed in real-time. However, the procedure is subject to variability, including differences in techniques between examiners, protocols across facilities, and discrepancies between equipment from different vendors. Recently, machine learning methods, particularly deep learning, have been increasingly employed in echocardiography1,2,3,4,5,6,7,8,9,10,11,12,13, raising concerns that such variations may affect the outcomes of these learning models. Specifically, in the context of machine learning, differences in image colors, the divergence angle of the echo region, image size and scale, and the inclusion of cardiac waveforms pose significant challenges. Therefore, robust preprocessing to ensure generalizability is crucial.
Several studies have been conducted to reduce image differences among protocols and vendor machines, particularly in computed tomography and magnetic resonance imaging14,15,16,17,18,19,20,21,22. For instance, generative adversarial networks (GANs)23 have been utilized for image conversion, demonstrating effectiveness in tasks such as diagnosis and segmentation24,25,26,27. In contrast, the investigation of inter-protocol and inter-vendor variability using echocardiography remains limited, despite numerous studies aimed at developing AI models. Deep learning models for echocardiographic interpretation, including view classification, have been successfully developed, and models predicting left ventricular ejection fraction (LVEF)—the percentage of blood pumped out of the left ventricle with each contraction—have achieved a mean absolute error (MAE) of 5% and a root mean square error (RMSE) of 8%28,29,30,31,32,33,34. However, most of these models rely on data from a single protocol and vendor machine28,30, and to our knowledge, no study has specifically addressed the variability between vendor machines in echocardiography using a multicenter database.
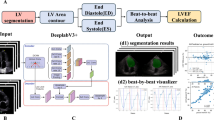
In this study, we developed a database composed of 3154 cohorts collected from five centers in Japan (Tokushima University, Tenri Hospital, Osaka University, Kobe University, and the National Cerebral and Cardiovascular Center). This database includes echocardiographic videos of five different chamber views from three major vendors (GE Healthcare: GE, Philips: PH, and Canon Medical Systems: CA). Using this dataset, we investigated inter-machine variability and its harmonization to improve the accuracy of LVEF prediction using a three-dimensional convolutional neural network (3D-CNN) model. Traditionally, preprocessing for such deep learning models involved manually registering reference images, aligning positions, cropping, and removing unnecessary information such as textual data. In this study, we automated these processes, resulting in standardized output images across different vendors. Additionally, we employed data augmentation (DA) techniques in the input images for LVEF model training. The LVEF model should be invariant to certain transformations of echocardiographic videos, such as GE-like images transformed from PH images using a GAN model. Instead of rigorously standardizing image statistics, we augmented the data with various image conversions to embed this invariance feature into the model. Techniques such as gamma correction, scaling, median filtering, unsharp masking, translation, rotation, noise corrections, and transformation using a cycle GAN24 were applied. We evaluated inter-machine variability among the three vendors, GE, PH, and CA, and observed significant improvements in LVEF prediction accuracy and a reduction in machine variability, particularly with gamma correction, scaling, and translation. Finally, we compared the LVEF prediction errors between the GE-based model with DA—including gamma correction, scaling, and translation corrections, which used only GE image data for training—and an all-vendor model that used image data from all vendors. An overview of this study is depicted in (Fig. 1).
Overview of this study. Five chamber views (long = parasternal long-axis view; short = parasternal short-axis view; 4ch = apical four-chamber view; 3ch = apical three-chamber view; 2ch = apical two-chamber view) of echocardiography were collected from five hospitals, creating datasets for three major vendor machines. These datasets were used to train: 1) the GE-based model, which utilizes only GE datasets, and 2) the all-vendor model, which incorporates datasets from all three vendors. For the GE-based model, various data augmentation techniques—including gamma correction, scaling, median filtering, unsharp masking, translation, rotation, noise correction, and image conversion using a generative adversarial network—were applied. GE general electric, PH Philips, CA canon, LVEF left ventricular ejection fraction.
Results
In this study, we collected 15,770 echocardiographic videos from 3154 patients across five centers, obtained using various vendor machines. The frames from these videos were preprocessed and standardized into 240 × 240 × 20 grayscale 3D images, also referred to as (2 + 1)-D images. The cropping process employed a shallow two-dimensional convolutional neural network (S2DCNN) model, which was trained on 1,600 randomly sampled images from a vendor- and center-mixed dataset. This approach reduced the variability among processed images compared to the original frames. Using these preprocessed images, we prepared training datasets for the LVEF prediction model and applied data augmentation (DA) with various techniques.
