Abstract
Breast cancer detection and diagnosis remain challenging due to the complexity of tumor tissues and image quality variations, which hinder early and accurate identification. Timely diagnosis is vital for initiating treatment and improving patient outcomes. This study presents a novel hybrid feature fusion method, combining deep features from pre-trained models (ResNet50, InceptionV3, and MobileNetV2) with traditional texture features from Gabor filters and wavelet transforms, applied separately to mammogram and ultrasound datasets for breast cancer detection. A robust pre-processing pipeline, including image resizing, scaling, normalization, and CLAHE for contrast enhancement, is used to improve model performance. Data augmentation strengthens model robustness, and tumor segmentation is performed using Otsu’s multi-thresholding to accurately localize high-intensity regions. The hybrid feature extraction method yields 600 features, which are optimized through statistical feature selection for enhanced classification accuracy. Machine learning algorithms–XGBoost, AdaBoost, and CatBoost–are utilized to classify breast lesions across datasets, including Mini-DDSM and INbreast for mammogram images, and Rodrigues and BUSI for ultrasound images. Unlike most prior work, this fusion is applied across both mammogram and ultrasound modalities within one framework, a combination that has not been widely explored. Our approach explicitly targets the multi-modal gap to enhance robustness and generalizability across imaging types. The performance of the ensemble classifiers is compared, demonstrating the effectiveness of the proposed approach. The models achieved high classification accuracies: 98.67% for Rodrigues, 97.06% for INbreast, 97.02% for BUSI, and 95.00% for Mini-DDSM. These results highlight the effectiveness of the method and its potential to improve breast cancer detection. Future research will focus on comparisons with state-of-the-art models and real-world clinical applications.
Similar content being viewed by others
Introduction
Breast cancer is a significant global health concern, affecting millions of women annually. According to the World Health Organization (WHO), it is the most frequently diagnosed cancer in women, whereas lung cancer remains the leading cause of cancer-related deaths. In the United States, approximately one in eight women will be diagnosed with invasive breast cancer during their lifetime. The American Cancer Society projects 2,041,910 new cancer cases in 2025, averaging 5600 cases per day including 1,053,250 in men and 988,660 in women, with 618,120 anticipated deaths1. Alarmingly, Black women are expected to experience a 41% higher incidence rate than White women2.
In Asia, Pakistan reports the highest incidence of breast cancer, with one in nine women at risk. In 2020 alone, 25,928 new cases and 13,725 deaths were recorded3. High mortality rates in Pakistan are primarily attributed to limited awareness and delayed diagnosis. Data from the Pakistan National Cancer Registry (NCR) shows a steady annual rise in cases, approaching 90,000 each year.
Breast cancer arises from uncontrolled cell growth due to genetic mutations that disrupt regulatory mechanisms4. The nucleus, which governs cell division, is often implicated, and mutations can lead to tumor formation5. The International Agency for Research on Cancer (IARC) predicts 35 million new cancer cases globally by 20506. Breast tumors are classified as benign or malignant: benign tumors grow slowly and remain localized, whereas malignant tumors are aggressive and metastasize if untreated7. Genetic abnormalities account for most cases8, with 5–10% linked to inherited mutations and 20–25% resulting from cellular changes acquired during development9.
Symptoms may include swelling, redness, palpable lumps, or nipple changes10. However, nearly 50% of cases present without noticeable symptoms or identifiable risk factors. Risk increases significantly with age, particularly after 40, necessitating regular mammographic or ultrasound screening every six months. Breastfeeding has been shown to reduce risk2. Breast cancer subtypes include invasive, non-invasive, and cancerous phyllodes tumors, with the latter being the most severe and incurable11.
Early detection is critical, as survival rates exceed 90% when cancer is identified and treated promptly12. Mammography, an X-ray imaging technique, is effective for detecting microcalcifications and other signs of breast cancer but is less effective in women with dense breast tissue13. Ultrasonography is useful for distinguishing between benign and malignant lesions, especially in dense tissue, but has limitations in terms of sensitivity14. Both methods have proven successful but are subject to limitations such as image quality, the need for radiologist expertise, and the risk of missing small lesions, especially in early stages15.
Manual diagnosis is time-consuming and prone to error, often leading to missed early-stage tumors. To overcome these challenges, Computer-aided Detection (CAD) systems16 have been developed to automate detection, improve diagnostic accuracy, and reduce oversight. Recent advances in Artificial Intelligence (AI), particularly Machine Learning (ML), Deep Learning (DL), and Transfer Learning (TL), have transformed medical imaging by simplifying analysis for radiologists and oncologists17. While ML relies on large labeled datasets and handcrafted feature, DL automates feature extraction but demands extensive computational resources. TL leverages pre-trained models to reduce training time and enhance accuracy, making it particularly useful in medical imaging applications18.
Several studies have explored these approaches. Hekmat et al.19 used a differential evolution-driven optimization technique to enhance ensemble learning models. The study demonstrated how combining optimization with deep learning models, such as Convolutional Neural Networks (CNNs), can improve the detection accuracy of complex medical conditions. Similarly, Khan et al.20 also used multiple CNN models in an ensemble, fine-tuned via grid search optimization. The combination of multiple models in an ensemble, along with fine-tuning through grid search, has been shown to improve classification performance significantly. TL models, pre-trained on extensive datasets such as ImageNet, CIFAR- 10, and CIFAR-100, for general tasks like image classification. In the context of breast cancer detection, these models can be fine-tuned to classify breast cancer images such as benign, malignant, and normal. These DL models initially capture broad features like shapes, edges, and textures, which help recognize and differentiate complex patterns in medical images21. Unlike traditional feature extraction methods that rely on manually crafted features based on expert knowledge, modern techniques like DL and TL automate the process22. While both approaches have value, selecting the most relevant features is key to reducing extraction time, optimizing computational resources, and improving efficiency.
Despite significant advancements existing methodologies often fail to exploit the complementary strengths of deep and handcrafted features. While DL models excel at capturing high-level semantics, traditional methods like Gabor filters and wavelet transforms provide valuable texture-based features. Addressing this gap, the present study proposes a hybrid feature fusion approach that integrates DL features from models such as ResNet50, InceptionV3, and MobileNetV2 with traditional texture descriptors. This combination provides a more comprehensive representation of breast cancer images. Another challenge in medical imaging is class imbalance, which can bias models and reduce accuracy. Our approach mitigates this by applying systematic statistical feature selection and employing ensemble classifiers capable of handling imbalanced datasets. The main contributions of this research are as follows:
-
Developed a unique framework that fuses both deep features from transfer learning models like ResNet50, InceptionV3, and MobileNetV2 with traditional features from Gabor filters and Wavelet transforms. This hybrid approach captures both high-level semantics and low-level textures, improving breast cancer image representation in mammograms and ultrasounds, making it one of the few methods to integrate multi-modal data for enhanced detection.
-
Applied image enhancement techniques like CLAHE, Wavelet Transform, and sharpening to improve mammogram quality. Used segmentation methods, including multi-threshold with Otsus method, to accurately isolate Region of Interest’s (ROIs). These enhancements help the model better detect abnormalities in noisy or low-contrast images, further improving the robustness of the model.
-
Applied a systematic statistical feature selection process to identify the most discriminative features, reducing the original pool of 218,072 to a robust set of 600, thereby optimizing classification performance by focusing on the most relevant features.
-
Utilized state-of-the-art ensemble learning algorithms (XGBoost, AdaBoost, and CatBoost) to classify breast cancer using the integrated feature set, demonstrating their effectiveness in improving classification accuracy and robustness when applied to both mammogram and ultrasound data.
-
The proposed hybrid feature fusion method is evaluated separately on publicly available mammogram (Mini-DDSM, INbreast) and ultrasound (Rodrigues, BUSI) datasets. This comprehensive evaluation across different clinical environments and imaging modalities highlights the generalizability and effectiveness of the method for breast cancer detection in real-world clinical settings.
In contrast to traditional DL models that rely solely on deep features from pre-trained models, our proposed approach introduces a hybrid feature fusion mechanism. This method combines deep learning features from models like ResNet50, InceptionV3, and MobileNetV2 with traditional texture-based features extracted using Gabor filters and wavelet transforms. Unlike models that focus only on high-level semantic features or rely solely on handcrafted features, our approach merges both types of features to provide a more comprehensive representation of breast cancer images, improving detection performance across diverse datasets. The remainder of this paper is organized as follows: Sect. 2 reviews the literature and related work, Sect. 3 outlines the materials and methods used in this research, Sect. 4 offers a detailed discussion of the experiments and results, and Sect. 5 concludes this paper with future directions.