Statistics of processed images
In echocardiographic images, the fan-shaped region of interest is a key feature, and extraneous elements such as characters, symbols, and the heartbeat signal are often superimposed on the image. Removing such unnecessary information during preprocessing is crucial. For this purpose, we employed the S2DCNN model. Examples of the resulting images are shown in Fig. S1 of the supplementary information, demonstrating successful removal of unnecessary image elements. We further evaluated the processed images by examining histograms of pixel intensities within the anatomical region of the ultrasound images (Fig. 2); grayscale pixel values were normalized to [0, 1]. The top row shows histograms for the input images, and the bottom row shows those for the processed outputs. In our dataset, the histograms indicate a reduction in the standard deviation of grayscale values: the global standard deviation decreased from 0.0642 to 0.0546 (left panel of Fig. 2). The right-hand panels summarize results by vendor, showing standard deviations comparable to or lower than those of the inputs (GE: 0.0489 → 0.0419; PH: 0.0643 → 0.0564; CA: 0.0419 → 0.0420). In addition, inter-vendor differences in mean grayscale values were reduced after processing. For example, the mean for CA increased from 0.157 (unprocessed) to 0.210 after processing, bringing it closer to the means for GE and PH (0.267 and 0.273, respectively).
Statistics in processed images. Left: Histograms of pixel intensities within the anatomical region of interest for the 400 test images, where grayscale pixel values were normalized to [0, 1]—input (top) and processed output (bottom). Right: Corresponding histograms stratified by vendor (GE, PH, CA). The standard deviation (STD) is comparable to or lower than in the inputs (GE: 0.0489 → 0.0419; PH: 0.0643 → 0.0564; CA: 0.0419 → 0.0420). The mean intensity for CA increases from 0.157 (input) to 0.210 (output), bringing it closer to GE and PH (0.267 and 0.273, respectively). GE general electric, PH philips, CA canon.
LVEF prediction accuracy with/without data augmentation
To evaluate the effect of DA on inter-machine variability, we assessed LVEF prediction accuracies using various DA techniques applied to GE training cohorts. The corrections used in DA are listed in the first column of Table 1, and the details of DA are described in the Methods section. Table 1 shows that gamma correction was particularly effective in reducing prediction errors (e.g., RMSE in the PH cohort and both MAE and RMSE in the CA cohort), as were scaling correction (MAE in the GE cohort) and translation correction (RMSE in the GE cohort and MAE in the PH cohort). The median filter also reduced prediction errors compared to the no data augmentation (NoDA-GE) model. Conversely, unsharp mask filtering and noise addition worsened accuracy. Image conversion techniques, such as transforming GE images to PH-like and CA-like images using cycle GAN, reduced prediction errors in the GE and PH cohorts but increased them in the CA cohort, indicating that effective image conversion using cycle GAN for data augmentation remains challenging.
Inter-machine harmonization in echocardiography
Based on the results above, we selected gamma correction, scaling, and translation corrections for inter-machine harmonization to predict LVEF from echocardiography. To assess the effectiveness of these DA techniques, we compared LVEF predictions using the model trained with GE data (DAfull-GE; 840 patients) to predictions using an all-vendor-based model (NoDA-all; 1,302 patients total, including 840 from GE, 270 from PH, and 192 from CA). Figure 3 shows the scatter plot of predicted LVEF (x-axis) versus ground truth (y-axis) for the test cohort. The prediction model performed well in describing the ground truth not only for the GE cohort but also for the PH and CA cohorts. Table 2 indicates that the DAfull-GE model yields results comparable to the NoDA-all model. Statistical tests reveal that the MAE difference between the DAfull-GE and NoDA-all models is not significant (p = 0.29). The prediction error in the GE cohort is lower than in the PH and CA cohorts. However, statistically, there is no significant difference among the GE, PH, and CA cohorts (GE-CA: p = 0.08, PH-CA: p = 0.22, and PH-GE: p = 0.89), suggesting that the DA techniques applied—gamma correction, scaling, and translation corrections—effectively harmonize inter-machine variability. The prediction accuracy is now sufficiently high, even when applying the model to different vendor machines, compared to previous reports.
Scatter plot of the predicted LVEF. The top row shows results from the model trained on the GE cohort with data augmentation (gamma, scaling, and translation) (DAfull-GE). The bottom row shows results from the model trained on all-vendor cohorts without data augmentation (NoDA-all). Both models were evaluated on test datasets comprising 1,071, 534, and 235 patients from GE, PH, and CA machines, respectively. GE general electric, PH philips, CA canon, LVEF left ventricular ejection fraction.
Comparison with existing vendor-independent software
We additionally evaluated LVEF measurements obtained using the US2.ai software, a cloud-based analysis tool developed for fully automated DICOM reading. This software is known for its speed and compatibility with various echocardiographic devices35. In this evaluation, twenty-four patients were randomly selected from the test cohort used in the present study, and LVEF predictions generated by US2.ai were compared with those from our model (Fig. 4). Our model outperformed the US2.ai software in terms of MAE (our model: 4.71; US2.ai software: 7.20). Statistical analysis demonstrated that our model significantly improved prediction accuracy (Paired t-test: t = 2.229, p = 0.036).
Comparison with existing software (US2.ai) for LVEF. The left panel shows results from the present model trained on the GE cohort with data augmentation (gamma, scaling, and translation) (DAfull-GE), while the right panel shows results from the US2.ai software. Twenty-four patients were randomly selected from the test cohort. MAE mean absolute error.
Discussion
In this study, we conducted a collaborative research project across five clinical facilities to explore DA techniques for echocardiographic movies. We employed both traditional image processing methods, such as gamma correction, affine transformation, and convolution operations, as well as an advanced image generation model based on cycle GAN. The primary goal of this study was to enhance the accuracy and efficacy of LVEF prediction models for datasets from different vendors. Even when using data acquired solely from GE machines for training, the application of gamma correction, scaling, and translation corrections in DA enabled us to achieve MAE of less than 5% when predicting LVEF for movies from machines not included in the training set. This level of accuracy is comparable to or better than that reported in previous studies focused on LVEF prediction using data from a single machine type28,30. Additionally, while many prior studies have developed models tailored to specific machines—resulting in high accuracy for that machine but significantly lower accuracy for others—our model demonstrated a novel and high degree of generalizability across multiple vendors. Moreover, the performance of the present model surpassed that of an existing vendor-independent solution, US2.ai software.
Recent advancements in self-supervised learning with foundational models are making it increasingly feasible to predict clinical indicators from large datasets29. These approaches are also improving the generalizability of LVEF prediction. However, our study suggests that, from an accuracy standpoint, dedicated deep learning models that incorporate established image processing techniques are preferable for specific tasks like LVEF estimation. The importance of DA using image correction techniques has been highlighted in the literature on medical images. However, available data is limited14, and no previous study has demonstrated the high performance in LVEF prediction achieved in our study.
By employing engineering processes in DA, we were able to accurately predict LVEF from images captured by machines not used during the training phase. Although we used a cycle GAN to generate PH-like and CA-like images from GE images for training, this approach did not yield as significant an improvement in performance as simpler engineering processes like gamma correction, scaling, and translation. We developed the cycle GAN image generation model to avoid learning image deformations or applying varying transformations on images within a single cardiac cycle, which can occur with models like U-net. To achieve this, we selected a model with shallow layers and a limited number of filters for image generation (see Fig. S2 of supplementary information). However, convolution-based image generation models, as used in this study, found it challenging to learn scaling and translation, and even learning gamma correction was inefficient, because this capability is constrained by the training data used for the cycle GAN. Consequently, the diversity of the generated images is limited compared to that required in actual engineering applications. This suggests that the inter-machine transformation of echocardiographic images is not overly complex, and simpler processing techniques were more effective.
The cross-vendor robustness of our LVEF prediction model is especially valuable when only a single-vendor dataset is available. Demand for highly accurate diagnostic AI is likely to grow in smaller clinics and home settings36, where simpler ultrasound devices are commonly used. In these contexts, harmonized preprocessing and standardized LVEF-related metrics enable practical use by non-specialists in education, emergency/point-of-care examinations, and home/tele-echocardiography. The tool assists—but does not replace—expert interpretation, and we plan prospective, workflow-integrated evaluations to quantify its clinical impact.