Literature review
Recent advancements in medical imaging have significantly improved breast cancer detection and classification. Automated computer vision techniques now support early diagnosis and treatment, providing time-efficient and reliable assistance for radiologists23. Various imaging modalities, including mammography (MG), ultrasound (US), computed tomography (CT), and magnetic resonance imaging (MRI), have been widely employed for breast cancer analysis24.
A DenseNet-II Neural Network model achieved 94.55% accuracy on the DDSM dataset using 10-fold cross-validation by integrating handcrafted features such as Gabor Initial Spatial Transform (GIST), Histogram of Oriented Gradients (HOG), Local Binary Patterns (LBP), and Scale-Invariant Feature Transform (SIFT)25. Deep transfer learning models, including ResNet, DenseNet, and VGG, demonstrated classification accuracies of 87.93% and 90.91% across INbreast and CBIS-DDSM datasets, while pre-trained CNNs like ResNet50 achieved 88% accuracy in detecting mammograms abnormalities26.
Further improvements are achieved with a deep CNN (ResNet50) that employed image enhancement, segmentation, and feature selection, achieving 93% accuracy on INbreast data27. Similarly, FV-Net processed bilateral MG images with CLAHE and truncated normalization, achieving 87.34% accuracy28. Data augmentation using Generative Adversarial Networks (GANs) enhanced ultrasound datasets and improved breast mass classification performance29. Traditional approaches, such as Support Vector Machines (SVM) combined with Gray-Level Co-Occurrence Matrix (GLCM) features and K-means segmentation, reported 94% accuracy in tumor classification30.
Ultrasound imaging also demonstrated promising results. Custom CNN models achieved 96% accuracy on Rodrigues dataset, with sensitivity of 97.3% and specificity of 94.12%31. Ensemble strategies combining VGGNet, ResNet, and DenseNet yielded 94.62% accuracy32, while statistical modeling using Rician Inverse Gaussian (RiIG) achieved 97.20% accuracy33. Feature selection techniques such as Minimum Redundancy Maximum Relevance (mRMR) combined with SVM, reached 95.6% accuracy34.
Several studies optimized pre-trained CNN architectures, including AlexNet, DenseNet201, MobileNetV2, ResNet18, ResNet50, VGG16, and Xception, using preprocessing methods such as resizing, augmentation, and standardization35. Deep representation scaling (DRS) layers integrated into CNN blocks further improved classification, achieving 91.5% accuracy and an Area Under the Curve (AUC) of 0.95536. Other approaches, involving ROI cropping, noise removal, and RGB image fusion with VGG19, attained 87.9% accuracy across benign and malignant classes37.
Hybrid and ensemble approaches also showed strong performance. Transfer learning models combined with ensemble techniques, such as InceptionV3 with stacking, achieved an AUC of 0.94738. Graph Convolutional Networks (GCN) analyzed malignancy patterns in ultrasound images, obtaining 94.37% accuracy by leveraging clinical features such as WH-ratio, Harris Corner, and Circularity39. Hybrid CNN classifiers integrating MG and US datasets demonstrated improved performance by combining handcrafted features (e.g., LBP, HOG) with deep features, classified using ML algorithms such as XGBoost, MLP, and AdaBoost22.
Classical ML classifiers, including SVM, Random Forest, K-Nearest Neighbor (KNN), and Naïve Bayes, achieved accuracies up to 95.6% with 10-fold cross-validation40. Transfer learning methods such as DenseNet201 and MobileNetV2 reported accuracies of 92.67% and 93.41% on MG and US datasets, respectively41. Pre-processing methods also played an essential role: for instance, the Modified High Boosting Laplacian of Gaussian (LoGMHP) filter42, when applied with AlexNet, ResNet18, and MobileNetV2, optimized performance on MG and US images.
One prominent work by Puttegowda et al.43 utilized MG and US images from the Mini-DDSM and BUSI datasets, incorporating DL models like ResNet-18, achieving an impressive 93.78% accuracy for the classification of benign and malignant cases. Similarly, another study focused on ultrasound images, using a multimodal deep learning approach for 290 breast masses from a Breast Ultrasound dataset, reporting 86% accuracy44. A key contribution from Ahmad et al.45 involved the application of YOLO for detection and ResUNet for segmentation, using the CBIS-DDSM mammogram dataset, achieving a stellar accuracy of 99.16%. Moreover, a hybrid CNN model employed with VGG16 and PCA for feature reduction46, achieving 99.5% accuracy for histopathological images. This is further complemented by47, which utilized BreakHis-400x and BUSI datasets, deploying DenseNet121 for classification, showing accuracy rates of 93.97% and 89.87%, respectively.
The study conducted by Qasrawi et al.48 applied a hybrid ensemble model and CLAHE for ultrasound image enhancement, achieving 94% overall accuracy, with higher accuracy for benign (91.4%) and malignant cases (84%). The paper discusses the use of a novel hybrid framework, the convolutional neural bidirectional feature pyramid network (CNBiFPN), designed for early detection and precise localization of breast cancer in CT scan images. The model achieves a significant classification accuracy of 96.11%, outperforming traditional models such as CNN and CNN-HD, which had accuracies of 94.44% and 95.01%, respectively49. Another, study demonstrated that optimizing the ENN model with different optimization algorithms, such as ADAM, RMSProp, and SGD, improved classification accuracy50. The ADAM optimizer achieved the highest accuracy of 98.99% on the training set, with a 95.00% accuracy on the test set. Several studies are conducted using various pre-trained CNN models, such as InceptionV3, ResNet152V2, MobileNetV2, VGG-16 and DenseNet-121, SVM, GCN and attention mechanisms51,52.
Table 1 summarizes the existing techniques, including models, methods used, reported limitations, and key observations for breast cancer detection and classification.
Material and methods
This section presents details of the datasets used for our research and the detailed workflow of the proposed approach.
Datasets
Mammogram and ultrasound images are used for multi-modal breast cancer detection, with datasets from publicly available repositories. The Mini-DDSM53 comprises 9684 X-ray images from 2,620 cases, including both Craniocaudal (CC) and Mediolateral Oblique (MLO) views. The MLO images are taken at 30 or 60 degrees, while CC images are captured from above. The INbreast dataset55, developed at the University of Porto, contains 410 images from 115 patients in 16-bit DICOM format, classified as normal, benign, or malignant, with both MLO and CC view. For ultrasound, the Rodrigues dataset38 from Mendeley Data includes 250 grayscale images (150 malignant, 100 benign). The BUSI dataset53 from Kaggle contains 780 grayscale images from 600 women (133 normal, 210 malignant, 437 benign).
To balance sizes, a subset of 600 Mini-DDSM images (200 each of normal, malignant, and benign) was used. INbreast was divided into two classes (67 normal, 269 abnormal) for comparison with the binary Rodrigues dataset. Table 2 summarizes all datasets, and Fig. 1 shows sample images: A–C (Mini-DDSM), D–E (INbreast), F–G (Rodrigues), H–J (BUSI).
Methodology
An efficient model for breast cancer identification and detection is presented, comprising several stages: image collection, pre-processing, data augmentation, image enhancement, segmentation, feature extraction, feature selection, feature fusion, performance evaluation, and final results. The detailed workflow is presented in Fig. 2.
Collection of images
The mammogram and ultrasound datasets used in this research are described in Table 2.
Pre-processing
Pre-processing was applied to all MG and US images to ensure compatibility with model requirements and computational efficiency before input to the pre-trained TL models (Resnet50, InceptionV3, and MobileNetV2). As datasets contained unequal image sizes, all images were resized to 224 \(\times\) 224. Pixel values were normalized to the range [0, 1] by dividing each by 255.0, improving convergence and robustness35. The scaling was performed using Eq. (1).
Here, original_image represents the pixel values of the original image, whereas Scaled_image represents the image after scaling, with each pixel value divided by 255.0. Mean subtraction and standard deviation normalization were applied to centralize pixel values and reduce illumination variations35. The mean and std were calculated per RGB channel, and images were normalized using Eq. (2):
Here, original_image is the input image, \(\mu\) represents the mean value for each channel and std denotes the standard deviation for each channel. This ensures numerical stability, faster convergence, and consistent pre-processing across MG and US datasets.