Although our model shows promise for applications in various echocardiographic devices and related equipment, it has not been evaluated using portable or low-end devices in the present study. Moreover, the dataset is limited to movies from only three ultrasound vendors. Within this dataset, GE-sourced images predominate, which may inherently favor model performance by increasing distributional similarity and biasing the learned parameters toward GE (particularly for the NoDA-all model). Accordingly, there remains a small possibility that the apparent parity between DAfull-GE and NoDA-all partly reflects a closer match between the training and test vendor distributions rather than true cross-vendor harmonization. Further validation with independent cohorts and a broader range of vendors is therefore warranted.
This study utilized a database comprising 15,770 echocardiographic movies from 3,154 patients. However, compared to AI development research in non-medical fields, this data volume is still relatively small. Moreover, our study compared a limited range of machines, highlighting the need for validation with more diverse data in the future. The LVEF values used for training and accuracy validation were measured by operators from echocardiographic movies following standardized protocols, which could include measurement errors or inaccuracies. For example, as detailed in the supplementary information (Fig. S3 and Table S1), most CA videos were contributed by a single clinical center (Kobe University Hospital), leading to smaller sample size and reduced diversity relative to GE and PH. Such site/vendor imbalance can introduce a domain shift that degrades generalization. In addition, the ground-truth LVEF values were derived from expert measurements of cine loops acquired under standardized protocols, yet inter- and intra-observer variability may be present. As we continue to expand the dataset, addressing these annotation challenges will be crucial. Combining our approach with self-training models might also prove beneficial.
Methods
Datasets
This study was approved by the Ethics Committee of Tokushima University Hospital (approval No. 3554-2). Informed consent was obtained from all participants using an opt-out procedure approved; accordingly, the requirement for written informed consent was waived. Notices describing the study and the opportunity to decline participation were posted on institutional websites and/or within the clinics, and individuals who opted out were excluded. All data were de-identified prior to analysis. All methods were performed in accordance with the relevant guidelines and regulations, including the Declaration of Helsinki, the applicable national, institutional research ethics policies, and site-specific privacy and data-handling requirements. The database is composed of 3,154 cohorts collected from five centers (Tokushima University, Tenri Hospital, Osaka University, Kobe University, and the National Cerebral and Cardiovascular Center) without personal information, including five different chamber views (long, short, 4ch, 3ch, and 2ch) of echo movies. Details of this database are described in Fig. S3 and Tables S1-S3 of the supplementary information. We prepared datasets for GE, PH, and CA vendors, which include 1,911, 804, and 427 cohorts, respectively. For DA analysis of LVEF prediction, the GE-based models with various DAs were developed using 840 GE cohorts (used as 80% for training and 20% for testing) and the developed model was applied to 270 PH cohorts and 192 CA cohorts. For inter-machine harmonization in echocardiography, another GE-based model was developed using 840 GE training cohorts, whereas the all-vendor model was developed using 840, 270, and 192 cohorts acquired with GE, PH, and CA machines, respectively. Both models were then tested with 1,071, 534, and 235 test cohorts acquired with GE, PH, and CA machines, respectively. To obtain the reference LVEF values, all studies were independently analyzed by five expert readers with more than 10 years’ experience with echocardiography as well as certification as Registered Medical Sonographers or Board Certificated Fellows by The Japan Society of Ultrasonics in Medicine. LVEF was calculated by the biplane method of disks using the 2ch and 4ch views, and then the measurement was confirmed on the other echocardiographic views (3ch, long and short).
Pre-processing in echocardiographic movies and a shallow two-dimensional convolutional neural network (S2DCNN)
The pre-processing used in this study is depicted in Fig. 5, where a single raw image masked for personal information is automatically cropped by a shallow (small layer) 2DCNN model. The S2DCNN was employed as this process only crops the images (not conversion or transformation, including deformation). The training and validation data were 1,600 and 400 manually cropped images with fan-shape, respectively, including five chamber views acquired by three vendors from five centers. The training conditions are found in Fig. S2 of the supplementary information, and representative output images are also shown. After this processing, images were then resized to 480 × 480, and the image signal intensity was normalized so that the maximum value is one in color. The developed S2DCNN model was applied to all frame-by-frame images in echocardiographic movies, and then 20 images per heartbeat were sampled at equal intervals using the heartbeat analysis model previously developed4,5,6,28, where If the difference between the heartbeat recorded in the DICOM tag and the heartbeat estimated from the actual data exceeded 20%, or if the heart rate was greater than 120 bpm or less than 40 bpm—which could indicate the presence of atrial fibrillation or irregular RR intervals—manual verification by visual inspection was conducted. Otherwise, 20 frames were uniformly selected under the assumption that the images were captured during a consistent cardiac cycle. For the visual verification, we employed a pixel summation algorithm that had previously been used for respiratory phase recognition37.