Data augmentation
Training data augmentation is used to address class imbalance and increase the size of the dataset without the need for additional labeled data. This approach makes the model more robust to variations and helps prevent overfitting21. The augmentation is applied only to the training data of all MG and US images. The applied augmentation technique is Shearing which is a transformation that shifts one part of an image in a fixed direction, keeping the rest of the image fixed. Both the horizontal and vertical shearing are applied to the images with the angles [−0.20, 0.20]. The augmentation ratio is set to 1:1, meaning the augmented dataset contained an equal number of samples as the original dataset. It enhances robustness by exposing the model to various perspectives. The shear angle range [−0.20, 0.20] was chosen based on empirical testing to balance image variation without distorting image quality. Here, \(-0.20\) means Minimum Absolute Shear and \(0.20\) means Maximum Absolute Shear. Shearing may create undefined regions in images. Setting fill mode to nearest resolves this by filling gaps with the nearest pixel values, preserving image consistency and ensuring augmented images remain representative of the original data. We also tested the ’reflect’ mode, but no significant performance improvement was observed, therefore, the ’nearest’ mode was selected for its simplicity and computational efficiency.
Image enhancement
The image enhancement is applied to MG images only from both datasets after the pre-processing of images. US images do not need to be enhanced as they are already small in size and ROI is extracted. However, MG images need to be enhanced due to low contrast and noise in the images. The image enhancement improves the contrast and removes the noise for better generalization of microcalcifications or masses. This stage is further effective in the image segmentation. Figures 3A, B, C, D and 4A, B, C, D show the image enhancement on Mini-DDSM dataset Images and INbreast dataset images, respectively. The images are enhanced by applying different techniques such as CLAHE, Wavelet Transform, and Sharpening of the images.
CLAHE
The CLAHE enhances image contrast by applying histogram equalization in even small parts or regions of an image and also helps to reduce noise. It improves visualization of breast tissues and structures for the detection of abnormalities. The CLAHE operates by dividing the image into small sections known as tiles (e.g., 8×8). It then applies histogram equalization to each tile independently. To limit the noise amplification, CLAHE applies a contrast limiting procedure, which redistributes the histogram values and clips it at a predefined contrast limit before redistributing the clipped pixels equally across the histogram56. Figures 3B and 4B show the CLAHE application on the original images of the Mini-DDSM and INbreast datasets, respectively, and Eq. (3) represents its mathematical formulation.
Here, \(C_{\text {new}}(x,y)\) is the new pixel value after CLAHE, \(N\) is the number of pixel values (usually 256 for 8-bit images), \(H(i)\) represents the histogram frequency of the pixel intensity \(i\) in the local region or tile, where \(I(x,y)\) is the original pixel value, and \(\max (H)\) is the maximum value of the histogram after clipping.
Wavelet transform
After applying CLAHE to enhance local contrast, the images are further processed using the wavelet transform to decompose them into different frequency components. This method focuses on the lower-frequency components while allowing both spatial and frequency localization. The Haar wavelet transform57 is used to calculate these low frequencies, which smooths the image and enhances its edges and textures. The 2D Discrete Wavelet Transform (DWT) decomposes an image into four sub-bands:
-
LL (Approximation) Captures the low-frequency components.
-
LH (Horizontal Detail) Captures horizontal edge information.
-
HL (Vertical Detail) Captures vertical edge information.
-
HH (Diagonal Detail) Captures diagonal detail components.
The wavelet transform is mathematically represented in Eq. (4).
Here, \(I\) is the input CLAHE_image, \(x\) and \(y\) are the image coordinates, denoting the horizontal and vertical pixel positions, \(LL = L\left( I\left( x,y\right) \right)\), \(LH = H\left( L\left( I\left( x,y\right) \right) \right)\), \(HL = L\left( H\left( I\left( x,y\right) \right) \right)\), and \(HH = H\left( H\left( I\left( x,y\right) \right) \right)\). The LL band provides a smoothed approximation of the image, while LH, HL, and HH capture various details in different orientations.
Once the CLAHE-enhanced image is decomposed into wavelet sub-bands, the original image can be reconstructed using the inverse DWT (IDWT). This restores the image representation in the spatial domain while retaining essential features extracted during decomposition. The process is mathematically represented in Eq. (5).
Here, \(I^{\prime }(x, y)\) is the input CLAHE_image. Figures 3C and 4C represent the Reconstructed Wavelet Transform images of Mini-DDSM and INbreast datasets, respectively.
Sharpening of the images
Following wavelet transform, images are further sharpened to enhance edge information and fine details. Each image is first decomposed into frequency components, after which a sharpening function is applied using a convolutional filter. This process highlights subtle features58, aiding in the detection of microcalcifications and other small abnormalities in the image. The sharpened wavelet-reconstructed images from the Mini-DDSM and INbreast datasets, are shown in Fig. 3D and 4D, respectively. The sharpening function is mathematically expressed in Eq. (6).
Here, \(I_{\text {sharpened}}\) is the sharpened image, \(I\) is the original image, \(\alpha\) is the scaling factor (set to −0.5), and \(\text {Blur}(I)\) denotes the Gaussian-blurred version of the original image.This operation enhances local contrast, emphasizing edges and fine structural patterns critical for breast cancer detection.
Image segmentation
After boosting contrast and sharpness in the image enhancement stage, the next stage involves segmenting the image into distinct regions. This process identifies and isolates region of interest (ROI) by leveraging the enhanced features, thereby enabling precise analysis and accurate detection of abnormalities59.
Image segmentation on Mini-DDSM MG dataset
The Mini-DDSM MG dataset contains tumor or lesion regions marked with a red circle in benign and malignant class images, while normal images appear clean with no visible abnormality. Creating a mask is essential for extracting the ROI from these images60. In this step, the mask image is generated and saved for further analysis. Figure 5A, B and C illustrates the segmentation process applied to the enhanced Mini-DDSM images.
For each image in the dataset, the standard Blue, Green, and Red (BGR) format is first converted into the Hue, Saturation, Value (HSV) format, which simplifies the detection of specific colors. In this case, red color detection was used to identify the highlighted tumor regions. A binary black-and-white mask was then created, corresponding to the largest red contour area within the image. The complete procedure of contour detection and mask generation is presented in Algorithm 1.
Image segmentation on INbreast MG dataset
Unlike the Mini-DDSM dataset, the INbreast dataset does not provide highlighted tumor regions or mask images. Therefore, the image enhancement process shown in Fig. 4A, B, C and D is is essential for segmenting tumor region or lesions in benign and malignant images. The ROI is segmented directly from the enhanced images. Since the images are already pre-processed, additional steps are required to isolate abnormal regions.
Otsu’s multi-thresholding method61 is applied to the grayscale images to classify them into three distinct intensity classes. Based on these thresholds, the images are digitized into separate regions. The brightest region, corresponding to the highest intensity class, is then segmented, as shown in Fig. 6B. For Otsu’s method with k thresholds, the between-class variance is defined in Eq. (7).
where, \(\omega _i\) is the weight (probability) of the i-th class, \(\mu _i\) is the mean intensity of the i-th clas, and \(\mu _T\) is the overall mean intensity of the image. The segmented region is then converted into a binary image. The binary conversion B (x,y) is expressed in Eq. (8).
where, I(x, y) represents the intensity value of the pixel located at coordinates (x, y) in the grayscale image, and t is the threshold value (0–255) distinguishing foreground from background. Morphological opening and closing operations are applied to the binary segmented image to remove noise and refine the segmented area. A 3 \(\times\) 3 kernel is used for these operations. The morphological operations are defined in Eqs. (9) and (10).
where K is the \(3 \times 3\) kernel, \(\min\) is the erosion operation, and \(\max\) is the dilation operation. Contours are then detected in the cleaned segmented image. The boundaries of connected components are identified within Bclosed. If multiple contours are present, the largest contour is selected, assuming it corresponds to the ROI. A bounding rectangle is computed around this contour, and the ROI is extracted from the original grayscale image. The processes of finding the largest contour, creating a bounding rectangle, and extracting the final ROI image are mathematically represented in Eqs. (11), (12), and (13).
where Area(C) is the number of pixels within contour C, the bounding box \(R\) is represented by its top-left \((x_{\text {min}}, y_{\text {min}})\) and bottom-right \((x_{\text {max}}, y_{\text {max}})\) coordinates, ROI is the final extracted Region of Interest.
Finally, binary thresholding is employed to generate a mask for the segmented image, as shown in Fig. 6C. Pixels with intensity values greater than or equal to 127 are assigned a value of 255 (white), while those below 127 are set to 0 (black). Once the binary mask is created, it is applied to the segmented image, as demonstrated in Fig. 6D. A bitwise AND operation is used, preserving pixel values where the mask is white (255) and setting all others to 0. Figure 6A, B, C and D provide a comprehensive overview of the image segmentation, mask creation, and application processes on the enhanced images. Additionally, Algorithm 2 outlines the step-by-step procedure for generating the binary mask and applying it to the original image.
Image segmentation process for rodrigues US dataset
The Rodrigues dataset is relatively small and the ROI is already cropped in the available images. Therefore, no additional segmentation is applied beyond the pre-processing steps. The dataset images are directly passed to the proposed models.
Image segmentation process for BUSI US dataset
The BUSI dataset includes a folder containing binary mask images that highlight tumor or lesion regions. For accurate classification of benign, malignant, and normal US images, the pre-processed images are combined with their corresponding masks to isolate the ROI. The mask-application process follows the same procedure described in Algorithm 2. The resulting segmentation steps for BUSI images are illustrated in Figure 7A, B and C.
Feature extraction
In the feature extraction process, both deep learning-based and traditional features are derived from the images to aid in breast cancer classification and detection. Traditional features, such as Gabor filters and wavelet transforms, captured low-level texture and spatial details62, while deep features provided high-level semantic representations through complex neural networks. Analyzing the correlation between these feature sets revealed their complementary nature–traditional features preserved fine-grained structural information that deep learning models might overlook, whereas deep features captured abstract patterns beyond the reach of conventional methods. By integrating both, we achieved a more comprehensive representation of the image data, enhancing classification accuracy and improving overall model performance.
Deep features
To enhance our feature set and leverage modern advancements in ML, deep features are extracted. TL allows the transfer of learned features from a large dataset, such as ImageNet, to a smaller, domain-specific dataset to improve performance and reduce training time. In this study, we use three TL models: InceptionV3, ResNet50, and MobileNetV2. These models are pretrained on ImageNet, which consists of natural images. While ImageNet images differ significantly from medical images in terms of modality and visual characteristics, the generic visual features (such as edges, textures, and shapes) learned from ImageNet have been shown to be transferable to a variety of tasks, including breast tumor classification. In our approach, the output from the penultimate layer of each model was used to extract the features, enabling us to capture high-level semantic information relevant to the task at hand. This approach enables us to capture high-level features that are critical for our specific task.
The feature extraction is completed by including the “GlobalAveragePooling2D” layer in each of these pre-trained models. This layer reduces the spatial dimensions of each feature map into a single vector, resulting in a compact representation of the extracted features. All layers of the pre-trained models are locked to maintain fixed weights throughout the feature extraction process.
For a fair comparison, the same hyperparameters values are maintained for both the US and MG image datasets. Hyperparameters and their corresponding values used in the model creation and compilation are provided in Table 4. A total of 167,872 deep features are extracted from these three models. A detailed description, structure, and working of each TL model are discussed below.
InceptionV3 transfer learning structure
In this research, the inceptionV3 deep CNN architecture is utilized, a model specifically designed for image classification. Developed by Google, InceptionV3 is capable of capturing complex patterns in images while maintaining computational efficiently. The model integrates multiple layers and operations within its architecture, comprising a total of 48 layers. Key components include convolutional layers, max pooling operations, inception modules, factorization, auxiliary classifiers, and batch normalization. A visual representation of the InceptionV3 model is provided in Fig. 8.
ResNet50 transfer learning structure
In this research, the ResNet50 model, a TL architecture introduced by Microsoft, was employed. ResNet50 leverages residual learning to effectively train very deep networks, addressing the common issue of vanishing gradients. The model comprises 50 layers, including convolutional, pooling, and fully connected layers, organized into blocks of residual units. Pre-trained on the large-scale ImageNet dataset - containing millions of images across thousands of categories - ResNet50 provides rich and diverse feature representations that can be fine-tuned for breast cancer image classification. A visual representation of the ResNet50 architecture is shown in Fig. 9.
MobileNetV2 transfer learning structure
MobileNetV2, a deep CNN model developed by Google, is also used in this research. Designed for efficient performance on mobile and embedded vision applications, MobileNetV2 achieves lightweight computation without compromising accuracy. Its core innovation lies in the use of depthwise separable convolutions, combined with linear bottlenecks, which streamline the architecture and improve efficiency. The model comprises multiple layers, including standard convolutions, depthwise separable convolutions, and bottleneck layers, which collectively enhance training robustness and prevent saturation. A visual representation of the MobileNetV2 architecture is shown in Fig. 10.
Traditional features
Traditional features are also extracted from the images, specifically focusing on Gabor and Wavelet features. Gabor features are well-regarded in image processing for texture analysis due to their ability to capture spatial frequency characteristics63, making them highly effective for edge detection and texture representation.
In contrast, wavelet features capture both spatial and frequency information at different scales, facilitating multi-resolution analysis of images. By decomposing images into different frequency components, wavelet transforms allow for the analysis of complex patterns and structures. The proposed approach is strengthened by using both types of features for breast cancer detection, improving the models robustness and accuracy.
Gabor filter features
Gabor filters are linear filters used for edge detection, texture representation, and disparity measurement64. In the context of the proposed research, Gabor filters features are extracted from US and MG images. The extraction process involves converting the image to grayscale and then applying Gabor filters with various orientations (0, \(\pi /4\), \(\pi /2\), \(3\pi /4\)) and frequencies (0.1, 0.3, 0.5). The real and imaginary parts of the filtered images are then computed, and their means are taken as the features. The texture information at different scales and orientations is captured in this method. A total of 24 Gabor features are extracted per image. The Gabor filter process applied to the images is shown in Fig. 11.
Wavelet transform features
Wavelet transforms are another powerful method for feature extraction, offering a multi-resolution analysis of images65. Wavelet transforms use a time-frequency representation, allowing both spatial and frequency information to be captured simultaneously. This is achieved by decomposing an image into a series of wavelets, which are small waves that vary in scale and position.
For the wavelet features extraction, all images are converted to grayscale. The Discrete Wavelet Transform (DWT) is then applied using the Haar wavelet, decomposing each image into four distinct sub-bands: the approximation coefficients (LL) and three detail coefficients (LH, HL, HH). The low-frequency content of the image is represented by the approximation coefficients, while variations in the horizontal, vertical, and diagonal directions are captured by the detail coefficients. The function then flattens and concatenates these coefficients into a single feature vector, which represents the images wavelet features.
Equations for wavelet transform features are presented in section 3.2.4. A pictorial overview of the complete DWT process is illustrated in Fig. 12. During this feature extraction phase, a total of 50,176 wavelet features are extracted per image.
Feature selection
The total number of features extracted from Gabor, wavelet, and deep models make a large feature set. However, not all features are equally significant regarding efficiency and accuracy. Therefore, feature selection is conducted to find out the most relevant features from a larger set, which improves model performance and decreases computational complexity. This process involves evaluating and ranking features according to their statistical significance concerning the target variable66. This univariate technique assesses the significance of each feature based on its statistical relationship with the target variable, specifically using the ANOVA F-value.
This metric assesses the variance between feature groups compared to the variance within each group, highlighting features that most effectively differentiate the target variable. A technique for selecting features involves scoring them using the ANOVA F-value and then choosing the top-scoring features based on this evaluation. This feature selection stage helps in balancing between retaining sufficient information and reducing dimensionality. The F-value is calculated using Eq. (14).
Between-Group Variance measures the variance of the feature values between different classes or groups and is calculated using Eq. (15).
where \(n_i\) = Number of samples in group \(i\), \(\overline{X_i}\) = Mean of the feature values in group \(i\), \(\overline{X}\) = Overall mean of the feature values, and \(k\) = Number of groups. Within-Group Variance measures the variance of the feature values within each class or group and is calculated using Eq. (16).
where \(X_{ij}\) represents the feature value of the \(j\)-th sample in group \(i\), and the total number of samples is represented by \(N\).
Before feeding the feature set into the classifiers, feature selection is performed using the ANOVA F-value method. This method ranked the features based on their ability to distinguish between the malignant, benign, and normal classes, and the top 600 most discriminative features by evaluating the variance between feature groups are selected. No additional dimensionality reduction techniques, such as Principal Component Analysis (PCA) or Linear Discriminant Analysis (LDA), are applied, as the feature selection process is sufficient for reducing the feature space to the most relevant features for classification.
Feature fusion
In this specific scenario, the top 10 Gabor features are selected out of 24. For the wavelet features, only the top 40 features are selected from the initial total of 50,176. Similarly, the top 550 deep features are retained from a total of 167,872. These features are concatenated into a single hybrid feature vector, integrating both high-level and low-level features for improved classification accuracy. Consequently, the final feature fusion set comprises 600 features selected from an original pool of 218,072, makeing a feature vector, which significantly reduces the number of features while preserving the most valuable ones for analysis. Figure 13 contains a detailed overview of feature fusion.