LVEF prediction model with/without data augmentations
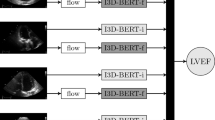
The LVEF prediction model is based on a 3D-CNN with an input 3D image of 240 × 240 × 20 grayscale (averaged over “RGB”) voxels, resizing the processed image described in the previous subsection. Through downsampling, a 3 × 3 × 3 convolution with a 2 × 2 × 2 stride was applied for each level of the encoding path. Further, each convolutional layer was batch-normalized and activated by LeakyReLU. The output (predicted LVEF) is obtained through ReLU function. The training conditions are found in Fig. S4 of the supplementary information.
A single echocardiographic movie can include multiple heartbeats, and the 3D images with all different heartbeat periods of the same patient are used both in the training and testing. During testing, the prediction was averaged over such heartbeats in the same patients. The 3D-CNN was trained with individual chamber views, resulting in five outputs for each patient. The predicted LVEF is determined by multiple regression, meaning the weighted mean of the five outputs. The weight is determined using test cohort data from GE, and the same weights were applied to the test cohorts from PH and CA.
The data used in DA analysis of LVEF prediction models is composed of 1,302 patients, with 840, 270, and 192 of them acquired by GE, PH, and CA, respectively. To obtain accurate LVEF even for rare cases such as low LVEFs (0–29) and high LVEFs (70–100), the data is balanced as seen in (Table 3). In this analysis, 672 echocardiography movies out of 840 acquired with GE machines were used in the training, and the rest were used in testing, along with the movies acquired with PH and CA machines.
To evaluate the effect of DA on inter-machine variability, the prediction accuracies with various DAs applied to GE training cohorts were estimated in PH and CA cohorts. We employed eight image corrections to GE data, including gamma correction (Gamma-GE), scaling correction (Scaling-GE), median filter correction (Median-GE), unsharp mask correction (Unsharp-GE), translation correction (Translation-GE), rotation correction (Rotation-GE), noise correction (Noise-GE), and cycle GAN correction (cGAN-GE). The training data was augmented three times with the above image corrections, resulting in a total of 672 × 3 = 2,016 training images.
Gamma correction
An image \(f\left({\varvec{x}}\right)\) is converted into \({f}_{\gamma }({\varvec{x}})\) by gamma correction,
where \(\gamma\) was randomly sampled in the range of [0.25, 2.0]. The images generated by gamma correction with GE images were included in the training of the Gamma-GE model.
Scaling correction
The scaling factor was selected as 1.2 for scaling up and 0.8 for scaling down. The nearest neighbor method was employed in the interpolation. For scaling down, the peripheral area was padded with zeros. The images generated by scaling correction with GE images were included in the training of the Scaling-GE model.
Median filter correction
Median filters with kernel sizes of both 2 × 2 and 3 × 3 were employed. The images generated by median filter correction with GE images were included in the training of the Median-GE model.
Unsharp mask correction
For unsharp mask correction, we employed a type of the difference of Gaussians method,
where zero for \(\sigma 1\) and 0.7 for \(\sigma 2\) were employed, and \(a\) was randomly sampled in the range of [0.3, 1.0]. The images generated by unsharp mask correction with GE images were included in the training of the Unsharp-GE model.
Translation correction
Translation was applied as,
where \({\varvec{c}}\) is the two-dimensional vector randomly sampled from [− 20, 20] in both the x and y directions. The images generated by translation correction with GE images were included in the training of the Translation-GE model.
Rotation correction
The rotation was applied as,
where \(\theta\) was randomly sampled from [− 20, 20] degrees. The images generated by rotation correction with GE images were included in the training of the Rotation-GE model.
Noise correction
Gaussian noise was added to the image as,
where \(\varepsilon\) means the value sampled from normal distribution \(N(0, \sigma )\). Here, \(\sigma\) was randomly sampled from [0.1, 0.25]. The images generated by noise correction with GE images were included in the training of the Noise-GE model.
Cycle GAN correction24
The cycle GAN is one of the unsupervised generative models for image generation. This can transform between images in different categories. In this study, we prepared three categories: GE, PH, and CA, and the cycle GAN was trained to transform between GE-PH and GE-CA, generating PH-like GE images and CA-like GE images. The details are described in Fig S5 of the supplementary information. The generated PH-like and CA-like GE images were included in the training of the cGAN-GE model.