Classification
The Best Features are now selected in the feature selection stage. Fusion of these features is also completed and a final feature vector is obtained successfully. This research utilizes several popular ensemble learning classifiers, including XGBoost, AdaBoost, and CatBoost. These methods are known for their ability to handle imbalanced datasets effectively by focusing on the minority classes during the classification process, thus improving the model’s performance on underrepresented classes. In this study, hyperparameters are selected based on preliminary experiments and established best practices, rather than through systematic optimization methods like grid search or random search. We empirically chose a n-estimators of 500, a learning rate of 0.1, and a random-state of 42 to balance performance and computational efficiency. The detailed description of each ensemble classifier is discussed below.
XGBoost In this research, the XGBoost classifier is utilized, a powerful ensemble algorithm that leverages gradient boosting to enhance model performance. An ensemble of weak learners, usually decision trees (DT), is created, where each subsequent tree is trained to rectify the mistakes of its predecessor67. This iterative process minimizes a loss function using gradient descent, resulting in a robust and optimized model. Predictions are refined at each boosting iteration by combining the previous prediction with adjustments from the new weak learner, scaled by a learning rate. XGBoost also incorporates a loss function to measure prediction errors and includes a regularization term to control model complexity, preventing overfitting and ensuring generalization.
AdaBoost AdaBoost combines weak classifiers into a strong classifier by focusing more on instances misclassified in previous iterations68.It adjusts weights to emphasize harder-to-classify examples, thereby improving accuracy.
The weight of each weak classifier is updated based on its performance, assigning higher importance to classifiers with lower error rates. The final strong classifier is then computed as a weighted sum of all weak classifiers.
CatBoost CatBoost is a gradient-boosting algorithm that is used in this research. It is designed to effectively manage categorical features and minimize overfitting69.It employs several techniques to enhance performance, including ordered boosting to address the problem of target leakage. It lessens the likelihood of overfitting, providing a more reliable model by building trees based on an ordered permutation of the data.
By integrating these advanced classification techniques; XGBoost, AdaBoost, and CatBoost significantly improve classification accuracy, demonstrating the effectiveness of boosting methods in handling complex datasets.
The methodology of this research proposes a hybrid feature fusion method that integrates deep features from pre-trained models (ResNet50, InceptionV3, and MobileNetV2) with traditional features (Gabor filters and wavelet transforms). The feature fusion process combines these features to capture both high-level semantics and low-level textures, ensuring enhanced breast cancer detection performance. The Methodology section outlines all necessary steps for replicating this process, including dataset preparation, image pre-processing, and the feature fusion and classification pipeline.
Results and discussions
The results of the proposed hybrid framework are presented and analyzed in this section. A comprehensive evaluation is carried out across mammogram and ultrasound datasets to demonstrate the robustness and generalizability of the approach. The subsections are organized to highlight different aspects of the analysis, including the performance of transfer learning models on individual datasets, the effectiveness of pre-processing and enhancement techniques, the role of segmentation in isolating regions of interest, the contribution of traditional and deep feature extraction methods, the impact of hybrid feature fusion, and the comparative performance of ensemble classifiers. Each subsection provides an in-depth discussion supported by experimental results, illustrating how the proposed methodology advances breast cancer detection and classification.
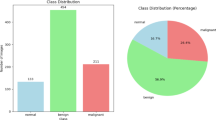
Distribution of the datasets
The MG and US images datasets are divided into training and testing sets with either two or three class distributions, are shown in Table 3. It is worth noting that a 90–10 split is adopted for the mammogram datasets (Mini-DDSM and INbreast) to preserve sufficient training samples in relatively balanced distributions, while a 70–30 split is used for the ultrasound datasets (BUSI and Rodrigues) to ensure adequate evaluation of their more imbalanced and heterogeneous data. For the Mini-DDSM dataset, a total of 600 images are used, with 540 images allocated for training and 60 for testing. The INbreast dataset consisted of 336 images, with 302 used for training and 34 for testing. The Rodrigues dataset consisted of 250 images, divided into 175 for training and 75 for testing. Lastly, the BUSI dataset included 780 images, with 545 images used for training and 235 for testing.
Environmental setup
The experiments are conducted using a Dell laptop with the following specifications: Intel Core i5-3337U CPU @ 1.80GHz, 8GB DDR3 RAM, x64-based processor, and a 512GB SSD running Windows 10 Pro. For model training and evaluation, Google Colab was emplyed, providing a cloud-based Jupyter Notebook environment with access to GPU and TPU resources. The environmental setup is configured with Python 3.7, TensorFlow 2.4.1, and scikit-learn 0.24.2. Table 4 contains the hyperparameters and their values that are used for TL model training and testing.
The models are trained using the Adam optimizer with a learning rate of 1e-3. This learning rate is chosen based on empirical testing. A batch size of 8 is used due to hardware limitations and memory constraints. We initially experimented with training for more than 12 epochs, but found that the results became stable after 12 epochs, with no significant improvement in training loss or validation accuracy. Therefore, 12 epochs are deemed sufficient for initial testing and model evaluation.
Evaluation metrics
In this study, widely accepted metrics commonly employed in breast cancer screening, such as accuracy, precision, specificity, sensitivity, and F1-Score are employed. These metrics are derived from the Confusion Matrix (CM), which includes the following parameters: True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN).
The CM offers an overview of the models predictions versus actual labels, detailing TP, TN, FP, and FN counts. It helps identify error types and provides insights for improvements. Model performance is further evaluated using the Area Under the Curve (AUC), which measures the classifiers ability to distinguish between benign, malignant, and normal classes–values closer to 1 indicate high effectiveness. AUCs are generated using the predicted probabilities obtained from the predict-proba method. The roc-curve function is used to compute the False Positive Rate (FPR) and True Positive Rate (TPR) for each class, and AUC values are calculated to evaluate model performance. Classification accuracy is also computed as the ratio of correctly classified cases to the total cases.
Experimental setup
The experiments are structured as follows:
Experiment 1: Evaluation using Mini-DDSM MG Dataset In this experiment, we evaluate the proposed method on the Mini-DDSM mammogram dataset. The focus is to assess the model’s ability to detect breast cancer in mammogram images, particularly concerning accuracy, precision, and recall.
Experiment 2: Evaluation using INBreast MG Dataset This experiment tests the model’s performance on the INBreast mammogram dataset, allowing a comparison with Experiment 1 and evaluating the model’s robustness across distinct mammogram images.
Experiment 3: Evaluation using Rodrigues US Dataset The third experiment shifts focus to the Rodrigues ultrasound (US) dataset. It evaluates the hybrid model’s performance on ultrasound images, highlighting its effectiveness in detecting breast cancer in a different imaging modality.
Experiment 4: Evaluation using BUSI US Dataset The final experiment assesses the model on the BUSI ultrasound dataset. This phase helps gauge the model’s generalization ability across ultrasound data and provides a comprehensive evaluation of performance across different modalities.
By structuring the experiments in this manner, we aim to clearly present the results for each dataset and modality, enabling a deeper understanding of the model’s capabilities and areas for future enhancement.
Experiment 1: evaluation using mini-DDSM MG dataset
Table 5 shows the performance of three hyper-tuned ensemble classifiers applied to the dataset. The first metric values are for benign class samples, the second metric values are for malignant class samples and the third metric values are for normal class samples. Among these classifiers, the XGBoost Classifier achieved the highest test accuracy at 95%, followed closely by the AdaBoost Classifier with 93.33% and the CatBoost classifier with a test accuracy is 86.67%.
Figure 14 offers an in-depth comparison between the models predictions and the actual ground truth labels, displaying the counts of TP, TN, FP, and FN.
To assess the performance of the proposed model, we employed the ROC curve illustrated in Fig. 14. This curve enables the evaluation of the trade-off between sensitivity (True Positive Rate) and specificity (False Positive Rate), offering valuable insights into the models diagnostic accuracy.
The model performs exceptionally well for both benign and malignant classes, achieving AUC values of 0.97 for XGBoost and CatBoost. In contrast, AdaBoost shows a slight decrease to 0.94, reflecting a minor variation in the models capacity to differentiate between these classes. Overall, the model exhibits high diagnostic accuracy with minimal performance fluctuations.
Experiment 2: evaluation using INBreast MG dataset
Among the already discussed ensemble classifiers, XGBoost Classifier achieved the highest test accuracy at 97.06%, followed by CatBoost with 97.06% and AdaBoost test accuracy is 94.12%. Table 6 shows the detailed results including accuracy, precision, recall, specificity, F1-score, and AUC values. The first metric values are for abnormal class samples and the second metric values are for normal class samples.