Inter-machine harmonization in echocardiography
The data used in inter-machine harmonization in echocardiography is shown in (Table 4). The data used in model training is balanced, whereas that used in model testing is unbalanced. Unbalanced data reflects the real LVEF distribution in a clinical situation and may be appropriate for evaluating the LVEF prediction model. Because the minimum number of the LVEF distribution histogram is used as the number of training data in each bin, there are blanks in Table 4 (“0–29” in GE, “70–100” in PH and CA).
Among the aforementioned corrections, gamma, translation, and scaling corrections were found to be effective in reducing prediction errors (Results section). Therefore, we finally compared the No DA with all vender data (NoDA-all) with DA with gamma, translation, scaling corrections of GE data (DAfull-GE) to evaluate its effectiveness in inter-machine harmonization in echocardiography. The DA created eight times from the original GE data. Other models, which additionally include median filter, rotation, etc., were also developed, and the results are found in Figs. S6-S27 and Tables S4-S14 of the supplementary information. Statistical difference between NoDA-all and DAfull-GE for absolute error was evaluated by random permutation test. The absolute error of predicted results for GE, PH, and CA by DAfull-GE were also compared with each other by Tukey’s multiple comparison test.
Data availability
Raw data were generated at Tokushima University. Derived data supporting the findings of this study are available from the corresponding author A.H. on request.
References
Qayyum, S. R. A comprehensive review of applications of artificial intelligence in echocardiography. Curr. Probl. Cardiol. 49, 102250 (2024).
Aziz, D. et al. The role of artificial intelligence in echocardiography: A clinical update. Curr. Cardiol. Rep. 25, 1897–1907 (2023).
Kusunose, K. Revolution of echocardiographic reporting: the new era of artificial intelligence and natural language processing. J. Echocardiogr. 21, 99–104 (2023).
Kusunose, K. et al. A deep learning approach for assessment of regional wall motion abnormality from echocardiographic images. Cardiovasc. Imaging 13, 374–381 (2020).
Morita, S. X. et al. Deep learning analysis of echocardiographic images to predict positive genotype in patients with hypertrophic cardiomyopathy. Front. Cardiovasc. Med. 8, 669860 (2021).
Kusunose, K., Haga, A., Abe, T. & Sata, M. Utilization of artificial intelligence in echocardiography. Circ. J. 83, 1623–1629 (2019).
Ghorbani, A. et al. Deep learning interpretation of echocardiograms. NPJ Digit. Med. 3, 10 (2020).
Madani, A. et al. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit. Med. 1, 6 (2018).
Madani, A. et al. Deep echocardiography: data-efficient supervised and semisupervised deep learning towards automated diagnosis of cardiac disease. NPJ Digit. Med. 1, 59 (2018).
Zhou, J., Du, M., Chang, S. & Chen, Z. Artificial intelligence in echocardiography: detection, functional evaluation, and disease diagnosis. Cardiovasc. Ultrasound 19, 29 (2021).
Zhang, J. et al. Fully automated echocardiogram interpretation in clinical practice. Circulation 138, 1623–1635 (2021).
Zhang, Z. et al. Artificial intelligence-enhanced echocardiography for systolic function assessment. J. Clin. Med. 11, 2893 (2022).
Fletcher, A. J., Lapidaire, W. & Leeson, P. Machine learning augmented echocardiography for diastolic function assessment. Front. Cardiovasc. Med. 8, 711611 (2021).
Zhang L, Wang X, Yang D, et al. When unseen domain generalization is unnecessary? Rethinking data augmentation. arXiv:1906.03347v2 (2019).
Zavala-Romero, O. et al. Segmentation of prostate and prostate zones using deep learning : A multi-MRI vendor analysis. Strahlenther. Onkol. 196, 932–942 (2020).
Hwang, H. J. et al. Generative adversarial network-based image conversion among different computed tomography protocols and vendors: effects on accuracy and variability in quantifying regional disease patterns of interstitial lung disease. Korean J. Radiol. 24, 807–820 (2023).
Verboom, S. D. et al. Deep learning-based breast region segmentation in raw and processed digital mammograms: generalization across views and vendors. J. Med. Imaging 11, 014001 (2024).