Figure 15 illustrates the CM and ROC curve corresponding to each ensemble classifier. The AUC for XGBoost and CatBoost classifiers are 1.00 for the normal class and 1.00 for the abnormal class, indicating that the model achieves complete discrimination without any false positives or false negatives. In these classifiers. Whereas for the AdaBoost classifier ROC curve demonstrates slightly reduced performance, with an AUC of 0.99 for both abnormal and normal classes. Overall, the classifiers demonstrate excellent diagnostic accuracy, with a minor fluctuation observed in the AdaBoost classifier.
Experiment 3: evaluation using rodrigues US dataset
Table 7 shows the detailed results, the first metric values are for benign class samples and the second metric values are for malignant class samples. Among these classifiers, XGBoost achieved the highest test accuracy at 98.67%, followed by AdaBoost with 98.67%. Whereas, the CatBoost classifier also performed well, with a test accuracy of 96%.
Figure 16 illustrated the CM and ROC curve corresponding to each ensemble classifier on Rodrigues Dataset. The AUC for all classifiers is 1.00 for the benign class and 1.00 for the malignant class, indicating that the model accurately distinguishes between these classes without any misclassifications. Overall, the classifiers demonstrate excellent diagnostic accuracy for all three classifiers.
Experiment 4: evaluation using BUSI US dataset
Table 8 shows the performance of the BUSI US dataset, the first metric values are for benign class samples, the second metric values are for malignant class samples and the third metric values are for normal class samples. Among these classifiers, XGBoost achieved the highest test accuracy at 97.02%, followed closely by CatBoost with 94.89% and AdaBoost test accuracy is 92.77%.
Figure 17 illustrates the CM and ROC curve for all three ensemble classifiers. AUC for the XGBoost classifier is 0.99 for benign, 0.99 for malignant, and 1.00 for normal, indicating that the model achieves high diagnostic performance across all classes, with perfect discrimination for the normal class. AUC for the AdaBoost classifier is 0.98 for benign, 0.97 for malignant, and 1.00 for the normal class. Whereas AUC for CatBoost is 0.99 for both benign, 0.98 for malignant, and 1.00 for the normal class. Overall, the model demonstrates high diagnostic accuracy with minimal fluctuations in performance.
Comparative analysis
In this section, the aforementioned ML classifiers with the datasets used in this research is compared. As shown in Fig. 18, XGBoost and AdaBoost generally performed better, while CatBoost consistently achieved comparatively lower accuracy. Similarly, Fig. 19, illustrates that the Rodrigues dataset achieved the highest accuracy, whereas other datasets demonstrated relatively lower performance.
From Figs. 18 and 19, it can be concluded that XGBoost, AdaBoost, and CatBoost exhibited varying performance across datasets, influenced by dataset characteristics. The Mini-DDSM dataset, being larger and more balanced, enabled XGBoost to capture more complex patterns, leading to higher accuracy. In contrast, AdaBoost faced challenges with Rodrigues, where the class imbalance (with fewer benign samples) led to suboptimal performance. CatBoost, designed to handle categorical data effectively, performed comparatively better on the BUSI dataset, where its robustness to noise and adaptability to smaller dataset proved advantageous. These findings emphasize that dataset-specific characteristics, such as size, class distribution, and feature diversity, play a crucial role in shaping classifier performance.
As discussed in Sect. 2, multiple pre-processing techniques, data augmentation strategies, and both TL and ML models are applied. Additionally, different testing percentages are emplyed in the classification process. Such variations can positively or negatively affect outcomes, making it important to evaluate each study within its own context. To provide a comprehensive comparison, Tables 9, 10, 11 and 12 present year-wise classification results for each dataset alongside the outcomes of the proposed model.
The Mini-DDSM dataset showed high performance, with XGBoost achieving 95.00% accuracy, further supported by the ROC curves and confusion matrices. For the INbreast dataset, both XGBoost and CatBoost achieved 97.06% accuracy, demonstrating strong performance with minimal misclassification. On the Rodrigues ultrasound dataset, XGBoost and AdaBoost achieved excellent accuracy of 98.67%, as illustrated by ROC analyses. Similarly, for the BUSI dataset, XGBoost also performed robustly with 97.02% accuracy.
Table 13, compares our proposed study with recent related works. The proposed approach demonstrates significant performance improvements across multiple datasets, achieving 95% accuracy on Mini-DDSM, 97.06% on INbreast, 98.67% on Rodrigues, and 97.02% on BUSI. These results are competitive with state-of-the-art methods, such as those achieving 99.16% with YOLO and ResUNet45 and 99.5% with hybrid CNN-VGG16 architectures46. However, our approach stands out due to its integration of traditional features (e.g., Gabor filters), which enhance robustness – particularly for ultrasound images, where deep learning models may fail to capture fine textural patterns. Furthermore, our ensemble framework, combining XGBoost, AdaBoost, and CatBoost, provides additional robustness by leveraging the strengths of multiple models. This is particularly beneficial in handling class imbalance, a common challenge in medical imaging datasets.
Unlike studies that focus on a single modality or dataset, our work encompasses both mammogram and ultrasound datasets, thereby offering broader generalization and applicability. While methods such as43 and48 demonstrate high performance with individual models, our hybrid approach—combining deep learning with traditional texture-based features—delivers a more versatile and effective solution for diverse clinical environments. The use of advanced preprocessing techniques, such as CLAHE, further enhances detection accuracy, especially in distinguishing malignant and benign cases across modalities. The integration of hybrid feature fusion with ensemble classifiers allows our approach to achieve consistently high accuracy while maintaining robustness, making it a promising candidate for real-world clinical applications.
Nevertheless, several limitations must be acknowledged. First, the proposed method is evaluated separately on mammograms and ultrasounds datasets. Second, fixed hyperparameters are employed for classifiers, which may not be optimal across all datasets; further fine-tuning could improve model performance. Third, while the proposed method shows potential, its scalability and real-world applicability remain uncertain, as it has not yet been tested on larger, more diverse datasets or in clinical environments. Although the model performed well overall, it may misclassify images with poor quality, such as noisy or low-contrast images, and struggles with class imbalance, particularly for less frequent classes like malignant tumors. Additionally, integrating multi-modal data (mammograms and ultrasound) can lead to challenges like misalignment and image quality variations.
Conclusion and future work
This research presented a novel approach based on deep and traditional breast cancer detection features. Combining features from both mammogram and ultrasound images enhanced detection accuracy and provided a more robust system. Multi-modal datasets are used in this research paper i.e., Mammograms and ultrasound images. Images are pre-processed, augmented, enhanced, and segmented using the aforementioned techniques. Deep features are extracted using InceptionV3, ResNet50, and MobileNetV2. Meanwhile, the Gabor filter and wavelet transform are the traditional extracted features. The top best-contributed features are selected using statistical ANOVA-F value. The final feature vector is comprised of 600 features only including both traditional and deep features. State-of-the art comparisons of the Ensemble classifiers, i.e., XGBoost, AdaBoost, and CatBoost are performed. For MG images, the proposed approach achieved highest accuracy of 95.00% using Mini-DDSM dataset on XGBoost and achieved 97.06% accuracy using INbreast dataset on XGBoost and CatBoost. Whereas for US images, 98.67% highest accuracy was achieved by XGBoost and AdaBoost on the Rodrigues dataset and for the BUSI dataset XGBoost gave the highest accuracy of 97.02%. The proposed framework outperforms others, even with a small dataset, and shows potential for improving breast cancer detection in mammograms and ultrasound images.
While the proposed hybrid feature fusion method demonstrates promising results, there are several limitations to consider. First, the method was evaluated on datasets for mammograms and ultrasounds, and future work should explore the integration of multi-modal data for a more comprehensive detection approach. Second, hyperparameters are used for the classifiers, which may not be optimal for all datasets, and further hyperparameter tuning is needed to improve model performance. Third, while the method shows potential, its scalability and real-world applicability remain uncertain, as it has not yet been tested on larger, more diverse datasets or in clinical environments.
To address these challenges, future work will focus on several enhancements to improve model performance and adaptability. We plan to explore techniques to reduce model complexity and improve computational efficiency, including model pruning, knowledge distillation, and the use of lightweight convolutional layers like depthwise separable convolutions. Additionally, we aim to enhance the adaptability of our hybrid model for resource-constrained hardware and real-time clinical applications. To address class imbalance and improve performance under varying image quality conditions, we will investigate the development of a custom loss function or specialized activation function. Future work will focus on addressing these issues with noise reduction, data augmentation, and improved multi-modal fusion to enhance performance and reduce misclassifications. We also plan to incorporate k-fold cross-validation and additional statistical analyses, such as confidence intervals and p-values, to provide further validation of reliable model performance and ensure a statistically significant assessment of the results. An ablation study will be conducted to assess the effects of modifying activation functions, experimenting with AdamW as an alternative optimizer, and increasing the number of convolutional layers to better understand model capacity and performance. We also plan to compare our approach with traditional machine learning classifiers such as SVM, KNN, and RF to investigate differences in feature representation and classification. Lastly, we will explore the Vision Transformer (ViT), a state-of-the-art model that has set new benchmarks in image classification, as it may offer further improvements in breast cancer detection.
Data availability
The datasets used and analyzed during this study are publicly available from the following sources: Mini-DDSM: Available from Kaggle at https://www.kaggle.com/datasets/cheddad/miniddsm2. INbreast: Available from Kaggle at https://www.kaggle.com/datasets/ramanathansp20/inbreast-dataset. Rodrigues: Available from Mendeley Data at https://data.mendeley.com/datasets/wmy84gzngw/1. BUSI: Available from Kaggle at https://www.kaggle.com/datasets/aryashah2k/breast-ultrasound-images-dataset. A subset of 600 images from the Mini-DDSM dataset was selected to approximately match the size of the ultrasound datasets. Additionally, the INbreast dataset was divided into two classes for evaluation purposes. The processed datasets are not publicly available but can be provided by the corresponding author upon reasonable request.
References
Siegel, R. L., Kratzer, T. B., Giaquinto, A. N., Sung, H. & Jemal, A. Cancer statistics. CA Cancer J. Clin. 55, 259 (2025).
Mubarik, S. et al. Breast cancer mortality trends and predictions to 2030 and its attributable risk factors in East and South Asian countries. Front. Nutr. 9, 847920 (2022).
Zaheer, S. & Yasmeen, F. Historical trends in breast cancer presentation among women in Pakistan from join-point regression analysis. Pak. J. Med. Sci. 40, 134 (2024).
Danaei, G., Vander Hoorn, S., Lopez, A. D., Murray, C. J. & Ezzati, M. Causes of cancer in the world: Comparative risk assessment of nine behavioural and environmental risk factors. Lancet 366, 1784–1793 (2005).
Bray, F. et al. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Bray, F. et al. Global cancer statistics 2022: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Chouhan, N., Khan, A., Shah, J. Z., Hussnain, M. & Khan, M. W. Deep convolutional neural network and emotional learning based breast cancer detection using digital mammography. Comput. Biol. Med. 132, 104318 (2021).
Javed, F. et al. Relevance of the human microbiome in breast cancer: A global bibliometric and visual analysis. Scientifica 2024(1), 5518835 (2024).
Khan, N. H., Duan, S.-F., Wu, D.-D. & Ji, X.-Y. Better reporting and awareness campaigns needed for breast cancer in Pakistani women. Cancer Manag. Res. 13, 2125–2129 (2021).
Davis, L. E. et al. Patient-reported symptoms after breast cancer diagnosis and treatment: A retrospective cohort study. Eur. J. Cancer 101, 1–11 (2018).
McCart Reed, A. E., Kalinowski, L., Simpson, P. T. & Lakhani, S. R. Invasive lobular carcinoma of the breast: The increasing importance of this special subtype. Breast Cancer Res. 23, 1–16 (2021).
Sun, L., Wang, J., Hu, Z., Xu, Y. & Cui, Z. Multi-view convolutional neural networks for mammographic image classification. IEEE Access 7, 126273–126282 (2019).
Tsochatzidis, L., Costaridou, L. & Pratikakis, I. Deep learning for breast cancer diagnosis from mammograms-a comparative study. J. Imaging 5, 37 (2019).
Awan, M. Z., Arif, M. S., Abideen, M. Z. U. & Abodayeh, K. Comparative analysis of machine learning models for breast cancer prediction and diagnosis: A dual-dataset approach. Indones. J. Electr. Eng. Comput. Sci. 34, 2032–2044 (2024).
Nicosia, L. et al. Automatic breast ultrasound: State of the art and future perspectives. ecancermedicalscience 14, 1062 (2020).
Moustafa, A. F. et al. Color doppler ultrasound improves machine learning diagnosis of breast cancer. Diagnostics 10, 631 (2020).
Elkorany, A. S., Marey, M., Almustafa, K. M. & Elsharkawy, Z. F. Breast cancer diagnosis using support vector machines optimized by whale optimization and dragonfly algorithms. IEEE Access 10, 69688–69699 (2022).
Raza, A. et al. A hybrid deep learning-based approach for brain tumor classification. Electronics 11, 1146 (2022).
Hekmat, A., Zuping, Z., Bilal, O. & Khan, S. U. R. Differential evolution-driven optimized ensemble network for brain tumor detection. Int. J. Mach. Learn. Cybern. 16, 6447 (2025).
Bilal, O., Hekmat, A. & Khan, S. U. R. Automated cervical cancer cell diagnosis via grid search-optimized multi-CNN ensemble networks. Netw. Model. Anal. Health Inform. Bioinform. 14, 67 (2025).
Raza, A. et al. Deepbreastcancernet: A novel deep learning model for breast cancer detection using ultrasound images. Appl. Sci. 13, 2082 (2023).
Cruz-Ramos, C. et al. Benign and malignant breast tumor classification in ultrasound and mammography images via fusion of deep learning and handcraft features. Entropy 25, 991 (2023).
Dhahri, H., Al Maghayreh, E., Mahmood, A., Elkilani, W. & Faisal Nagi, M. Automated breast cancer diagnosis based on machine learning algorithms. J. Healthc. Eng. 2019, 4253641 (2019).
Houssein, E. H., Emam, M. M., Ali, A. A. & Suganthan, P. N. Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review. Expert Syst. Appl. 167, 114161 (2021).
Li, H., Zhuang, S., Li, D.-A., Zhao, J. & Ma, Y. Benign and malignant classification of mammogram images based on deep learning. Biomed. Signal Process. Control 51, 347–354 (2019).
Heenaye-Mamode Khan, M. et al. Multi-class classification of breast cancer abnormalities using deep convolutional neural network (cnn). PLoS ONE 16, e0256500 (2021).
Rahman, H., Naik Bukht, T. F., Ahmad, R., Almadhor, A. & Javed, A. R. Efficient breast cancer diagnosis from complex mammographic images using deep convolutional neural network. Comput. Intell. Neurosci. 2023, 7717712 (2023).
Wen, X., Li, J. & Yang, L. Breast cancer diagnosis method based on cross-mammogram four-view interactive learning. Tomography 10, 848–868 (2024).
Al-Dhabyani, W., Gomaa, M., Khaled, H. & Aly, F. Deep learning approaches for data augmentation and classification of breast masses using ultrasound images. Int. J. Adv. Comput. Sci. Appl. 10, 1–11 (2019).
Acevedo, P. & Vazquez, M. Classification of tumors in breast echography using a svm algorithm. In 2019 International Conference on Computational Science and Computational Intelligence (CSCI), 686–689 (IEEE, 2019).
Kabir, S. M., Shihavuddin, A., Tanveer, M. S. & Bhuiyan, M. I. H. Parametric image-based breast tumor classification using convolutional neural network in the contourlet transform domain. In 2020 11th International Conference on Electrical and Computer Engineering (ICECE), 439–442 (IEEE, 2020).
Moon, W. K. et al. Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks. Comput. Methods Programs Biomed. 190, 105361 (2020).
Kabir, S. M., Tanveer, M. S., Shihavuddin, A. & Bhuiyan, M. I. H. Weighted contourlet parametric (wcp) feature based breast tumor classification from b-mode ultrasound image. Preprint at arXiv:2102.05896 (2021).
Eroğlu, Y., Yildirim, M. & Çinar, A. Convolutional neural networks based classification of breast ultrasonography images by hybrid method with respect to benign, malignant, and normal using mrmr. Comput. Biol. Med. 133, 104407 (2021).
Masud, M. et al. Pre-trained convolutional neural networks for breast cancer detection using ultrasound images. ACM Trans. Internet Technol. (TOIT) 21, 1–17 (2021).
Byra, M. Breast mass classification with transfer learning based on scaling of deep representations. Biomed. Signal Process. Control 69, 102828 (2021).
Alotaibi, M. et al. Breast cancer classification based on convolutional neural network and image fusion approaches using ultrasound images. Heliyon9 (2023).
Rao, K. S. et al. Intelligent ultrasound imaging for enhanced breast cancer diagnosis: Ensemble transfer learning strategies. IEEE Access (2024).