Haoliang Li, Wang Y, Wan R, et. al. Domain Generalization for Medical Imaging Classification with Linear-Dependency Regularization. 34th Conference on Neural Information Processing Systems (NeurIPS 2020), Vancouver, Canada. (2020)
Dou Q, Castro DC, Kamnitsas K, Glocker B. "Domain generalization via model-agnostic learning of semantic features. In: 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, Canada. (2019).
Li D, Yang Y, Song YZ, Hospedales TM. Learning to Generalize: Meta-Learning for Domain Generalization. In: The Thirty-Second AAAI Conference on Artificial Intelligence (AAAI-18) (2018).
Yoon, C., Hamarneh, G. & Garbi, R. Generalizable feature learning in the presence of data bias and domain class imbalance with application to skin lesion classification. Med. Image Comput. Comput. Assist. Intervent. MICCAI 2019, 365–373 (2019).
Wang, S. et al. DoFE: Domain-oriented feature embedding for generalizable fundus image segmentation on unseen datasets. IEEE Trans. Med. Imaging 39, 4237–4248 (2020).
Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. arXiv:1406.2661v1 (2014).
Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. In: IEEE International Conference on Computer Vision 2223–2232. (2017)
Jafari, M. H. et al. Cardiac point-of-care to cart-based ultrasound translation using constrained CycleGAN. Int. J. Comput. Assist. Radiol. Surg. 15, 877–886 (2020).
Teng, L., Fu, Z. & Yao, Y. Interactive translation in echocardiography training system with enhanced cycle-GAN. IEEE English Med. Biol. Soc. Sect. 8, 106147–106156 (2020).
Khan, S., Huh, J. & Ye, J. C. Variational formulation of unsupervised deep learning for ultrasound image artifact removal. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 68, 2086–2100 (2021).
Kusunose, K. et al. Deep learning for assessment of left ventricular ejection fraction from echocardiographic images. J. Am. Soc. Echocardiogr. 33, 632–635 (2020).
Christensen, M. et al. Vision–language foundation model for echocardiogram interpretation. Nat. Med. 30, 1481–1488 (2024).
Ouyang, D. et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 580, 252–256 (2020).
Asch, F. M. et al. Automated echocardiographic quantification of left ventricular ejection fraction without volume measurements using a machine learning algorithm mimicking a human expert. Circul. Cardiovasc. Imaging 12, e009303 (2019).
Kusunose, K., Zheng, R., Yamada, H. & Sata, M. How to standardize the measurement of left ventricular ejection fraction. Japan Soc. Ultrason. Med. 49, 35–43 (2021).
Reynaud, H. et al. Ultrasound video transformers for cardiac ejection fraction estimation. Med. Image Comput. Comput. Assist. Intervent. MICCAI 2021, 495–505 (2021).
Tromp, J. et al. Automated interpretation of systolic and diastolic function on the echocardiogram: a multicohort study. Lancet Digit. Health 4, e46-54 (2022).
https://us2.ai/wp-content/uploads/2022/01/FDA-clearance-release-2.0.pdf
Kusunose, K. Transforming echocardiography: The role of artificial intelligence in enhancing diagnostic accuracy and accessibility. Intern Med. 64, 331–336 (2024).
Kavanagh, A., Evans, P. M., Hansen, V. N. & Webb, S. Obtaining breathing patterns from any sequential thoracic x-ray image set. Phys. Med. Biol. 54, 4879 (2009).
Acknowledgements
R.I. and A.H. thank Kohei Torii, Tokushima University, for fruitful discussion in medical image processing. This work was supported by JSPS KAKENHI Grant Number 23K07084 and the Research Cluster program of Tokushima University.
Author information
Authors and Affiliations
Contributions
K.K and A.H. conceived and designed the study. A.H. wrote the main manuscript. H.T., M.M., K.M., Y.T., K.K., and A.H. designed the framework. H.T., M.M., K.M., Y.T., and K.K. prepared the clinical data. A.H. and R.I. developed the programing code used in this study. R.I. performed the model development and the analyses. All authors, including those mentioned above, contributed to the discussion and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Iwasaki, R., Kusunose, K., Tanaka, H. et al. Inter-machine harmonization of multicenter echocardiographic images for improvement of left ventricular ejection fraction prediction model. Sci Rep 15, 37434 (2025). https://doi.org/10.1038/s41598-025-21277-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-21277-y