Montaha, S. et al. Malignancy pattern analysis of breast ultrasound images using clinical features and a graph convolutional network. Digit. Health 10, 20552076241251660 (2024).
Zhang, B., Shi, H. & Wang, H. Machine learning and ai in cancer prognosis, prediction, and treatment selection: a critical approach. J. Multidiscip. Healthc. 16, 1779–1791 (2023).
Atrey, K., Singh, B. K., Bodhey, N. K. & Pachori, R. B. Mammography and ultrasound based dual modality classification of breast cancer using a hybrid deep learning approach. Biomed. Signal Process. Control 86, 104919 (2023).
Sahu, A., Das, P. K. & Meher, S. An efficient deep learning scheme to detect breast cancer using mammogram and ultrasound breast images. Biomed. Signal Process. Control 87, 105377 (2024).
Puttegowda, K. et al. Enhanced machine learning models for accurate breast cancer mammogram classification. Glob. Transit. (2025).
Wei, T.-R. et al. Multimodal deep learning for enhanced breast cancer diagnosis on sonography. Comput. Biol. Med. 194, 110466 (2025).
Ahmad, J. et al. Deep learning empowered breast cancer diagnosis: Advancements in detection and classification. PLoS ONE 19, e0304757 (2024).
Singh, A. K. et al. Transforming early breast cancer detection: A deep learning approach using convolutional neural networks and advanced classification techniques. Int. J. Comput. Intell. Syst. 18, 134 (2025).
Alom, M. R. et al. An explainable ai-driven deep neural network for accurate breast cancer detection from histopathological and ultrasound images. Sci. Rep. 15, 17531 (2025).
Qasrawi, R. et al. Advancing breast cancer detection in ultrasound images using a novel hybrid ensemble deep learning model. Intelli.-Based Med. 11, 100222 (2025).
Alahmadi, T. J. et al. Early breast cancer detection in CT scans using convolutional neural bidirectional feature pyramid network. PeerJ Comput. Sci. 11, e2994 (2025).
Arshad, W. et al. Cancer unveiled: A deep dive into breast tumor detection using cutting-edge deep learning models. IEEE Access 11, 133804–133824 (2023).
Malik, W., Javed, R., Tahir, F. & Rasheed, M. A. Covid-19 detection by chest x-ray images through efficient neural network techniques. Int. J. Theor. Appl. Comput. Intell. 35–56 (2025).
Ali, H. A meta-review of computational intelligence techniques for early autism disorder diagnosis. Int. J. Theor. Appl. Comput. Intell. 1–21 (2025).
Muduli, D., Dash, R. & Majhi, B. Automated diagnosis of breast cancer using multi-modal datasets: A deep convolution neural network based approach. Biomed. Signal Process. Control 71, 102825 (2022).
Sahu, A., Das, P. K. & Meher, S. High accuracy hybrid CNN classifiers for breast cancer detection using mammogram and ultrasound datasets. Biomed. Signal Process. Control 80, 104292 (2023).
Moreira, I. C. et al. Inbreast: Toward a full-field digital mammographic database. Acad. Radiol. 19, 236–248 (2012).
Ragab, D. A., Attallah, O., Sharkas, M., Ren, J. & Marshall, S. A framework for breast cancer classification using Multi-DCNNs. Comput. Biol. Med. 131, 104245 (2021).
Yousefi, P. Mammographic image enhancement for breast cancer detection applying wavelet transform. In 2015 IEEE Student Symposium in Biomedical Engineering & Sciences (ISSBES), 82–86 (IEEE, 2015).
Sharma, J., Rai, J. & Tewari, R. Identification of pre-processing technique for enhancement of mammogram images. In 2014 International Conference on Medical Imaging, m-Health and Emerging Communication Systems (MedCom), 115–119 (IEEE, 2014).
Ali, M., Wu, T., Hu, H. & Mahmood, T. Breast tumor segmentation using neural cellular automata and shape guided segmentation in mammography images. PLoS ONE 19, e0309421 (2024).
Tsietso, D., Yahya, A., Samikannu, R., Qureshi, B. & Babar, M. Computational approach for automated segmentation and classification of region of interest in lateral breast thermograms. Computers, Materials & Continua80 (2024).
Raj, S. S., Raja, N. S. M., Madhumitha, M. & Rajinikanth, V. Examination of digital mammogram using otsu’s function and watershed segmentation. In 2018 Fourth International Conference on Biosignals, Images and Instrumentation (ICBSII), 206–212 (IEEE, 2018).
Shaukat, A. Automated classification of hair care plants using geometrical and textural features from leaf images: A pattern recognition based approach. Pakistan Journal of Science68 (2016).
Shaukat, A. et al. Textural and geometrical features based approach for identification of individuals using palmprint and hand shape images from multiple multimodal datasets. J. Test. Eval. 46, 2281–2298 (2018).
Khan, S., Hussain, M., Aboalsamh, H. & Bebis, G. A comparison of different gabor feature extraction approaches for mass classification in mammography. Multimed. Tools Appl. 76, 33–57 (2017).
Zhao, M., Chai, Q. & Zhang, S. A method of image feature extraction using wavelet transforms. In Emerging Intelligent Computing Technology and Applications: 5th International Conference on Intelligent Computing, ICIC 2009, Ulsan, South Korea, September 16-19, 2009. Proceedings 5, 187–192 (Springer, 2009).
Khalid, N. H. M., Ismail, A. R., Aziz, N. A. & Hussin, A. A. A. Performance comparison of feature selection methods for prediction in medical data. In International Conference on Soft Computing in Data Science, 92–106 (Springer, 2023).
Chen, T. & Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, 785–794 (2016).
Zhang, Y. & Wang, L. An adaboost method with k’ k-means bayes classifier for imbalanced data. Mathematics 11, 1878 (2023).
Bentéjac, C., Csörgő, A. & Martínez-Muñoz, G. A comparative analysis of gradient boosting algorithms. Artif. Intell. Rev. 54, 1937–1967 (2021).
Rahman, A. S. A. et al. Breast mass tumor classification using deep learning. In 2020 IEEE International Conference on Informatics, IoT, and Enabling Technologies (ICIoT), 271–276 (IEEE, 2020).
Arora, R., Rai, P. K. & Raman, B. Deep feature-based automatic classification of mammograms. Med. Biol. Eng. Comput. 58, 1199–1211 (2020).
Zhang, H. et al. De-ada*: A novel model for breast mass classification using cross-modal pathological semantic mining and organic integration of multi-feature fusions. Inf. Sci. 539, 461–486 (2020).
Shakeel, S. & Raja, G. Classification of breast cancer from mammogram images using deep convolution neural networks. In 2021 International Bhurban Conference on Applied Sciences and Technologies (IBCAST), 595–599 (IEEE, 2021).
Dehghan Rouzi, M. et al. Breast cancer detection with an ensemble of deep learning networks using a consensus-adaptive weighting method. J. Imaging 9, 247 (2023).
Carneiro, G., Nascimento, J. & Bradley, A. P. Automated analysis of unregistered multi-view mammograms with deep learning. IEEE Trans. Med. Imaging 36, 2355–2365 (2017).
Aslan, M. F. A hybrid end-to-end learning approach for breast cancer diagnosis: Convolutional recurrent network. Comput. Electr. Eng. 105, 108562 (2023).
Deb, S. D. & Jha, R. K. Breast ultrasound image classification using fuzzy-rank-based ensemble network. Biomed. Signal Process. Control 85, 104871 (2023).
Chen, J. et al. A deep learning-based multimodal medical imaging model for breast cancer screening. Sci. Rep. 15, 14696 (2025).
Acknowledgements
The authors want to acknowledge the fund by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R346). The authors would like also to acknowledge the support of AIDA Lab CCIS Prince Sultan University, Riyadh Saudi Arabia for APC support.
Funding
This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R346), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
All authors have read and agreed to the published version of the manuscript.
A.S. and S.F. conceptualized the study and designed the methodology. A.S., S.F., R.J. and S.I. conducted the experiments and analyzed the results. S.F. supervised the research and contributed to manuscript refinement. A.S., F.S.A, A.A. assisted in data preprocessing and feature extraction. S.F. and T.S. provided critical insights on model evaluation and validation. A.S. and S.F. wrote the main manuscript text. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shaukat, A., Farhan, S., Javed, R. et al. HyFusion-X: hybrid deep and traditional feature fusion with ensemble classifiers for breast cancer detection using mammogram and ultrasound images. Sci Rep 15, 38314 (2025). https://doi.org/10.1038/s41598-025-22262-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-22262-1